Mismatch Repair (MMR) Gene Mutation Carriers Have Favorable Outcome in Colorectal and Endometrial Cancer: A Prospective Cohort Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Study Cohort and Data Collection
2.2. Genetic Testing for Cancer Susceptibility Genes
2.3. Immunohistochemistry (IHC) for Tumor Samples
2.4. Clinical Correlation and Statistical Analyses
3. Results
3.1. Study Population and Germline MMR Pathogenic Variants (gMMRd)
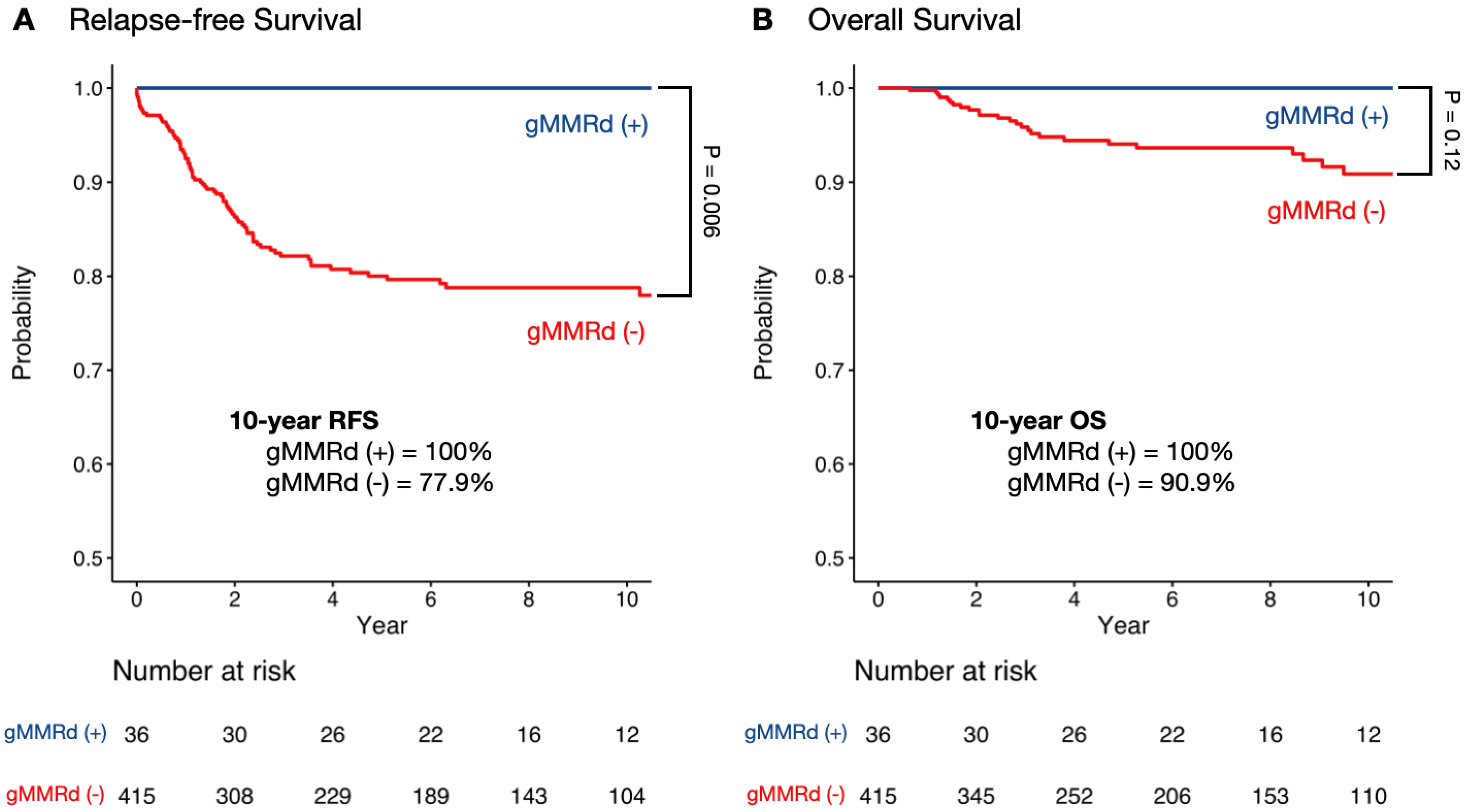
3.2. Survival Analysis in Colorectal and Endometrial Cancer by gMMRd
3.3. Characteristics and Survival for Colorectal and Endometrial Cancer Patients Based on gMMRd and pMMRd Status
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviations
References
- Lynch, H.T.; Lynch, P.M.; Lanspa, S.J.; Snyder, C.L.; Lynch, J.F.; Boland, C.R. Review of the Lynch syndrome: History, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin. Genet. 2009, 76, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Ijsselsteijn, R.; Jansen, J.G.; de Wind, N. DNA mismatch repair-dependent DNA damage responses and cancer. DNA Repair 2020, 93, 102923. [Google Scholar] [CrossRef] [PubMed]
- Cox, V.L.; Saeed Bamashmos, A.A.; Foo, W.C.; Gupta, S.; Yedururi, S.; Garg, N.; Kang, H.C. Lynch Syndrome: Genomics Update and Imaging Review. Radiographics 2018, 38, 483–499. [Google Scholar] [CrossRef] [PubMed]
- Cerretelli, G.; Ager, A.; Arends, M.J.; Frayling, I.M. Molecular pathology of Lynch syndrome. J. Pathol. 2020, 250, 518–531. [Google Scholar] [CrossRef] [PubMed]
- Hampel, H.; Frankel, W.; Panescu, J.; Lockman, J.; Sotamaa, K.; Fix, D.; Comeras, I.; La Jeunesse, J.; Nakagawa, H.; Westman, J.A.; et al. Screening for Lynch syndrome (hereditary nonpolyposis colorectal cancer) among endometrial cancer patients. Cancer Res. 2006, 66, 7810–7817. [Google Scholar] [CrossRef]
- Hampel, H.; Frankel, W.L.; Martin, E.; Arnold, M.; Khanduja, K.; Kuebler, P.; Nakagawa, H.; Sotamaa, K.; Prior, T.W.; Westman, J.; et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N. Engl. J. Med. 2005, 352, 1851–1860. [Google Scholar] [CrossRef] [PubMed]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Engel, C.; Ahadova, A.; Seppälä, T.T.; Aretz, S.; Bigirwamungu-Bargeman, M.; Bläker, H.; Bucksch, K.; Büttner, R.; de Vos Tot Nederveen Cappel, W.T.; Endris, V.; et al. Associations of Pathogenic Variants in MLH1, MSH2, and MSH6 With Risk of Colorectal Adenomas and Tumors and With Somatic Mutations in Patients With Lynch Syndrome. Gastroenterology 2020, 158, 1326–1333. [Google Scholar] [CrossRef] [PubMed]
- Møller, P.; Seppälä, T.T.; Bernstein, I.; Holinski-Feder, E.; Sala, P.; Gareth Evans, D.; Lindblom, A.; Macrae, F.; Blanco, I.; Sijmons, R.H.; et al. Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: A report from the Prospective Lynch Syndrome Database. Gut 2018, 67, 1306–1316. [Google Scholar] [CrossRef]
- ten Broeke, S.W.; Brohet, R.M.; Tops, C.M.; van der Klift, H.M.; Velthuizen, M.E.; Bernstein, I.; Capellá Munar, G.; Gomez Garcia, E.; Hoogerbrugge, N.; Letteboer, T.G.; et al. Lynch syndrome caused by germline PMS2 mutations: Delineating the cancer risk. J. Clin. Oncol. 2015, 33, 319–325. [Google Scholar] [CrossRef]
- Haraldsdottir, S.; Hampel, H.; Wu, C.; Weng, D.Y.; Shields, P.G.; Frankel, W.L.; Pan, X.; de la Chapelle, A.; Goldberg, R.M.; Bekaii-Saab, T. Patients with colorectal cancer associated with Lynch syndrome and MLH1 promoter hypermethylation have similar prognoses. Genet. Med. 2016, 18, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.C.; Liu, R.Y.; Yan, J.P.; An, X.; Jiang, W.; Ling, Y.H.; Chen, J.W.; Bei, J.X.; Zuo, X.Y.; Cai, M.Y.; et al. The Heterogeneity Between Lynch-Associated and Sporadic MMR Deficiency in Colorectal Cancers. J. Natl. Cancer Inst. 2018, 110, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.; Buhard, O.; Cervera, P.; Hain, E.; Dumont, S.; Bardier, A.; Bachet, J.B.; Gornet, J.M.; Lopez-Trabada, D.; Dumont, S.; et al. Clinical and molecular characterisation of hereditary and sporadic metastatic colorectal cancers harbouring microsatellite instability/DNA mismatch repair deficiency. Eur. J. Cancer 2017, 86, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Ribic, C.M.; Sargent, D.J.; Moore, M.J.; Thibodeau, S.N.; French, A.J.; Goldberg, R.M.; Hamilton, S.R.; Laurent-Puig, P.; Gryfe, R.; Shepherd, L.E.; et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N. Engl. J. Med. 2003, 349, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Gryfe, R.; Kim, H.; Hsieh, E.T.; Aronson, M.D.; Holowaty, E.J.; Bull, S.B.; Redston, M.; Gallinger, S. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N. Engl. J. Med. 2000, 342, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, C.; Zheng, C.Z.; Zhang, Y.Q.; Guo, T.A.; Liu, F.Q.; Xu, Y. Comparison of long-term outcomes between Lynch sydrome and sporadic colorectal cancer: A propensity score matching analysis. BMC Cancer 2021, 21, 45. [Google Scholar] [CrossRef] [PubMed]
- Popat, S.; Hubner, R.; Houlston, R.S. Systematic review of microsatellite instability and colorectal cancer prognosis. J. Clin. Oncol. 2005, 23, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Soler, M.; Perez-Carbonell, L.; Guarinos, C.; Zapater, P.; Castillejo, A.; Barbera, V.M.; Juarez, M.; Bessa, X.; Xicola, R.M.; Clofent, J.; et al. Risk of cancer in cases of suspected lynch syndrome without germline mutation. Gastroenterology 2013, 144, 926–932.e1. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Roca, A.; Giner-Calabuig, M.; Murcia, O.; Castillejo, A.; Soto, J.L.; Garcia-Heredia, A.; Jover, R. Lynch-like Syndrome: Potential Mechanisms and Management. Cancers 2022, 14, 1115. [Google Scholar] [CrossRef]
- Umar, A.; Boland, C.R.; Terdiman, J.P.; Syngal, S.; de la Chapelle, A.; Ruschoff, J.; Fishel, R.; Lindor, N.M.; Burgart, L.J.; Hamelin, R.; et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J. Natl. Cancer Inst. 2004, 96, 261–268. [Google Scholar] [CrossRef]
- Wang, Y.A.; Jian, J.W.; Hung, C.F.; Peng, H.P.; Yang, C.F.; Cheng, H.S.; Yang, A.S. Germline breast cancer susceptibility gene mutations and breast cancer outcomes. BMC Cancer 2018, 18, 315. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Gu, X.; Padmanabhan, R.; Wu, Z.; Peng, Q.; DiCarlo, J.; Wang, Y. smCounter2: An accurate low-frequency variant caller for targeted sequencing data with unique molecular identifiers. Bioinformatics 2018, 35, 1299–1309. [Google Scholar] [CrossRef] [PubMed]
- Reinecke, F.; Satya, R.V.; DiCarlo, J. Quantitative analysis of differences in copy numbers using read depth obtained from PCR-enriched samples and controls. BMC Bioinform. 2015, 16, 17. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Boeva, V.; Popova, T.; Lienard, M.; Toffoli, S.; Kamal, M.; Le Tourneau, C.; Gentien, D.; Servant, N.; Gestraud, P.; Rio Frio, T.; et al. Multi-factor data normalization enables the detection of copy number aberrations in amplicon sequencing data. Bioinformatics 2014, 30, 3443–3450. [Google Scholar] [CrossRef] [PubMed]
- Johansson, L.F.; van Dijk, F.; de Boer, E.N.; van Dijk-Bos, K.K.; Jongbloed, J.D.H.; van der Hout, A.H.; Westers, H.; Sinke, R.J.; Swertz, M.A.; Sijmons, R.H.; et al. CoNVaDING: Single Exon Variation Detection in Targeted NGS Data. Hum. Mutat. 2016, 37, 457–464. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Xavier, A.; Scott, R.J.; Talseth-Palmer, B.A. TAPES: A tool for assessment and prioritisation in exome studies. PLoS Comput. Biol. 2019, 15, e1007453. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Dominguez-Valentin, M.; Haupt, S.; Seppala, T.T.; Sampson, J.R.; Sunde, L.; Bernstein, I.; Jenkins, M.A.; Engel, C.; Aretz, S.; Nielsen, M.; et al. Mortality by age, gene and gender in carriers of pathogenic mismatch repair gene variants receiving surveillance for early cancer diagnosis and treatment: A report from the prospective Lynch syndrome database. EClinicalMedicine 2023, 58, 101909. [Google Scholar] [CrossRef]
- Pasanen, A.; Loukovaara, M.; Butzow, R. Clinicopathological significance of deficient DNA mismatch repair and MLH1 promoter methylation in endometrioid endometrial carcinoma. Mod. Pathol. 2020, 33, 1443–1452. [Google Scholar] [CrossRef]
- Chika, N.; Eguchi, H.; Kumamoto, K.; Suzuki, O.; Ishibashi, K.; Tachikawa, T.; Akagi, K.; Tamaru, J.I.; Okazaki, Y.; Ishida, H. Prevalence of Lynch syndrome and Lynch-like syndrome among patients with colorectal cancer in a Japanese hospital-based population. Jpn. J. Clin. Oncol. 2017, 47, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Nakamori, S.; Takao, M.; Takao, A.; Natsume, S.; Iijima, T.; Kojika, E.; Nakano, D.; Kawai, K.; Inokuchi, T.; Fujimoto, A.; et al. Clinicopathological characteristics of Lynch-like syndrome. Int. J. Clin. Oncol. 2024, 29, 944–952. [Google Scholar] [CrossRef] [PubMed]

| gMMRd (−) n = 415 | gMMRd (+) n = 36 | p-Value | |
|---|---|---|---|
| Female, n (%) | 272 (65.5%) | 23 (63.9%) | 0.84 |
| Number of cancers | <0.0001 | ||
| 1 | 367 (88.4%) | 25 (69.4%) | |
| 2 | 45 (10.8%) | 8 (22.2%) | |
| ≥3 | 3 (0.7%) | 3 (8.3%) | |
| Colorectal cancer | n = 309 | n = 25 | |
| Age of onset, mean (SD) | 42.4 (5.3) | 42.7 (7.1) | 0.81 |
| Stage | 0.22 | ||
| 0 | 9 (2.9%) | 2 (8.0%) | |
| 1 | 49 (15.9%) | 2 (8.0%) | |
| 2 | 81 (26.2%) | 9 (36.0%) | |
| 3 | 125 (40.5%) | 8 (32.0%) | |
| 4 | 43 (13.9%) | 3 (12.0%) | |
| Unknown | 2 (0.7%) | 1 (4.0%) | |
| Location in colon | 0.0002 | ||
| Proximal * | 53 (17.2%) | 12 (48.0%) | |
| Distal * | 255 (82.8%) | 13 (52.0%) | |
| Chemotherapy | 192 (62.1%) | 14 (56.0%) | 0.54 |
| 5-FU based | 188 (97.9%) | 14 (100.0%) | 0.59 |
| Radiotherapy | 63 (20.4%) | 3 (12.0%) | 0.31 |
| Endometrial cancer | n = 111 | n = 14 | |
| Age of onset, mean (SD) | 43.2 (7.1) | 44.5 (8.1) | 0.53 |
| Stage | 0.014 | ||
| 1 | 85 (76.6%) | 7 (50.0%) | |
| 2 | 3 (2.7%) | 3 (21.4%) | |
| 3 | 16 (14.4%) | 3 (21.4%) | |
| 4 | 7 (6.3%) | 1 (7.1%) | |
| Chemotherapy | 25 (22.5%) | 5 (35.7%) | 0.28 |
| Platinum-based | 25 (100.0%) | 5 (100.0%) | |
| Radiotherapy | 26 (23.4%) | 5 (35.7%) | 0.32 |
| Patients | Events | 10-Year RFS (%) | p-Value | ||
|---|---|---|---|---|---|
| Gender | |||||
| Female | gMMRd (+) | 23 | 0 | 100 | 0.063 |
| gMMRd (−) | 272 | 39 | 83.9 | ||
| Male | gMMRd (+) | 13 | 0 | 100 | 0.032 |
| gMMRd (−) | 143 | 40 | 68.1 | ||
| Age of onset | |||||
| <50 | gMMRd (+) | 31 | 0 | 100 | 0.0099 |
| gMMRd (−) | 499 | 74 | 74.0 | ||
| ≥50 | gMMRd (+) | 5 | 0 | 100 | 0.19 |
| gMMRd (−) | 16 | 5 | 67.5 | ||
| Cancer type | |||||
| Colorectal | gMMRd (+) | 24 | 0 | 100 | 0.01 |
| gMMRd (−) | 304 | 72 | 73.3 | ||
| Endometrial | gMMRd (+) | 12 | 0 | 100 | 0.38 |
| gMMRd (−) | 111 | 7 | 92.0 | ||
| Cancer stage | |||||
| Stage I | gMMRd (+) | 9 | 0 | 100 | 0.45 |
| gMMRd (−) | 134 | 8 | 92.2 | ||
| Stage II | gMMRd (+) | 10 | 0 | 100 | 0.26 |
| gMMRd (−) | 82 | 9 | 86.8 | ||
| Stage III | gMMRd (+) | 11 | 0 | 100 | 0.12 |
| gMMRd (−) | 140 | 29 | 76.0 | ||
| Stage IV | gMMRd (+) | 4 | 0 | 100 | 0.032 |
| gMMRd (−) | 49 | 32 | 30.6 | ||
| Treatment | |||||
| Chemotherapy | gMMRd (+) | 18 | 0 | 100 | 0.026 |
| gMMRd (−) | 207 | 49 | 74.4 | ||
| No chemotherapy | gMMRd (+) | 18 | 0 | 100 | 0.094 |
| gMMRd (−) | 208 | 30 | 81.5 | ||
| Radiotherapy | gMMRd (+) | 9 | 0 | 100 | 0.18 |
| gMMRd (−) | 78 | 16 | 77.8 | ||
| No radiotherapy | gMMRd (+) | 27 | 0 | 100 | 0.016 |
| gMMRd (−) | 337 | 63 | 77.9 | ||
| Location of colon | |||||
| Distal | gMMRd (+) | 13 | 0 | 100 | 0.057 |
| gMMRd (−) | 255 | 64 | 71.7 | ||
| Proximal | gMMRd (+) | 12 | 0 | 100 | 0.12 |
| gMMRd (−) | 53 | 9 | 81 |
| gMMRd (+) | gMMRd(−) pMMRd (+) | gMMRd(−) pMMRd (−) | p-Value | |
|---|---|---|---|---|
| n | 36 | 14 | 88 | |
| Age of onset, mean (SD) | 43.6 (7.0) | 53.2 (13.7) | 44.1 (6.6) | 0.0002 |
| Stage | 0.3257 | |||
| 0 | 2 (5.6%) | 0 (0%) | 1 (1.1%) | |
| 1 | 9 (25.0%) | 2 (14.3%) | 13 (14.8%) | |
| 2 | 10 (27.8%) | 5 (35.7%) | 19 (21.6%) | |
| 3 | 11 (30.6%) | 4 (28.6%) | 29 (33.0%) | |
| 4 | 4 (11.1%) | 3 (21.4%) | 26 (29.5%) | |
| Chemotherapy | 18 (50.0%) | 9 (64.2%) | 42 (47.7%) | 0.5156 |
| Radiotherapy | 9 (25.0%) | 4 (28.6%) | 11 (12.5%) | 0.1266 |
| Cancer type | 0.1698 | |||
| Endometrial cancer | 12 (33.3%) | 4 (28.6%) | 16 (18.2%) | |
| Colorectal cancer | 24 (66.7%) | 10 (71.4%) | 72 (81.8%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeh, J.-T.; Peng, H.-P.; Hung, F.-H.; Hung, C.-F.; Hsieh, L.-L.; Yang, A.-S.; Wang, Y.A. Mismatch Repair (MMR) Gene Mutation Carriers Have Favorable Outcome in Colorectal and Endometrial Cancer: A Prospective Cohort Study. Cancers 2024, 16, 2342. https://doi.org/10.3390/cancers16132342
Yeh J-T, Peng H-P, Hung F-H, Hung C-F, Hsieh L-L, Yang A-S, Wang YA. Mismatch Repair (MMR) Gene Mutation Carriers Have Favorable Outcome in Colorectal and Endometrial Cancer: A Prospective Cohort Study. Cancers. 2024; 16(13):2342. https://doi.org/10.3390/cancers16132342
Chicago/Turabian StyleYeh, Jiunn-Tyng, Hung-Pin Peng, Fei-Hung Hung, Chen-Fang Hung, Ling-Ling Hsieh, An-Suei Yang, and Yong Alison Wang. 2024. "Mismatch Repair (MMR) Gene Mutation Carriers Have Favorable Outcome in Colorectal and Endometrial Cancer: A Prospective Cohort Study" Cancers 16, no. 13: 2342. https://doi.org/10.3390/cancers16132342
APA StyleYeh, J.-T., Peng, H.-P., Hung, F.-H., Hung, C.-F., Hsieh, L.-L., Yang, A.-S., & Wang, Y. A. (2024). Mismatch Repair (MMR) Gene Mutation Carriers Have Favorable Outcome in Colorectal and Endometrial Cancer: A Prospective Cohort Study. Cancers, 16(13), 2342. https://doi.org/10.3390/cancers16132342






