Simple Summary
Genetic information is essential before starting the treatment of advanced-stage non-small-cell lung cancer (NSCLC) and in the adjuvant setting (EGFR mutation status only) after pulmonary resection of early-stage NSCLC. Several genetic tests for NSCLC are available, which vary in turnaround time and cost (usually higher costs for multi-gene tests). The Idylla™ EGFR Mutation Test is an ultra-rapid single-gene test used to detect EGFR mutations. In this study, we compared the performance of the Idylla EGFR Mutation Test with the current standard EGFR single-gene test (Cobas® EGFR Mutation Test v2) and demonstrate the accuracy of the Idylla EGFR Mutation Test as a molecular screening platform. From these data, we propose genetic testing strategies that may reduce the costs and shorten the turnaround time in advanced-stage NSCLC and in the adjuvant setting after pulmonary resection of early-stage NSCLC.
Abstract
Background: The Idylla™ EGFR Mutation Test is an ultra-rapid single-gene test that detects epidermal growth factor receptor (EGFR) mutations using formalin-fixed paraffin-embedded specimens. Here, we compared the performance of the Idylla EGFR Mutation Test with the Cobas® EGFR Mutation Test v2. Methods: Surgically resected NSCLC specimens obtained at two Japanese institutions (N = 170) were examined. The Idylla EGFR Mutation Test and the Cobas EGFR Mutation Test v2 were performed independently and the results were compared. For discordant cases, the Ion AmpliSeq Colon and Lung Cancer Research Panel V2 was performed. Results: After the exclusion of five inadequate/invalid samples, 165 cases were evaluated. EGFR mutation analysis revealed 52 were positive and 107 were negative for EGFR mutation in both assays (overall concordance rate: 96.4%). Analyses of the six discordant cases revealed that the Idylla EGFR Mutation Test was correct in four and the Cobas EGFR Mutation Test v2 was correct in two. In a trial calculation, the combination of the Idylla EGFR Mutation Test followed by a multi-gene panel test will reduce molecular screening expenses if applied to a cohort with EGFR mutation frequency >17.9%. Conclusions: We demonstrated the accuracy and potential clinical utility of the Idylla EGFR Mutation Test as a molecular screening platform in terms of turnaround time and molecular testing cost if applied to a cohort with a high EGFR mutation incidence (>17.9%).
1. Introduction
Lung cancer is the leading cause of cancer-related mortality in the world. Driver mutation testing is an essential element of diagnostic procedures for advanced-stage non-small cell lung cancer (NSCLC) patients [1] and some surgically resected NSCLC patients. Multiplex genetic tests, such as the Oncomine Dx Target Test (Thermo Fisher Scientific, Wilmington, DE, USA), meet the latest recommendations for NSCLC screening in the advanced-stage setting, because multiplex analyses are time-saving compared with performing a series of single gene analyses [2,3]. However, it is also true that multiplex genetic tests are usually expensive. Because the reduction of medical costs is an urgent economic issues in many countries and as NSCLC is one of the most frequent malignancies in many countries [4], a strategy to reduce the cost of molecular testing for NSCLCs [5] without extending the turnaround time (TAT) is required. In addition, in the setting of surgically resectable NSCLC, EGFR mutation analysis is the only clinically required genetic test, because osimertinib, an EGFR inhibitor, is the only approved molecular targeted drug in the adjuvant setting in many countries including Japan for surgically resected pathological-stage (pStage) IB-III NSCLC patients with activating EGFR mutations [6].
The Idylla™ system (Biocartis, Mechelen, Belgium) is a simple, fully automated, real-time PCR (qPCR)-based platform that uses unextracted formalin-fixed, paraffin-embedded (FFPE) tissue sections as input material [7,8]. The Idylla EGFR Mutation Test has some advantages over other EGFR single gene tests, including an ultrarapid TAT (150 min for a single EGFR test), no requirement of specific technical skill, and a closed cartridge during the entire workflow (reducing the risk of contamination). Therefore, the Idylla system can be used in most laboratories with minimal infrastructure. These advantages of the Idylla system have accelerated the development of several other molecular testing platforms, including the Idylla GeneFusion assay, which assesses gene fusions in ALK, ROS1, RET, MET exon 14 skipping, and NTRK1/2/3, the Idylla BRAF Mutation Test, the Idylla KRAS Mutation Test, and the Idylla NRAS-BRAF Mutation Test. Several groups have performed comparison studies between the novel Idylla assays and conventional, clinically approved molecular testing platforms.
In this study, we performed a comparison of the Idylla EGFR mutation test with the Cobas® EGFR Mutation Test v2 (Roche Diagnostics, Basel, Switzerland) in terms of the accuracy. We evaluated the performance of the Idylla EGFR Mutation Test and the Cobas EGFR Mutation Test v2 using FFPE specimens obtained from NSCLC patients who underwent surgical resection at two institutions in Japan. We also discuss a molecular testing strategy that incorporates the Idylla EGFR Mutation Test with the aim of reducing the molecular screening cost without extending the TAT.
2. Materials and Methods
2.1. Patients
A series of FFPE specimens were obtained from NSCLC patients who underwent surgical resection at the Kindai University Hospital (N = 100) or Tokyo Medical University Hospital (N = 70). After exclusion of four cases with scant/no tumor cells in the specimens (Figure 1), a total of 166 specimens were initially registered for the study. Patient characteristics are summarized in Table 1. The cohort consisted of 122 lung adenocarcinomas (73%), 6 adenosquamous carcinomas (4%), 35 squamous cell carcinomas (21%), and 3 pleomorphic carcinomas (2%). Following the 8th edition of the TNM classification, there were 100 pStage IA (60%), 20 pStage IB (12%), 37 pStage II–III (22%), and 9 pStage IV (5%) patients.
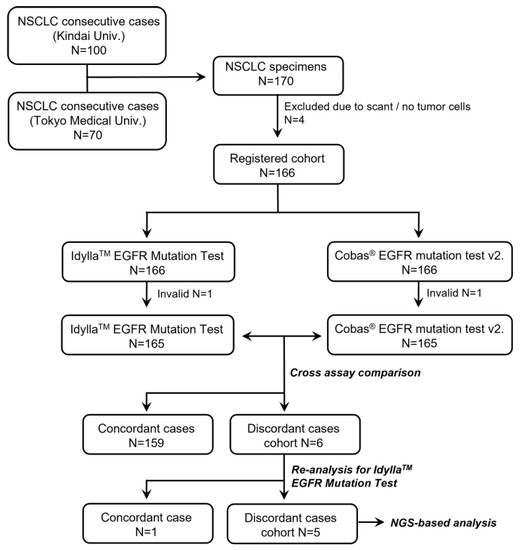
Figure 1.
Flowchart of the study.

Table 1.
Clinical and pathological characteristics of patients.
Serial FFPE sections from FFPE specimens used for DNA extraction were stained with hematoxylin and eosin to assess the proportion of tumor cells. Macro-dissection was used on 85% of specimens to enrich tumor cells (median proportion of tumor cells was 25%, range 20–70%). This study was approved by the ethical committees of the Kindai University Faculty of Medicine and the Tokyo Medical University Hospital (R03-191 and T2021-0281, respectively). Written informed consent was waived because of the retrospective nature of this study.
2.2. Study Design
The unstained FFPE specimens from the 166 NSCLC patients were analyzed by the Idylla EGFR Mutation Test at the Department of Genome Biology, Kindai University Faculty of Medicine. The Idylla EGFR Mutation Test is a single-use cartridge-based test designed for the qualitative detection of 39 different EGFR mutations. In this study, the Idylla investigator-use-only (IUO) assay was used.
The Cobas EGFR Mutation Test v2 was performed independently at LSI Medience Corporation (Tokyo, Japan) and SRL, Inc. (Tokyo, Japan). The Cobas EGFR Mutation Test v2 was used as the control because it is widely used in clinical practice and has shown a high concordance rate and ĸ value (97.5% and 0.938, respectively) compared with an NGS-based multiplex genetic test (Oncomine Dx Target test) [9].
For discordant cases between the Idylla vs. Cobas assays, the sample was reanalyzed with the Idylla EGFR Mutation Test. For discordant cases after the second Idylla EGFR Mutation Test, the Ion AmpliSeq Colon and Lung Cancer Research Panel V2 (CLV2, Thermo Fisher Scientific), which includes a single primer pool to amplify hotspots and targeted regions of 22 cancer genes frequently mutated in colorectal cancers and NSCLCs [10], was performed.
2.3. DNA Extraction and NGS-Based Panel Test
The Ion AmpliSeq Colon and Lung Cancer Research Panel V2 (CLV2) was performed as an NGS-based panel test. Briefly, DNA was isolated from FFPE specimens with the AllPrep DNA/RNA FFPE Kit (Qiagen, Venlo, Netherlands). The quality and quantity of the nucleic acid were verified with a NanoDrop 2000 device and PicoGreen dsDNA Reagent (both from Thermo Fisher Scientific). For library preparation, DNA was subjected to multiplex PCR amplification using the Ion AmpliSeq Library Kit 2.0 (Thermo Fisher Scientific) following the manufacturer’s protocol. Pooled libraries were subjected to the Ion Chef System (Thermo Fisher Scientific) for template preparation. Libraries were then loaded onto an Ion 550 chip and sequenced with the Ion S5 sequencing system. DNA sequencing data were accessed through the Torrent Suite version 5.12 program (Thermo Fisher Scientific). Reads were aligned with the hg19 human reference genome, and potential mutations were identified using Variant Caller version 5.12. Raw variant calls were filtered with a quality score of <100 and depth of coverage of <19 and were manually checked using the integrative genomics viewer (IGV; Broad Institute, Cambridge, MA, USA).
2.4. Statistical Analysis
Kappa statistics was used to compare the results of the Idylla EGFR Mutation Test with the Cobas EGFR Mutation Test v2. Statistical analyses were performed using GraphPad Prism software (Version 8; GraphPad Software Inc., La Jolla, CA, USA).
2.5. Total Expense Calculation for Molecular Testing
The total costs for molecular testing were calculated on the basis of medical insurance scores designated by the Japanese Pharmaceuticals and Medical Devices Agency (PMDA) in March 2023. The amount that medical institutions can claim for the molecular testing was determined by the PMDA for each examination. The total cost of a potential testing strategy that incorporates the Idylla EGFR mutation test was calculated on the basis of frequencies of EGFR mutations, as the cost of a single-plex EGFR test for patients with EGFR mutation and the sum of the costs of a single-plex EGFR test plus a following multiplex genetic test for patients without EGFR mutation. We calculated the EGFR mutation frequency at which the cost of molecular testing that incorporates the Idylla EGFR mutation test becomes less than the cost of molecular testing with a multiplex genetic test for all patients.
3. Results
3.1. Performance of the Idylla EGFR Mutation Test
As shown in Figure 1, both the Idylla and the Cobas assays were successfully performed in 165 cases out of 166 (success rates: 99.4%). Because one specimen was invalid for both assays, the quality of the sample was assumed to be low. The detailed results of the 165 specimens are summarized in Table 2. Among the 165 samples, 52 were positive for EGFR mutation (21 samples had exon 19 deletion and 28 had L858R, 2 had G719X, and 1 sample had L861Q point mutations) and 107 were negative for EGFR mutation in both assays (overall concordance rate: 96.4%). The ĸ value between the two assays was 0.920 (95% confidential interval: 0.857–0.983). Discordant results were seen in six cases (Table 2).

Table 2.
Concordance summary in all patients (N = 165) with valid results for EGFR testing.
The six cases were subjected to a second Idylla assay. One case (exon 19 deletion by the Cobas assay but a negative result in the initial Idylla assay) showed exon 19 deletion in the second Idylla assay. The other five cases showed the same results at the second Idylla EGFR Mutation Test. An NGS-based panel test was performed on the five discordant cases between the Idylla vs. Cobas assays.
3.2. Detailed Examination of the Discordant Cases
An NGS-based panel test (CLv2) was performed for the five discordant samples. In the cases with positive results by the Idylla assay (L858R or exon 20 insertion) but negative results by the Cobas assay, the NGS results were concordant with the Idylla assay. The detected exon 20 insertion was Asn771_Pro772insThr, which was not covered by the Cobas assay. The reason why the Cobas assay could not detect L858R EGFR point mutation is not clear. The NGS-based panel test revealed that the specimen had compound EGFR mutations (L858R plus L776H). The L776H EGFR mutation is not covered by the Idylla assay or Cobas assay, which may clarify why the sample was determined as L858R point mutation by the Idylla assay.
Three cases had positive results shown by the Cobas assay (all showed exon 19 deletion) but negative results by the Idylla assay. In the NGS-based analysis, one had a rare exon 19 deletion (Leu747_Lys754delinsSerThr) that is not covered by the Idylla assay, one had an EGFR exon 19 insertion mutation (Lys745_Glu746insIleProValAlaIleLys), and the other had EGFR L747P point mutation of the exon 19. Therefore, we concluded that the Idylla assay was correct in two cases and the Cobas assay was correct in one case.
Because primer sequences of the Cobas EGFR Mutation Test v2 are not available, it was not possible to elucidate the reason for the false positive results. However, we noticed a shared sequence (… AAG GAA CCA…) between our case with the EGFR L747P point mutation (K-38) and one of the detectable exon 19 deletion mutations (Leu747_Thr751insPro) by the Cobas assay (Figure 2). Because the clinical genetic test for this patient (K-38), which was performed independently using the Cobas EGFR Mutation Test v2, also detected the exon 19 deletion, it is possible that the Cobas assay may call a false positive result for this rare EGFR exon 19 point mutation (L747P). However, this phenomenon should be confirmed in future studies using other NSCLC specimens with this rare EGFR mutation. The reason for the false-positive results by the Cobas EGFR Mutation Test v2 in the specimen with EGFR exon 19 insertion mutation (Lys745_Glu746insIleProValAlaIleLys) is unclear.
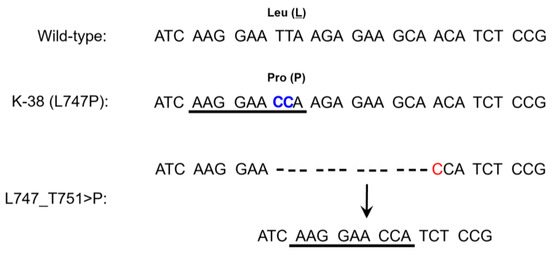
Figure 2.
Hypothesis for a false-positive result by the Cobas EGFR mutation test v2 in a tumor with EGFR L747P point mutation (K-38). Similarity of sequences between L747P point mutation and a detectable EGFR exon 19 deletion (L747_T751>P) by the Cobas EGFR mutation test v2 were highlighted by the underline (AAG GAA CCA). The mutated nucleotides are highlighted in color including the double thymine-to-cytosine (TT > CC) transition in L747P mutation.
4. Discussion
In this study, we found that the performance of the Idylla EGFR Mutation Test is comparable with that of the Cobas EGFR Mutation Test v2. The overall concordance rate of both assays was 96.4% and the ĸ value between the two assays was 0.920. This concordance rate was comparable with the result of a recently reported FACILITATE study, a real-world, prospective, multicenter European study that evaluated the performance of the Idylla EGFR Mutation Test with local reference methods (either NGS-based, Cobas, Therascreen EGFR RGQ PCR, Sanger sequencing, or other methods) in 16 sites [11]. In the FACILITATE study, the overall percentage agreement between the Idylla assay and local reference methods was 97.7%; the positive agreement was 87.4%, the negative agreement was 99.2%, and there were 38 (2.6%) discordant cases. In the analysis of discordant cases using a third method analysis such as digital droplet PCR (ddPCR), the mutant allele frequencies were quite low (0.4–4%) in cases with discordant-negative cases for the Idylla assay. In a third method analysis for discordant-positive cases, the Idylla assay was correct in three cases and the local reference method was correct in five cases. In our study, we compared PCR-based EGFR single gene analyses (Idylla EGFR Mutation Test vs. Cobas EGFR Mutation Test v2); after re-testing some discordant cases, the Idylla assay was found to achieve the correct results (indicating the Cobas assay had false negative or false positive results) in a few specimens. These data, together with the results of the FACILITATE study, support the clinical application of the Idylla EGFR Mutation Test as a single-plex EGFR mutation test for NSCLC patients.
The most Important advantage of the Idylla EGFR Mutation Test is the ultra-rapid TAT (~150 min). We thus consider how to apply this test in the era of multiplex genetic tests (Figure 3A). Because the EGFR mutation is one of the most frequent driver mutations in NSCLC, and as driver mutations in NSCLCs are usually present in mutually exclusive fashion, performing the Idylla EGFR Mutation Test prior to a multiplex genetic test may be a reasonable strategy to reduce the medical expenses of molecular profiling for NSCLC patients (Figure 3B). In fact, several previous studies reported the feasibility of performing the Idylla EGFR Mutation Test prior to NGS-based assays [12,13], although these studies focused on prompt EGFR mutation testing for patients under oncological emergency.
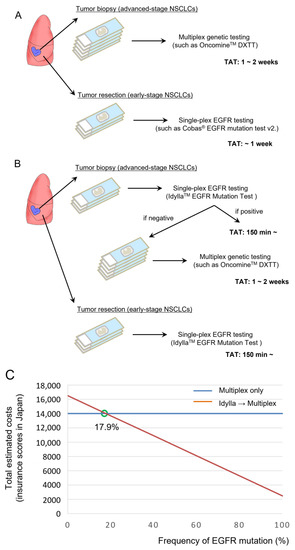
Figure 3.
Incorporation of the Idylla EGFR mutation test before multiplex genetic testing in a cohort with high EGFR mutation frequency. (A) Current standard of care for molecular profiling of advanced-stage NSCLCs and early-stage NSCLCs. (B) Potential strategies to incorporate the Idylla EGFR mutation test. (C) Total estimated costs of multiplex testing vs. the Idylla EGFR mutation test followed by multiplex testing (if EGFR is negative) in cohorts with various EGFR mutation frequencies.
Here we calculate the total expenses for molecular profiling using the medical insurance scores designated by the Japanese PMDA in March 2023; the insurance score for a single-plex EGFR test is 2500 (=25,000 yen) and the insurance score for a multiplex genetic test is 14,000 (=140,000 yen) for the Oncomine Dx Target Test. As shown in Figure 3C, the total estimated cost will become smaller in the combined Idylla assay plus multiplex test (Figure 3B) compared with the use of multiplex test only (Figure 3A), if applied for a cohort with an EGFR mutation frequency higher than 17.9%. Therefore, the combination test will reduce the total cost of molecular profiling, at least in Japan, if used for lung adenocarcinoma patients, in which the frequency of the EGFR mutation is approximately 30% or higher [14]. A recent case study reported that the Idylla EGFR Mutation Test failed to detect a rare EGFR exon 19 deletion (EGFR L747_A755delinsSS), which is not covered by the Idylla EGFR Mutation Test [15]. This is in line with the application of Idylla cartridges, which are intended to identify common, clinically relevant Tier 1 and 2 mutations and not rare or complex variants [16]. In our proposed molecular testing strategy, patients with a potentially targetable rare EGFR mutation will be identified by a subsequent NGS-based multiplex genetic testing such as the Oncomine Dx Target Test (Figure 3B).
In the early-stage setting after curative pulmonary resection, the current essential biomarker testing involves EGFR mutation analysis and PD-L1 staining to decide the adaptation of adjuvant treatment using osimertinib [6] or atezolizumab [17]. Because ASCO guideline updates [18], and ESMO consensus statements [19] indicate that adjuvant osimertinib is recommended for NSCLC patients with an EGFR sensitizing mutation regardless of the PD-L1 expression status, the ultra-rapid Idylla EGFR Mutation Test will provide a potential to avoid unessential PD-L1 testing if the examined specimen had an EGFR-sensitizing mutation.
Notably, PCR-based single-plex EGFR mutation analyses, including the Idylla and Cobas assays, have a disadvantage of EGFR mutation coverages (87–98%) compared with NGS [12,13,20]. These data support the usefulness of comprehensive genomic profiling during the treatment course of NSCLC patients [21]. In this analysis, we incidentally observed that the Cobas assay reported false-positive results (calling EGFR exon 19 deletion in specimens with EGFR exon 19 L747P point mutation or exon 19 insertion). The detailed mechanisms and the incidence of such false positive results is unclear because of the lack of sufficient data [22]. Clinicians should be aware that patients may lose the opportunity to receive adequate treatment because of false-positive or false-negative biomarker testing results.
EGFR L747P point mutation, which results from a double thymine-to-cytosine (TT > CC) transition (Figure 2), comprises less than 1% of EGFR mutations [23]. The current standard of care for NSCLC patients with EGFR exon 19 deletion is osimertinib [24]. However, a recent structure-function analysis of various EGFR mutation variants [25] indicated that the L747P mutation is classified into “P-loop and C-helix compressing group” that will show inherent resistance to 1st- or 3rd-generation TKIs but is sensitive to 2nd-generation TKIs such as afatinib or dacomitinib. Several case studies have reported on the efficacy of afatinib or dacomitinib and low efficacy of gefitinib or osimertinib in NSCLC patients with EGFR L747P point mutation [23,26,27].
Another EGFR mutation that led to a false-positive result in the Cobas assay was a rare EGFR exon 19 insertion mutation (Leu747_Lys754delinsSerThr). Several studies have found that tumors with EGFR exon 19 insertion mutation are usually sensitive to EGFR-TKIs [28,29], and therefore these mutations should be included in the list of detectable driver mutations in the near future. Additionally, compound EGFR mutations, which are often missed through the use of mutation-specific assays, may lead to varying responses to EGFR-TKIs. These observations also highlight the importance of comprehensive genomic profiling in NSCLC patients at least once during the treatment course.
On the basis of several advantages of the Idylla assays, other molecular testing platforms including Idylla GeneFusion assay, the Idylla BRAF Mutation Test, the Idylla KRAS Mutation Test, and the Idylla NRAS-BRAF Mutation Test have been developed. In a comparison analysis of the Idylla GeneFusion assay for previously characterized fusion-positive tumors (37 NSCLCs and 2 parotid gland carcinomas), the Idylla GeneFusion assay successfully detected 36 fusions (overall agreement: 92.3%) [30]. Another group compared the performance of two ultrafast gene fusion assays (the Idylla GeneFusion assay and the NGS-based Genexus assay) in 195 NSCLC cases (113 known gene fusions and 82 wild-type tumors). The accuracy was 92.3% and 93.1% for the Idylla assay and the Genexus assay, respectively [31].
Comparisons of the Idylla assay with other genetic aberrations have been performed in solid tumors other than NSCLCs. For example, in the analysis of KRAS, NRAS, and BRAF mutations in 850 colorectal cancer (CRC) cases, the concordance rate was 88.6%, accounting for all mutations including those not in the Idylla cartridge by design [32]. The colorectal team in our department also performed a comparison study between the Idylla assay and the MEBGEN RASKET-B assay for KRAS, NRAS, and BRAF mutations using 253 CRC specimens. The authors observed the potential clinical usefulness of the Idylla assay, showing a high concordance rate of 97.4%, a negative concordance rate of 95.7%, and overall concordance rate of 95.3% (κ = 0.919, 95% CI 0.871–0.967) [33]. Another group compared the Idylla assay with anti-BRAF V600E (clone VE1) immunohistochemistry in 90 melanoma samples. The agreement rate of the assays was 91% (72/79) [34]. Thus, these comparison studies, including the current study, have revealed the accuracy and clinical usefulness of various Idylla assays.
5. Conclusions
We demonstrated the comparability of the ultra-rapid Idylla EGFR Mutation Test compared with the Cobas EGFR mutation test v2. The Idylla EGFR Mutation Test may have the potential to reduce the cost of driver mutation testing if used in a cohort with high EGFR mutation rates without extending the TAT; furthermore, it may dramatically reduce the TAT if patients have EGFR mutation. We also observed the limitations of the PCR-based EGFR genetic tests in terms of false-positive results (especially for the Cobas EGFR mutation test v2) and false-negative results, including the detection of the EGFR compound mutations. While these phenomena might be rare, clinicians should keep the possibility of false-positive/negative results in mind when treating NSCLC patients.
Author Contributions
Conceptualization, K.S. (Kazuko Sakai) and K.N.; methodology, K.S. (Kazuko Sakai) and K.N.; validation, K.S. (Kenichi Suda); formal analysis, K.S. (Kazuko Sakai) and K.N.; investigation, K.S. (Kenichi Suda), K.S. (Kazuko Sakai) and K.N.; resources, K.S. (Kenichi Suda), T.O., T.C., T.S., J.M., T.N., N.I., Y.T. and T.M.; data curation, K.S. (Kazuko Sakai); writing—original draft preparation, K.S. (Kenichi Suda) and K.S. (Kazuko Sakai); writing—review and editing, all authors; supervision, K.N.; project administration, K.N.; funding acquisition, K.S. (Kenichi Suda) and K.N. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a research grant from Nichirei Bioscience Inc. and a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science [grant number 22K07291 to K. Suda].
Institutional Review Board Statement
This study was approved by the ethical committees of the Kindai University Faculty of Medicine and the Tokyo Medical University Hospital (R03-191 and T2021-0281, respectively).
Informed Consent Statement
Written informed consent from each patient was waived by the ethical committees because of the retrospective nature of this study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The authors thank Keiko Obata and Kumiko Nagase for their technical assistance. The authors also thank all the participants of this study.
Conflicts of Interest
K. Suda reports a research grant from AstraZeneca and honoraria from AstraZeneca, Chugai, and Taiho. K. Sakai reports honoraria from Roche Diagnostics, Bio–Rad, AstraZeneca, Chugai Pharma and Hitachi. N. Ikeda reports receiving grants from Astra Zeneca, Chugai Pharma, Boehringer Ingelheim, Pfizer, Taiho Pharma, Eli Lilly, Ono Pharma, Bristol Meyers, MSD, Nihon Mediphysics, Teijin Pharma, Kyowa Kirin, Sanofi, Eizai, Astellas, Shionogi, Daiichi-Sankyo, Roche Diagnostic, Fuji film and Johnson & Johnson and honoraria from Astra Zeneca, Chugai Pharma, Boehringer Ingelheim, Taiho Pharma, Eli Lilly, Ono Pharma, Bristol Meyers, Olympus, MSD, Johnson & Johnson, Nihon Mediphysics, Medtronics and Teijin Pharma. Y. Tsutani reports receiving grants and personal fees from Boehringer Ingelheim, Chugai, Taiho, and Medtronic; grants from Daiichi Sankyo; and personal fees from AstraZeneca, Bristol-Myers Squibb, Ono Pharmaceutical, MSD, Novartis, Nihon Mediphysics, Japan Blood Products Organization, Takeda Pharmaceutical, and Johnson and Johnson. T. Mitsudomi reports receiving grants and personal fees from Boehringer Ingelheim, Chugai, MSD, and Ono; grants from AstraZeneca, Taiho, Daiichi Sankyo, and Ethicon; and personal fees from Pfizer, Eli Lilly, Novartis, Thermo Fisher, BMS, Janssen, Amgen, Guardant Health, Invitae, and Merck Biopharma. K. Nishio reports grants and personal fees from Eli Lilly and Nippon Boehringer Ingelheim, grants from West Japan Oncology Group, Ignyta, Inc., Korea Otsuka Pharmaceutical, Thoracic Oncology Research Group, North East Japan Study Group, Clinical Research Support Center Kyushu, Nichirei Biosciences, and Osakaminami Hospital, and lecture fees from Chugai Pharma, Eisai, Pfizer, Boehringer Ingelheim, Novartis Pharma, MSD, Ono Pharmaceutical, Bristol-Myers Squibb, SymBio Pharmaceuticals, Life Technologies, Solasia Pharma, Eli Lilly, Yakult Honsha, Roche Diagnostics, Life Technologies, AstraZeneca, Otsuka Pharmaceutical, Sanofi, Guardant Health, Amgen, and Merck Biopharma. The other authors have declared no conflict of interest. These funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Hirsch, F.R.; Suda, K.; Wiens, J.; Bunn, P.A., Jr. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet 2016, 388, 1012–1024. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment with Targeted Tyrosine Kinase Inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J. Thorac. Oncol. 2018, 13, 323–358. [Google Scholar] [CrossRef] [PubMed]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Loong, H.H.; Wong, C.K.H.; Chan, C.P.K.; Chang, A.; Zhou, Z.Y.; Tang, W.; Gibbs, M. Clinical and Economic Impact of Upfront Next-Generation Sequencing for Metastatic NSCLC in East Asia. JTO Clin. Res. Rep. 2022, 3, 100290. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.L.; Tsuboi, M.; He, J.; John, T.; Grohe, C.; Majem, M.; Goldman, J.W.; Laktionov, K.; Kim, S.W.; Kato, T.; et al. Osimertinib in Resected EGFR-Mutated Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 1711–1723. [Google Scholar] [CrossRef]
- De Luca, C.; Gragnano, G.; Pisapia, P.; Vigliar, E.; Malapelle, U.; Bellevicine, C.; Troncone, G. EGFR mutation detection on lung cancer cytological specimens by the novel fully automated PCR-based Idylla EGFR Mutation Assay. J. Clin. Pathol. 2017, 70, 295–300. [Google Scholar] [CrossRef]
- Lambros, L.; Caumont, C.; Guibourg, B.; Barel, F.; Quintin-Roue, I.; Marcorelles, P.; Merlio, J.P.; Uguen, A. Evaluation of a fast and fully automated platform to diagnose EGFR and KRAS mutations in formalin-fixed and paraffin-embedded non-small cell lung cancer samples in less than one day. J. Clin. Pathol. 2017, 70, 544–549. [Google Scholar] [CrossRef]
- Murakami, S.; Yokose, T.; Shinada, K.; Isaka, T.; Katakura, K.; Ushio, R.; Kondo, T.; Kato, T.; Ito, H.; Saito, H. Comparison of next-generation sequencing and cobas EGFR mutation test v2 in detecting EGFR mutations. Thorac. Cancer 2022, 13, 3217–3224. [Google Scholar] [CrossRef]
- Belardinilli, F.; Capalbo, C.; Buffone, A.; Petroni, M.; Colicchia, V.; Ferraro, S.; Zani, M.; Nicolussi, A.; D’Inzeo, S.; Coppa, A.; et al. Validation of the Ion Torrent PGM sequencing for the prospective routine molecular diagnostic of colorectal cancer. Clin. Biochem. 2015, 48, 908–910. [Google Scholar] [CrossRef]
- Behnke, A.; Cayre, A.; De Maglio, G.; Giannini, G.; Habran, L.; Tarsitano, M.; Chetta, M.; Cappellen, D.; Lespagnol, A.; Le Naoures, C.; et al. FACILITATE: A real-world, multicenter, prospective study investigating the utility of a rapid, fully automated real-time PCR assay versus local reference methods for detecting epidermal growth factor receptor variants in NSCLC. Pathol. Oncol. Res. 2023, 29, 1610707. [Google Scholar] [CrossRef] [PubMed]
- Arcila, M.E.; Yang, S.R.; Momeni, A.; Mata, D.A.; Salazar, P.; Chan, R.; Elezovic, D.; Benayed, R.; Zehir, A.; Buonocore, D.J.; et al. Ultrarapid EGFR Mutation Screening Followed by Comprehensive Next-Generation Sequencing: A Feasible, Informative Approach for Lung Carcinoma Cytology Specimens with a High Success Rate. JTO Clin. Res. Rep. 2020, 1, 100077. [Google Scholar] [CrossRef]
- Momeni-Boroujeni, A.; Salazar, P.; Zheng, T.; Mensah, N.; Rijo, I.; Dogan, S.; Yao, J.; Moung, C.; Vanderbilt, C.; Benhamida, J.; et al. Rapid EGFR Mutation Detection Using the Idylla Platform: Single-Institution Experience of 1200 Cases Analyzed by an In-House Developed Pipeline and Comparison with Concurrent Next-Generation Sequencing Results. J. Mol. Diagn. 2021, 23, 310–322. [Google Scholar] [CrossRef]
- Suda, K.; Mitsudomi, T.; Shintani, Y.; Okami, J.; Ito, H.; Ohtsuka, T.; Toyooka, S.; Mori, T.; Watanabe, S.I.; Asamura, H.; et al. Clinical Impacts of EGFR Mutation Status: Analysis of 5780 Surgically Resected Lung Cancer Cases. Ann. Thorac. Surg. 2021, 111, 269–276. [Google Scholar] [CrossRef]
- O’Sullivan, H.; d’Arienzo, P.D.; Yousaf, N.; Cui, W.; Popat, S. Inadequacy of PCR genotyping in advanced non-small cell lung cancer: EGFR L747_A755delinsSS exon 19 deletion is not detected by the real-time PCR Idylla EGFR mutation test but is detected by ctDNA next generation sequencing and responds to osimertinib. Eur. J. Cancer 2022, 166, 38–40. [Google Scholar] [CrossRef]
- Nkosi, D.; Casler, V.L.; Syposs, C.R.; Oltvai, Z.N. Utility of Select Gene Mutation Detection in Tumors by the Idylla Rapid Multiplex PCR Platform in Comparison to Next-Generation Sequencing. Genes 2022, 13, 799. [Google Scholar] [CrossRef] [PubMed]
- Felip, E.; Altorki, N.; Zhou, C.; Csoszi, T.; Vynnychenko, I.; Goloborodko, O.; Luft, A.; Akopov, A.; Martinez-Marti, A.; Kenmotsu, H.; et al. Adjuvant atezolizumab after adjuvant chemotherapy in resected stage IB-IIIA non-small-cell lung cancer (IMpower010): A randomised, multicentre, open-label, phase 3 trial. Lancet 2021, 398, 1344–1357. [Google Scholar] [CrossRef] [PubMed]
- Pisters, K.; Kris, M.G.; Gaspar, L.E.; Ismaila, N.; Adjuvant Systemic, T.; Adjuvant Radiation Therapy for Stage, I.t.I.N.G.E.P. Adjuvant Systemic Therapy and Adjuvant Radiation Therapy for Stage I-IIIA Completely Resected Non-Small-Cell Lung Cancer: ASCO Guideline Rapid Recommendation Update. J. Clin. Oncol. 2022, 40, 1127–1129. [Google Scholar] [CrossRef]
- Passaro, A.; Leighl, N.; Blackhall, F.; Popat, S.; Kerr, K.; Ahn, M.J.; Arcila, M.E.; Arrieta, O.; Planchard, D.; de Marinis, F.; et al. ESMO expert consensus statements on the management of EGFR mutant non-small-cell lung cancer. Ann. Oncol. 2022, 33, 466–487. [Google Scholar] [CrossRef]
- Khalifa, E.; Chapusot, C.; Tournier, B.; Sentis, J.; Marion, E.; Remond, A.; Aubry, M.; Pioche, C.; Bergeron, A.; Primois, C.; et al. Idylla EGFR assay on extracted DNA: Advantages, limits and place in molecular screening according to the latest guidelines for non-small-cell lung cancer (NSCLC) patients. J. Clin. Pathol. 2022. [Google Scholar] [CrossRef]
- Kunimasa, K.; Sugimoto, N.; Kawamura, T.; Yamasaki, T.; Honma, K.; Nagata, S.; Kukita, Y.; Fujisawa, F.; Inoue, T.; Yamaguchi, Y.; et al. Clinical application of comprehensive genomic profiling panel to thoracic malignancies: A single-center retrospective study. Thorac. Cancer 2022, 13, 2970–2977. [Google Scholar] [CrossRef]
- Shepherd, P.; Sheath, K.L.; Tin, S.T.; Khwaounjoo, P.; Aye, P.S.; Li, A.; Laking, G.R.; Kingston, N.J.; Lewis, C.A.; Elwood, J.M.; et al. Lung cancer mutation testing: A clinical retesting study of agreement between a real-time PCR and a mass spectrometry test. Oncotarget 2017, 8, 101437–101451. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.K.; Ko, J.C.; Yang, J.C.; Shih, J.Y. Afatinib is effective in the treatment of lung adenocarcinoma with uncommon EGFR p.L747P and p.L747S mutations. Lung Cancer 2019, 133, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Soria, J.C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef]
- Robichaux, J.P.; Le, X.; Vijayan, R.S.K.; Hicks, J.K.; Heeke, S.; Elamin, Y.Y.; Lin, H.Y.; Udagawa, H.; Skoulidis, F.; Tran, H.; et al. Structure-based classification predicts drug response in EGFR-mutant NSCLC. Nature 2021, 597, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, Y.; Zhai, Y.; Wang, J. Non-small cell lung cancer harboring a rare EGFR L747P mutation showing intrinsic resistance to both gefitinib and osimertinib (AZD9291): A case report. Thorac. Cancer 2018, 9, 745–749. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Zhou, X.; Li, P.; Qi, C.; Ling, Y. EGFR L747P mutation in one lung adenocarcinoma patient responded to afatinib treatment: A case report. J. Thorac. Dis. 2018, 10, E802–E805. [Google Scholar] [CrossRef]
- He, M.; Capelletti, M.; Nafa, K.; Yun, C.H.; Arcila, M.E.; Miller, V.A.; Ginsberg, M.S.; Zhao, B.; Kris, M.G.; Eck, M.J.; et al. EGFR exon 19 insertions: A new family of sensitizing EGFR mutations in lung adenocarcinoma. Clin. Cancer Res. 2012, 18, 1790–1797. [Google Scholar] [CrossRef]
- Janning, M.; Suptitz, J.; Albers-Leischner, C.; Delpy, P.; Tufman, A.; Velthaus-Rusik, J.L.; Reck, M.; Jung, A.; Kauffmann-Guerrero, D.; Bonzheim, I.; et al. Treatment outcome of atypical EGFR mutations in the German National Network Genomic Medicine Lung Cancer (nNGM). Ann. Oncol. 2022, 33, 602–615. [Google Scholar] [CrossRef]
- Leone, A.; Muscarella, L.A.; Graziano, P.; Tornese, A.; Grillo, L.R.; Di Lorenzo, A.; Bronzini, M.; Scarpino, S.; Sparaneo, A.; Rossi, G. Robust Performance of the Novel Research-Use-Only Idylla GeneFusion Assay Using a Diverse Set of Pathological Samples with a Proposed 1-Day Workflow for Advanced NSCLC Evaluation. Cancers 2022, 15, 292. [Google Scholar] [CrossRef]
- Hofman, V.; Heeke, S.; Bontoux, C.; Chalabreysse, L.; Barritault, M.; Bringuier, P.P.; Fenouil, T.; Benzerdjeb, N.; Begueret, H.; Merlio, J.P.; et al. Ultrafast Gene Fusion Assessment for Nonsquamous NSCLC. JTO Clin. Res. Rep. 2023, 4, 100457. [Google Scholar] [CrossRef] [PubMed]
- Tsongalis, G.J.; Al Turkmani, M.R.; Suriawinata, M.; Babcock, M.J.; Mitchell, K.; Ding, Y.; Scicchitano, L.; Tira, A.; Buckingham, L.; Atkinson, S.; et al. Comparison of Tissue Molecular Biomarker Testing Turnaround Times and Concordance Between Standard of Care and the Biocartis Idylla Platform in Patients with Colorectal Cancer. Am. J. Clin. Pathol. 2020, 154, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Makutani, Y.; Sakai, K.; Yamada, M.; Wada, T.; Chikugo, T.; Satou, T.; Iwasa, Y.; Yamamoto, H.; de Velasco, M.A.; Nishio, K.; et al. Performance of Idylla() RAS-BRAF mutation test for formalin-fixed paraffin-embedded tissues of colorectal cancer. Int. J. Clin. Oncol. 2022, 27, 1180–1187. [Google Scholar] [CrossRef]
- Long-Mira, E.; Picard-Gauci, A.; Lassalle, S.; Hofman, V.; Lalvee, S.; Tanga, V.; Zahaf, K.; Bonnetaud, C.; Lespinet, V.; Camuzard, O.; et al. Comparison of Two Rapid Assays for the Detection of BRAF V600 Mutations in Metastatic Melanoma including Positive Sentinel Lymph Nodes. Diagnostics 2022, 12, 751. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).