Harnessing the Microbiome to Reduce Pancreatic Cancer Burden
Abstract
Simple Summary
Abstract
1. Introduction
2. Pancreatic Cancer Risk Factors
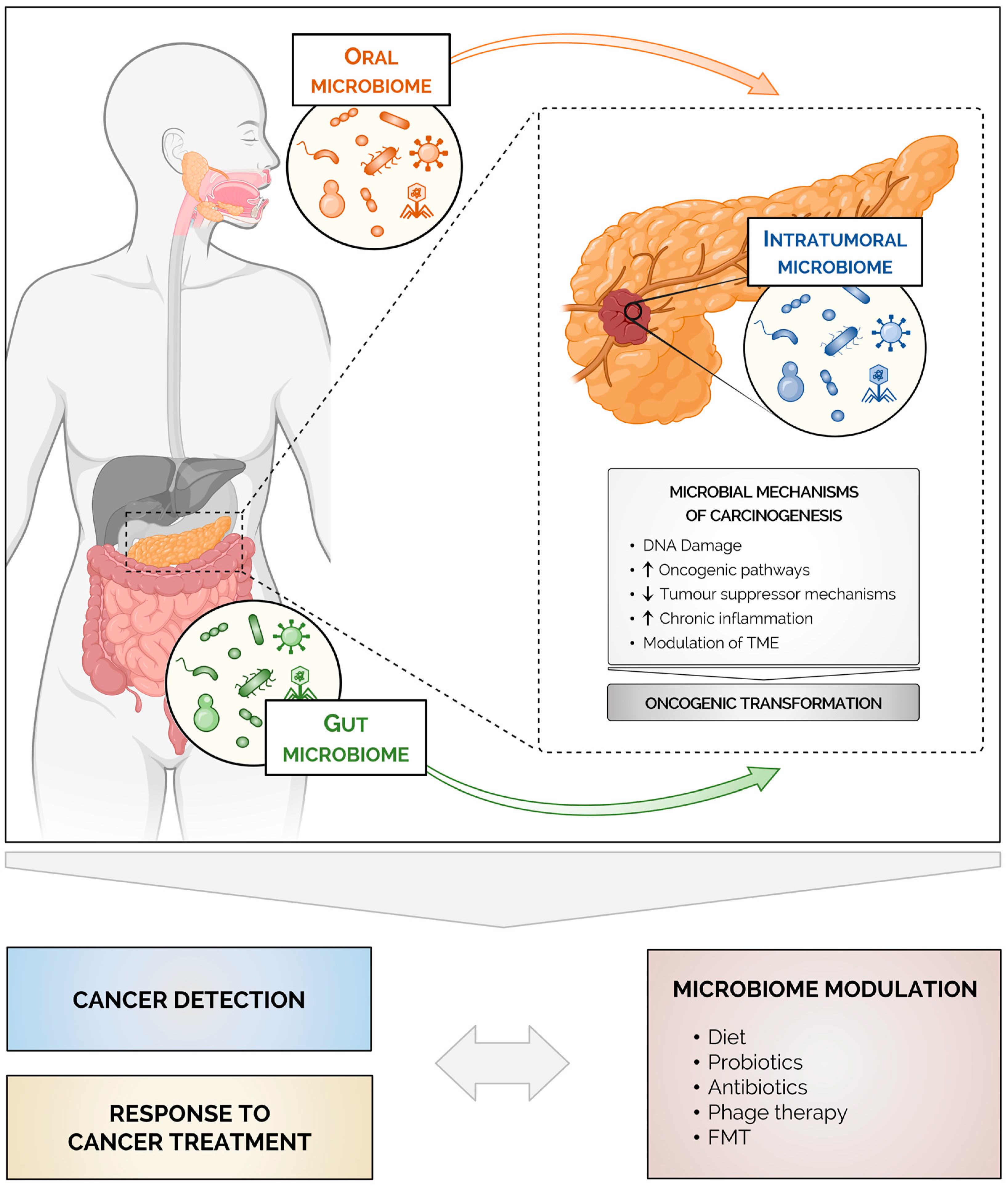
3. The Human Microbiome
4. The Microbiome in Pancreatic Cancer
4.1. The Intratumoral Microbiome
4.2. The Gut and the Oral Microbiomes in Pancreatic Cancer
4.2.1. The Gut Microbiome and Pancreatic Cancer
4.2.2. The Oral Microbiome and Pancreatic Cancer
5. Mechanisms Linking the Microbiome to Pancreatic Cancer
6. The Microbiome and Treatment Responses in Pancreatic Cancer
6.1. Chemotherapy
6.2. Immunotherapy
7. Microbiome Modulation in Pancreatic Cancer
7.1. Diet
7.2. Probiotics
7.3. Antibiotics
7.4. Fecal Microbiota Transplantation
8. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Laversanne, M.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I. Global Cancer Observatory: Cancer Tomorrow. Lyon, France: International Agency for Research on Cancer. 2020. Available online: https://gco.iarc.fr/tomorrow (accessed on 16 November 2022).
- Mizrahi, J.D.; Surana, R.; Valle, J.W.; Shroff, R.T. Pancreatic Cancer. Lancet 2020, 395, 2008–2020. [Google Scholar] [CrossRef]
- Zambirinis, C.P.; Pushalkar, S.; Saxena, D.; Miller, G. Pancreatic Cancer, Inflammation, and Microbiome. Cancer J. 2014, 20, 195–202. [Google Scholar] [CrossRef]
- Guerra, C.; Schuhmacher, A.J.; Cañamero, M.; Grippo, P.J.; Verdaguer, L.; Pérez-Gallego, L.; Dubus, P.; Sandgren, E.P.; Barbacid, M. Chronic Pancreatitis Is Essential for Induction of Pancreatic Ductal Adenocarcinoma by K-Ras Oncogenes in Adult Mice. Cancer Cell 2007, 11, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Löhr, M.; Klöppel, G.; Maisonneuve, P.; Lowenfels, A.B.; Lüttges, J. Frequency of K-Ras Mutations in Pancreatic Intraductal Neoplasias Associated with Pancreatic Ductal Adenocarcinoma and Chronic Pancreatitis: A Meta-Analysis. Neoplasia 2005, 7, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Midha, S.; Chawla, S.; Garg, P.K. Modifiable and Non-Modifiable Risk Factors for Pancreatic Cancer: A Review. Cancer Lett. 2016, 381, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Carreras-Torres, R.; Johansson, M.; Gaborieau, V.; Haycock, P.C.; Wade, K.H.; Relton, C.L.; Martin, R.M.; Davey Smith, G.; Brennan, P. The Role of Obesity, Type 2 Diabetes, and Metabolic Factors in Pancreatic Cancer: A Mendelian Randomization Study. J. Natl. Cancer Inst. 2017, 109, djx012. [Google Scholar] [CrossRef]
- Hujoel, P.P.; Drangsholt, M.; Spiekerman, C.; Weiss, N.S. An Exploration of the Periodontitis-Cancer Association. Ann. Epidemiol. 2003, 13, 312–316. [Google Scholar] [CrossRef]
- Michaud, D.S.; Izard, J.; Wilhelm-Benartzi, C.S.; You, D.-H.; Grote, V.A.; Tjønneland, A.; Dahm, C.C.; Overvad, K.; Jenab, M.; Fedirko, V.; et al. Plasma Antibodies to Oral Bacteria and Risk of Pancreatic Cancer in a Large European Prospective Cohort Study. Gut 2013, 62, 1764–1770. [Google Scholar] [CrossRef]
- Ahn, J.; Segers, S.; Hayes, R.B. Periodontal Disease, Porphyromonas Gingivalis Serum Antibody Levels and Orodigestive Cancer Mortality. Carcinogenesis 2012, 33, 1055–1058. [Google Scholar] [CrossRef]
- Tijeras-Raballand, A.; Hilmi, M.; Astorgues-Xerri, L.; Nicolle, R.; Bièche, I.; Neuzillet, C. Microbiome and Pancreatic Ductal Adenocarcinoma. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101589. [Google Scholar] [CrossRef] [PubMed]
- Cullin, N.; Azevedo Antunes, C.; Straussman, R.; Stein-Thoeringer, C.K.; Elinav, E. Microbiome and Cancer. Cancer Cell 2021, 39, 1317–1341. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Gao, H.-L.; Wang, W.-Q.; Yu, X.-J.; Liu, L. Bidirectional and Dynamic Interaction between the Microbiota and Therapeutic Resistance in Pancreatic Cancer. Biochim. Biophys. acta. Rev. cancer 2021, 1875, 188484. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current Understanding of the Human Microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Glover, T. Progress in Cancer Research. Canada Lancet Pract. 1926, 67, 161–216. [Google Scholar]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J.; et al. Microbiome Analyses of Blood and Tissues Suggest Cancer Diagnostic Approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The Human Tumor Microbiome Is Composed of Tumor Type-Specific Intracellular Bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef]
- Kartal, E.; Schmidt, T.S.B.; Molina-Montes, E.; Rodríguez-Perales, S.; Wirbel, J.; Maistrenko, O.M.; Akanni, W.A.; Alashkar Alhamwe, B.; Alves, R.J.; Carrato, A.; et al. A Faecal Microbiota Signature with High Specificity for Pancreatic Cancer. Gut 2022, 71, 1359–1372. [Google Scholar] [CrossRef]
- Tan, Q.; Ma, X.; Yang, B.; Liu, Y.; Xie, Y.; Wang, X.; Yuan, W.; Ma, J. Periodontitis Pathogen Porphyromonas Gingivalis Promotes Pancreatic Tumorigenesis via Neutrophil Elastase from Tumor-Associated Neutrophils. Gut Microbes 2022, 14, 2073785. [Google Scholar] [CrossRef]
- Huang, Y.; Zhu, N.; Zheng, X.; Liu, Y.; Lu, H.; Yin, X.; Hao, H.; Tan, Y.; Wang, D.; Hu, H.; et al. Intratumor Microbiome Analysis Identifies Positive Association Between Megasphaera and Survival of Chinese Patients with Pancreatic Ductal Adenocarcinomas. Front. Immunol. 2022, 13, 785422. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; San Lucas, A.; et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 2019, 178, 795–806.e12. [Google Scholar] [CrossRef] [PubMed]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential Role of Intratumor Bacteria in Mediating Tumor Resistance to the Chemotherapeutic Drug Gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef]
- Del Castillo, E.; Meier, R.; Chung, M.; Koestler, D.C.; Chen, T.; Paster, B.J.; Charpentier, K.P.; Kelsey, K.T.; Izard, J.; Michaud, D.S. The Microbiomes of Pancreatic and Duodenum Tissue Overlap and Are Highly Subject Specific but Differ between Pancreatic Cancer and Noncancer Subjects. Cancer Epidemiol. Biomarkers Prev. 2019, 28, 370–383. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Hu, Y. Microbiome Harbored within Tumors: A New Chance to Revisit Our Understanding of Cancer Pathogenesis and Treatment. Signal Transduct. Target. Ther. 2020, 5, 136. [Google Scholar] [CrossRef] [PubMed]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef]
- Sethi, V.; Vitiello, G.A.; Saxena, D.; Miller, G.; Dudeja, V. The Role of the Microbiome in Immunologic Development and Its Implication For Pancreatic Cancer Immunotherapy. Gastroenterology 2019, 156, 2097–2115.e2. [Google Scholar] [CrossRef]
- Elaskandrany, M.; Patel, R.; Patel, M.; Miller, G.; Saxena, D.; Saxena, A. Fungi, Host Immune Response, and Tumorigenesis. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 321, G213–G222. [Google Scholar] [CrossRef]
- Aykut, B.; Pushalkar, S.; Chen, R.; Li, Q.; Abengozar, R.; Kim, J.I.; Shadaloey, S.A.; Wu, D.; Preiss, P.; Verma, N.; et al. The Fungal Mycobiome Promotes Pancreatic Oncogenesis via Activation of MBL. Nature 2019, 574, 264–267. [Google Scholar] [CrossRef]
- Dambuza, I.M.; Brown, G.D. Fungi Accelerate Pancreatic Cancer. Nature 2019, 574, 184–185. [Google Scholar] [CrossRef]
- Huang, J.; Roosaar, A.; Axéll, T.; Ye, W. A Prospective Cohort Study on Poor Oral Hygiene and Pancreatic Cancer Risk. Int. J. Cancer 2016, 138, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Chung, L.-M.; Liang, J.-A.; Lin, C.-L.; Sun, L.-M.; Kao, C.-H. Cancer Risk in Patients with Candidiasis: A Nationwide Population-Based Cohort Study. Oncotarget 2017, 8, 63562–63573. [Google Scholar] [CrossRef] [PubMed]
- Makohon-Moore, A.P.; Zhang, M.; Reiter, J.G.; Bozic, I.; Allen, B.; Kundu, D.; Chatterjee, K.; Wong, F.; Jiao, Y.; Kohutek, Z.A.; et al. Limited Heterogeneity of Known Driver Gene Mutations among the Metastases of Individual Patients with Pancreatic Cancer. Nat. Genet. 2017, 49, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Xiao, L.; Gao, Y.; Wang, G.; Gao, H.; Peng, Y.; Zhu, X.; Wei, J.; Miao, Y.; Jiang, K.; et al. Comparative Bioinformatical Analysis of Pancreatic Head Cancer and Pancreatic Body/Tail Cancer. Med. Oncol. 2020, 37, 46. [Google Scholar] [CrossRef]
- Gleeson, F.C.; Jeraldo, P.; Levy, M.J.; Murphy, S.J.; Mendes-Soares, H.; Karagouga, G.; Mccune, A.F.; Garcia Garcia Deparedes, A.; Kipp, B.R.; Song, S.D.; et al. Composition, Diversity and Potential Utility of Intervention-Naïve Pancreatic Cancer Intratumoral Microbiome Signature Profiling via Endoscopic Ultrasound. Gut 2022, 71, 441–443. [Google Scholar] [CrossRef]
- Mitsuhashi, K.; Nosho, K.; Sukawa, Y.; Matsunaga, Y.; Ito, M.; Kurihara, H.; Kanno, S.; Igarashi, H.; Naito, T.; Adachi, Y.; et al. Association of Fusobacterium Species in Pancreatic Cancer Tissues with Molecular Features and Prognosis. Oncotarget 2015, 6, 7209–7220. [Google Scholar] [CrossRef]
- Nagata, N.; Nishijima, S.; Kojima, Y.; Hisada, Y.; Imbe, K.; Miyoshi-Akiyama, T.; Suda, W.; Kimura, M.; Aoki, R.; Sekine, K.; et al. Metagenomic Identification of Microbial Signatures Predicting Pancreatic Cancer From a Multinational Study. Gastroenterology 2022, 163, 222–238. [Google Scholar] [CrossRef]
- Matsukawa, H.; Iida, N.; Kitamura, K.; Terashima, T.; Seishima, J.; Makino, I.; Kannon, T.; Hosomichi, K.; Yamashita, T.; Sakai, Y.; et al. Dysbiotic Gut Microbiota in Pancreatic Cancer Patients Form Correlation Networks with the Oral Microbiota and Prognostic Factors. Am. J. Cancer Res. 2021, 11, 3163–3175. [Google Scholar] [PubMed]
- Hashimoto, S.; Tochio, T.; Funasaka, K.; Funahashi, K.; Hartanto, T.; Togashi, Y.; Saito, M.; Nishimoto, Y.; Yoshinori, M.; Nakaoka, K.; et al. Changes in Intestinal Bacteria and Imbalances of Metabolites Induced in the Intestines of Pancreatic Ductal Adenocarcinoma Patients in a Japanese Population: A Preliminary Result. Scand. J. Gastroenterol. 2023, 58, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zhang, D.; Li, Z.; Jiang, H.; Li, J.; Ren, R.; Gao, X.; Li, J.; Wang, X.; Wang, W.; et al. The Fecal Microbiota of Patients with Pancreatic Ductal Adenocarcinoma and Autoimmune Pancreatitis Characterized by Metagenomic Sequencing. J. Transl. Med. 2021, 19, 215. [Google Scholar] [CrossRef]
- Half, E.; Keren, N.; Reshef, L.; Dorfman, T.; Lachter, I.; Kluger, Y.; Reshef, N.; Knobler, H.; Maor, Y.; Stein, A.; et al. Fecal Microbiome Signatures of Pancreatic Cancer Patients. Sci. Rep. 2019, 9, 16801. [Google Scholar] [CrossRef]
- Ren, Z.; Jiang, J.; Xie, H.; Li, A.; Lu, H.; Xu, S.; Zhou, L.; Zhang, H.; Cui, G.; Chen, X.; et al. Gut Microbial Profile Analysis by MiSeq Sequencing of Pancreatic Carcinoma Patients in China. Oncotarget 2017, 8, 95176–95191. [Google Scholar] [CrossRef]
- Petrick, J.L.; Wilkinson, J.E.; Michaud, D.S.; Cai, Q.; Gerlovin, H.; Signorello, L.B.; Wolpin, B.M.; Ruiz-Narváez, E.A.; Long, J.; Yang, Y.; et al. The Oral Microbiome in Relation to Pancreatic Cancer Risk in African Americans. Br. J. Cancer 2022, 126, 287–296. [Google Scholar] [CrossRef]
- Sun, H.; Zhao, X.; Zhou, Y.; Wang, J.; Ma, R.; Ren, X.; Wang, H.; Zou, L. Characterization of Oral Microbiome and Exploration of Potential Biomarkers in Patients with Pancreatic Cancer. Biomed Res. Int. 2020, 2020, 4712498. [Google Scholar] [CrossRef] [PubMed]
- Vogtmann, E.; Han, Y.; Caporaso, J.G.; Bokulich, N.; Mohamadkhani, A.; Moayyedkazemi, A.; Hua, X.; Kamangar, F.; Wan, Y.; Suman, S.; et al. Oral Microbial Community Composition Is Associated with Pancreatic Cancer: A Case-Control Study in Iran. Cancer Med. 2020, 9, 797–806. [Google Scholar] [CrossRef]
- Wei, A.-L.; Li, M.; Li, G.-Q.; Wang, X.; Hu, W.-M.; Li, Z.-L.; Yuan, J.; Liu, H.-Y.; Zhou, L.-L.; Li, K.; et al. Oral Microbiome and Pancreatic Cancer. World J. Gastroenterol. 2020, 26, 7679–7692. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Alekseyenko, A.V.; Wu, J.; Peters, B.A.; Jacobs, E.J.; Gapstur, S.M.; Purdue, M.P.; Abnet, C.C.; Stolzenberg-Solomon, R.; Miller, G.; et al. Human Oral Microbiome and Prospective Risk for Pancreatic Cancer: A Population-Based Nested Case-Control Study. Gut 2018, 67, 120–127. [Google Scholar] [CrossRef]
- Olson, S.H.; Satagopan, J.; Xu, Y.; Ling, L.; Leong, S.; Orlow, I.; Saldia, A.; Li, P.; Nunes, P.; Madonia, V.; et al. The Oral Microbiota in Patients with Pancreatic Cancer, Patients with IPMNs, and Controls: A Pilot Study. Cancer Causes Control 2017, 28, 959–969. [Google Scholar] [CrossRef]
- Torres, P.J.; Fletcher, E.M.; Gibbons, S.M.; Bouvet, M.; Doran, K.S.; Kelley, S.T. Characterization of the Salivary Microbiome in Patients with Pancreatic Cancer. PeerJ 2015, 3, e1373. [Google Scholar] [CrossRef]
- Farrell, J.J.; Zhang, L.; Zhou, H.; Chia, D.; Elashoff, D.; Akin, D.; Paster, B.J.; Joshipura, K.; Wong, D.T.W. Variations of Oral Microbiota Are Associated with Pancreatic Diseases Including Pancreatic Cancer. Gut 2012, 61, 582–588. [Google Scholar] [CrossRef]
- Mei, Q.-X.; Huang, C.-L.; Luo, S.-Z.; Zhang, X.-M.; Zeng, Y.; Lu, Y.-Y. Characterization of the Duodenal Bacterial Microbiota in Patients with Pancreatic Head Cancer vs. Healthy Controls. Pancreatol. Off. J. Int. Assoc. Pancreatol. 2018, 18, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Kohi, S.; Macgregor-Das, A.; Dbouk, M.; Yoshida, T.; Chuidian, M.; Abe, T.; Borges, M.; Lennon, A.M.; Shin, E.J.; Canto, M.I.; et al. Alterations in the Duodenal Fluid Microbiome of Patients With Pancreatic Cancer. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2022, 20, e196–e227. [Google Scholar] [CrossRef] [PubMed]
- Panthangi, V.; Cyril Kurupp, A.R.; Raju, A.; Luthra, G.; Shahbaz, M.; Almatooq, H.; Foucambert, P.; Esbrand, F.D.; Zafar, S.; Khan, S. Association Between Helicobacter Pylori Infection and the Risk of Pancreatic Cancer: A Systematic Review Based on Observational Studies. Cureus 2022, 14, e28543. [Google Scholar] [CrossRef]
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gesell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A.; et al. The Gut Mycobiome of the Human Microbiome Project Healthy Cohort. Microbiome 2017, 5, 153. [Google Scholar] [CrossRef] [PubMed]
- Shkoporov, A.N.; Clooney, A.G.; Sutton, T.D.S.; Ryan, F.J.; Daly, K.M.; Nolan, J.A.; McDonnell, S.A.; Khokhlova, E.V.; Draper, L.A.; Forde, A.; et al. The Human Gut Virome Is Highly Diverse, Stable, and Individual Specific. Cell Host Microbe 2019, 26, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Zella, D.; Gallo, R.C. Viruses and Bacteria Associated with Cancer: An Overview. Viruses 2021, 13, 1039. [Google Scholar] [CrossRef]
- Seo, S.-U.; Kweon, M.-N. Virome-Host Interactions in Intestinal Health and Disease. Curr. Opin. Virol. 2019, 37, 63–71. [Google Scholar] [CrossRef]
- Abreu, M.T.; Peek, R.M.J. Gastrointestinal Malignancy and the Microbiome. Gastroenterology 2014, 146, 1534–1546. [Google Scholar] [CrossRef]
- Cuevas-Ramos, G.; Petit, C.R.; Marcq, I.; Boury, M.; Oswald, E.; Nougayrède, J.-P. Escherichia Coli Induces DNA Damage In Vivo and Triggers Genomic Instability in Mammalian Cells. Proc. Natl. Acad. Sci. USA 2010, 107, 11537–11542. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium Nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via Its FadA Adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef]
- Liu, Y.; Fu, K.; Wier, E.M.; Lei, Y.; Hodgson, A.; Xu, D.; Xia, X.; Zheng, D.; Ding, H.; Sears, C.L.; et al. Bacterial Genotoxin Accelerates Transient Infection-Driven Murine Colon Tumorigenesis. Cancer Discov. 2022, 12, 236–249. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, A.; Wang, Y.; Zhang, Y. Intratumoral Microbiota: Roles in Cancer Initiation, Development and Therapeutic Efficacy. Signal Transduct. Target. Ther. 2023, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Mendez, R.; Kesh, K.; Arora, N.; Di Martino, L.; McAllister, F.; Merchant, N.; Banerjee, S.; Banerjee, S. Microbial Dysbiosis and Polyamine Metabolism as Predictive Markers for Early Detection of Pancreatic Cancer. Carcinogenesis 2020, 41, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Daillère, R.; Roberti, M.P.; Routy, B.; Kroemer, G. Anticancer Effects of the Microbiome and Its Products. Nat. Rev. Microbiol. 2017, 15, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Panebianco, C.; Villani, A.; Pisati, F.; Orsenigo, F.; Ulaszewska, M.; Latiano, T.P.; Potenza, A.; Andolfo, A.; Terracciano, F.; Tripodo, C.; et al. Butyrate, a Postbiotic of Intestinal Bacteria, Affects Pancreatic Cancer and Gemcitabine Response in in Vitro and in Vivo Models. Biomed. Pharmacother. 2022, 151, 113163. [Google Scholar] [CrossRef] [PubMed]
- Kanika, G.; Khan, S.; Jena, G. Sodium Butyrate Ameliorates L-Arginine-Induced Pancreatitis and Associated Fibrosis in Wistar Rat: Role of Inflammation and Nitrosative Stress. J. Biochem. Mol. Toxicol. 2015, 29, 349–359. [Google Scholar] [CrossRef]
- Leinwand, J.; Miller, G. Regulation and Modulation of Antitumor Immunity in Pancreatic Cancer. Nat. Immunol. 2020, 21, 1152–1159. [Google Scholar] [CrossRef]
- Morgillo, F.; Dallio, M.; Della Corte, C.M.; Gravina, A.G.; Viscardi, G.; Loguercio, C.; Ciardiello, F.; Federico, A. Carcinogenesis as a Result of Multiple Inflammatory and Oxidative Hits: A Comprehensive Review from Tumor Microenvironment to Gut Microbiota. Neoplasia 2018, 20, 721–733. [Google Scholar] [CrossRef]
- Vitiello, G.A.; Cohen, D.J.; Miller, G. Harnessing the Microbiome for Pancreatic Cancer Immunotherapy. Trends in cancer 2019, 5, 670–676. [Google Scholar] [CrossRef]
- Ochi, A.; Nguyen, A.H.; Bedrosian, A.S.; Mushlin, H.M.; Zarbakhsh, S.; Barilla, R.; Zambirinis, C.P.; Fallon, N.C.; Rehman, A.; Pylayeva-Gupta, Y.; et al. MyD88 Inhibition Amplifies Dendritic Cell Capacity to Promote Pancreatic Carcinogenesis via Th2 Cells. J. Exp. Med. 2012, 209, 1671–1687. [Google Scholar] [CrossRef]
- Ochi, A.; Graffeo, C.S.; Zambirinis, C.P.; Rehman, A.; Hackman, M.; Fallon, N.; Barilla, R.M.; Henning, J.R.; Jamal, M.; Rao, R.; et al. Toll-like Receptor 7 Regulates Pancreatic Carcinogenesis in Mice and Humans. J. Clin. Investig. 2012, 122, 4118–4129. [Google Scholar] [CrossRef] [PubMed]
- Daley, D.; Mani, V.R.; Mohan, N.; Akkad, N.; Pandian, G.S.D.B.; Savadkar, S.; Lee, K.B.; Torres-Hernandez, A.; Aykut, B.; Diskin, B.; et al. NLRP3 Signaling Drives Macrophage-Induced Adaptive Immune Suppression in Pancreatic Carcinoma. J. Exp. Med. 2017, 214, 1711–1724. [Google Scholar] [CrossRef] [PubMed]
- Daley, D.; Mani, V.R.; Mohan, N.; Akkad, N.; Ochi, A.; Heindel, D.W.; Lee, K.B.; Zambirinis, C.P.; Pandian, G.S.D.B.; Savadkar, S.; et al. Dectin 1 Activation on Macrophages by Galectin 9 Promotes Pancreatic Carcinoma and Peritumoral Immune Tolerance. Nat. Med. 2017, 23, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Jobin, C.; Thomas, R.M. Implications of the Microbiome in the Development and Treatment of Pancreatic Cancer: Thinking Outside of the Box by Looking inside the Gut. Neoplasia 2021, 23, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Zambirinis, C.P.; Levie, E.; Nguy, S.; Avanzi, A.; Barilla, R.; Xu, Y.; Seifert, L.; Daley, D.; Greco, S.H.; Deutsch, M.; et al. TLR9 Ligation in Pancreatic Stellate Cells Promotes Tumorigenesis. J. Exp. Med. 2015, 212, 2077–2094. [Google Scholar] [CrossRef]
- Balachandran, V.P.; Łuksza, M.; Zhao, J.N.; Makarov, V.; Moral, J.A.; Remark, R.; Herbst, B.; Askan, G.; Bhanot, U.; Senbabaoglu, Y.; et al. Identification of Unique Neoantigen Qualities in Long-Term Survivors of Pancreatic Cancer. Nature 2017, 551, 512–516. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Forget, M.-A.; Lucas, F.A.S.; Alvarez, H.A.; Haymaker, C.; Chattopadhyay, C.; Kim, S.-H.; Ekmekcioglu, S.; Grimm, E.A.; et al. Exploiting the Neoantigen Landscape for Immunotherapy of Pancreatic Ductal Adenocarcinoma. Sci. Rep. 2016, 6, 35848. [Google Scholar] [CrossRef]
- Neesse, A.; Bauer, C.A.; Öhlund, D.; Lauth, M.; Buchholz, M.; Michl, P.; Tuveson, D.A.; Gress, T.M. Stromal Biology and Therapy in Pancreatic Cancer: Ready for Clinical Translation? Gut 2019, 68, 159–171. [Google Scholar] [CrossRef]
- Ghaddar, B.; Biswas, A.; Harris, C.; Omary, M.B.; Carpizo, D.R.; Blaser, M.J.; De, S. Tumor Microbiome Links Cellular Programs and Immunity in Pancreatic Cancer. Cancer Cell 2022, 40, 1240–1253.e5. [Google Scholar] [CrossRef]
- Panebianco, C.; Adamberg, K.; Jaagura, M.; Copetti, M.; Fontana, A.; Adamberg, S.; Kolk, K.; Vilu, R.; Andriulli, A.; Pazienza, V. Influence of Gemcitabine Chemotherapy on the Microbiota of Pancreatic Cancer Xenografted Mice. Cancer Chemother. Pharmacol. 2018, 81, 773–782. [Google Scholar] [CrossRef]
- Conroy, T.; Hammel, P.; Hebbar, M.; Ben Abdelghani, M.; Wei, A.C.; Raoul, J.-L.; Choné, L.; Francois, E.; Artru, P.; Biagi, J.J.; et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2395–2406. [Google Scholar] [CrossRef] [PubMed]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal Bacteria Control Cancer Response to Therapy by Modulating the Tumor Microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef]
- Stringer, A.M.; Gibson, R.J.; Logan, R.M.; Bowen, J.M.; Yeoh, A.S.J.; Keefe, D.M.K. Faecal Microflora and Beta-Glucuronidase Expression Are Altered in an Irinotecan-Induced Diarrhea Model in Rats. Cancer Biol. Ther. 2008, 7, 1919–1925.e6. [Google Scholar] [CrossRef] [PubMed]
- Goel, N.; Nadler, A.; Reddy, S.; Hoffman, J.P.; Pitt, H.A. Biliary Microbiome in Pancreatic Cancer: Alterations with Neoadjuvant Therapy. HPB Off. J. Int. Hepato Pancreato Biliary Assoc. 2019, 21, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, S.O.; Jajja, M.R.; Maxwell, D.W.; Pouch, S.M.; Sarmiento, J.M. Neoadjuvant Chemotherapy for Pancreatic Cancer and Changes in the Biliary Microbiome. Am. J. Surg. 2021, 222, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Hank, T.; Sandini, M.; Ferrone, C.R.; Rodrigues, C.; Weniger, M.; Qadan, M.; Warshaw, A.L.; Lillemoe, K.D.; Fernández-Del Castillo, C. Association Between Pancreatic Fistula and Long-Term Survival in the Era of Neoadjuvant Chemotherapy. JAMA Surg. 2019, 154, 943–951. [Google Scholar] [CrossRef]
- Shrader, H.R.; Miller, A.M.; Tomanek-Chalkley, A.; McCarthy, A.; Coleman, K.L.; Ear, P.H.; Mangalam, A.K.; Salem, A.K.; Chan, C.H.F. Effect of Bacterial Contamination in Bile on Pancreatic Cancer Cell Survival. Surgery 2021, 169, 617–622. [Google Scholar] [CrossRef]
- Hilmi, M.; Bartholin, L.; Neuzillet, C. Immune Therapies in Pancreatic Ductal Adenocarcinoma: Where Are We Now? World J. Gastroenterol. 2018, 24, 2137–2151. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut Microbiome Influences Efficacy of PD-1-Based Immunotherapy against Epithelial Tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.-L.; et al. Commensal Bifidobacterium Promotes Antitumor Immunity and Facilitates Anti-PD-L1 Efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Sethi, V.; Kurtom, S.; Tarique, M.; Lavania, S.; Malchiodi, Z.; Hellmund, L.; Zhang, L.; Sharma, U.; Giri, B.; Garg, B.; et al. Gut Microbiota Promotes Tumor Growth in Mice by Modulating Immune Response. Gastroenterology 2018, 155, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.M.; Gharaibeh, R.Z.; Gauthier, J.; Beveridge, M.; Pope, J.L.; Guijarro, M.V.; Yu, Q.; He, Z.; Ohland, C.; Newsome, R.; et al. Intestinal Microbiota Enhances Pancreatic Carcinogenesis in Preclinical Models. Carcinogenesis 2018, 39, 1068–1078. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, V.P.; Beatty, G.L.; Dougan, S.K. Broadening the Impact of Immunotherapy to Pancreatic Cancer: Challenges and Opportunities. Gastroenterology 2019, 156, 2056–2072. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Gong, Y.; Fan, Z.; Luo, G.; Huang, Q.; Deng, S.; Cheng, H.; Jin, K.; Ni, Q.; Yu, X.; et al. Molecular Alterations and Targeted Therapy in Pancreatic Ductal Adenocarcinoma. J. Hematol. Oncol. 2020, 13, 130. [Google Scholar] [CrossRef]
- Brandi, G.; Turroni, S.; McAllister, F.; Frega, G. The Human Microbiomes in Pancreatic Cancer: Towards Evidence-Based Manipulation Strategies? Int. J. Mol. Sci. 2021, 22, 9914. [Google Scholar] [CrossRef]
- Malesza, I.J.; Malesza, M.; Walkowiak, J.; Mussin, N.; Walkowiak, D.; Aringazina, R.; Bartkowiak-Wieczorek, J.; Mądry, E. High-Fat, Western-Style Diet, Systemic Inflammation, and Gut Microbiota: A Narrative Review. Cells 2021, 10, 3164. [Google Scholar] [CrossRef]
- Bultman, S.J. Molecular Pathways: Gene-Environment Interactions Regulating Dietary Fiber Induction of Proliferation and Apoptosis via Butyrate for Cancer Prevention. Clin. cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 799–803. [Google Scholar] [CrossRef]
- DeMartino, P.; Cockburn, D.W. Resistant Starch: Impact on the Gut Microbiome and Health. Curr. Opin. Biotechnol. 2020, 61, 66–71. [Google Scholar] [CrossRef]
- Panebianco, C.; Adamberg, K.; Adamberg, S.; Saracino, C.; Jaagura, M.; Kolk, K.; Di Chio, A.G.; Graziano, P.; Vilu, R.; Pazienza, V. Engineered Resistant-Starch (ERS) Diet Shapes Colon Microbiota Profile in Parallel with the Retardation of Tumor Growth in In Vitro and In Vivo Pancreatic Cancer Models. Nutrients 2017, 9, 331. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Sobocki, B.K.; Kaźmierczak-Siedlecka, K.; Folwarski, M.; Hawryłkowicz, V.; Makarewicz, W.; Stachowska, E. Pancreatic Cancer and Gut Microbiome-Related Aspects: A Comprehensive Review and Dietary Recommendations. Nutrients 2021, 13, 4425. [Google Scholar] [CrossRef] [PubMed]
- Panebianco, C.; Pisati, F.; Ulaszewska, M.; Andolfo, A.; Villani, A.; Federici, F.; Laura, M.; Rizzi, E.; Potenza, A.; Latiano, T.P.; et al. Tuning Gut Microbiota through a Probiotic Blend in Gemcitabine-Treated Pancreatic Cancer Xenografted Mice. Clin. Transl. Med. 2021, 11, e580. [Google Scholar] [CrossRef] [PubMed]
- Mohindroo, C.; Hasanov, M.; Rogers, J.E.; Dong, W.; Prakash, L.R.; Baydogan, S.; Mizrahi, J.D.; Overman, M.J.; Varadhachary, G.R.; Wolff, R.A.; et al. Antibiotic Use Influences Outcomes in Advanced Pancreatic Adenocarcinoma Patients. Cancer Med. 2021, 10, 5041–5050. [Google Scholar] [CrossRef] [PubMed]
- Boursi, B.; Mamtani, R.; Haynes, K.; Yang, Y.-X. Recurrent Antibiotic Exposure May Promote Cancer Formation--Another Step in Understanding the Role of the Human Microbiota? Eur. J. Cancer 2015, 51, 2655–2664. [Google Scholar] [CrossRef]
- Galloway-Peña, J.R.; Kontoyiannis, D.P. The Gut Mycobiome: The Overlooked Constituent of Clinical Outcomes and Treatment Complications in Patients with Cancer and Other Immunosuppressive Conditions. PLoS Pathog. 2020, 16, e1008353. [Google Scholar] [CrossRef]
- Harada, L.K.; Silva, E.C.; Campos, W.F.; Del Fiol, F.S.; Vila, M.; Dąbrowska, K.; Krylov, V.N.; Balcão, V.M. Biotechnological Applications of Bacteriophages: State of the Art. Microbiol. Res. 2018, 212–213, 38–58. [Google Scholar] [CrossRef]
- Derosa, L.; Routy, B.; Desilets, A.; Daillère, R.; Terrisse, S.; Kroemer, G.; Zitvogel, L. Microbiota-Centered Interventions: The Next Breakthrough in Immuno-Oncology? Cancer Discov. 2021, 11, 2396–2412. [Google Scholar] [CrossRef]
| Reference | Country Disease (N) | Microbiota and Measurement | Major Findings |
|---|---|---|---|
| Kartal et al. 2022 [20] | Spain PDAC (50) | Tumor and adjacent normal tissue 16S rRNA gene sequencing | Lactobacillus spp., Akkermansia muciniphila and Bacteroides spp. enriched in tumor vs. adjacent normal tissue. |
| Tan et al. 2022 [21] | China PC (20) | Tumor and adjacent normal tissue 16S rRNA gene sequencing | No differences in alpha diversity and beta diversity between tumor and adjacent normal tissue; Forty-four genera were significantly different between PC and NAT; Pseudomonas, Herbaspirillum and Sphingomonas enriched in PC tissues. |
| Huang et al. 2022 [22] | China LTS-PDAC (17) STS-PDAC (13) | Tumor tissue 16S rRNA gene sequencing | No difference in Shannon index, but a higher number of species between LTS and STS; beta diversity analysis distinguished between LTS and STS; Sphingomonas, Megasphaera, Bradyrhizobium, Desulfovibrio, Flavobacterium, Enhydrobacter, and Megamonas were enriched in LTS, and Clostridium_sensu stricto 1, Actinomyces, Porphyromonas, Aggregatibacter, and Neisseria were enriched in STS. |
| Nejman et al. 2020 [19] | USA and Israel PC (67) | Tumor tissue 16S rRNA gene sequencing | Bacteria DNA in 68% of PC tumors; Proteobacteria dominated the microbiome of PC, followed by Firmicutes; Enterobacter asburiae, Klebsiella pneumoniae, Citrobacter freundii, and Fusobacterium nucleatum were the most prevalent species. |
| Riquelme et al. 2019 [23] | USA Discovery: LTS-PDAC (21) STS-PDAC (22) Validation: LTS-PDAC (36) STS-PDAC (32) | Tumor tissue 16S rRNA gene sequencing | Alpha diversity was higher in LTS vs. STS in both cohorts; Beta diversity distinguished LTS and STS in both cohorts; Patients with high microbial alpha diversity had significantly more prolonged OS than those with low alpha diversity did; Saccharopolyspora, Pseudoxanthomonas, and Streptomyces were significantly more abundant in LTS vs. STS, and were associated with significantly better outcomes. |
| Geller et al. 2017 [24] | USA and Israel NP (20) PDAC (113) | Tumor tissue and normal pancreatic tissue 16S rRNA gene qPCR and sequencing | Bacterial DNA in 76% of PDAC and in 15% of NP controls; the most common species were from Gammaproteobacteria, and most were members of the Enterobacteriaceae and Pseudomonadaceae, with Pseudomonas, Citrobacter, Klebsiella, Streptococcus, and Acinetobacter having highest mean relative abundances. |
| Reference | Country Disease (N) | Microbiota and Measurement | Major Findings |
|---|---|---|---|
| Nagata et al. 2022 [38] | Japan, Spain, and Germany HC (235) PDAC (37) | Gut and oral Metagenomics | Alpha diversity decreased in the gut microbiome and increased in the oral microbiome of PDAC vs. HC; Significant differences in beta diversity for both the gut and oral microbiome between PDAC and HC; Streptococcus oralis, Streptococcus vestibularis, Streptococcus anginosus, Veillonella atypica, Veillonella parvula and Actinomyces sp. were enriched, and species of the order Clostridiales, such as Lachnospiraceae, Eubacterium ventriosum, and Faecalibacterium prausnitzii were depleted in the gut of PDAC; Unknown species of Firmicutes, Dialister sp., Solobacterium sp., Prevotella pallens, and Prevotella sp. C561 were enriched, and Streptococcus salivarius, Streptococcus thermophilus, and Streptococcus australis was depleted in saliva of PDAC. |
| Kartal et al. 2022 [20] | Discovery: Spain HC (50) CP (29) PDAC (57) Validation: Germany HC (32) PDAC (44) | Gut and oral Metagenomics | Fecal microbiome composition of PDAC different from that of HC and from that of CP; Veillonella atypica, Fusobacterium nucleatum/hwasookii and Alloscardovia omnicolens enriched and Romboutsia timonensis, Faecalibacterium prausnitzii, Bacteroides coprocola and Bifidobacterium bifidum were depleted in fecal microbiome of PDAC; No significant differences in taxa abundance in salivary microbiome. Fecal metagenomic classifiers were better than saliva-based classifiers in identifying PDAC patients in early- and late-disease stages; accuracy improved in combination with serum levels of CA 19-9. |
| Matsukawa et al. 2021 [39] | Japan HC (18) PC (24) | Gut and oral Metagenomics | Significant differences in gut bacterial and viral microbiomes between PC and HC; 26 species were significantly different between PC and HC, including Klebsiella pneumoniae, Clostridium bolteae, C. symbiosum, Streptococcus mutans, Alistipes shahii, Bacteroides spp., Parabacteroides spp., and Lactobacillus spp. being enriched, and Collinsella aerofaciens, Blautia obeum, Bifidobacterium pseudocatenulatum, Eubacterium hallii, Lachnospiraceae, Coprococcus sp., and Streptococcus sanguinis being depleted in PC; Nineteen viral genera were significantly different in PC vs. HC. |
| Hashimoto et al. 2023 [40] | Japan HC (68) PDAC-bt (5) | Gut 16S rRNA gene sequencing | No differences in alpha diversity and beta diversity between PDAC-bt and HC; Actinomyces, Streptococcus, Veillonella, and Lactobacillus were enriched, and Anaerostipes was depleted in the fecal microbiome of PDAC-bt patients. |
| Zhou et al. 2021 [41] | China HC (32) PDAC (32) | Gut Metagenomics | Beta diversity analysis distinguished PDAC and HC; Proteobacteria (Gammaproteobacteria) and Fusobacteria were enriched and Firmicutes was depleted in PDAC; 24 species were differentially abundant in PDAC vs. HC, including Escherichia coli, Fusobacterium nucleatum, Clostridium spp., Megamonas spp., Veillonella atypica, Veillonella parvula, and Prevotella stercorea, which were enriched in PDAC, and Faecalibacterium prausnitzii, Eubacterium rectale, Roseburia intestinalis, and Ruminococcus sp., which were depleted in PDAC. |
| Half et al. 2019 [42] | Israel HC (13) PCL (6) PC (30) NAFLD (16) | Gut 16S rRNA gene sequencing | No differences in alpha diversity between HC, PCL and PC; beta diversity analysis distinguished PC and HC; Bacteroidales, Odoribacter, Lachnospiraceae, Veillonellaceae, Megasphaera, and Akkermansia were enriched, and Clostridiales, Clostridium, Anaerostipes, Ruminococcaceae, Faecalibacterium, and Subdoligranulum were depleted in PC. |
| Pushalkar et al. 2018 [27] | USA HC (31) PDAC (32) | Gut 16S rRNA gene sequencing | Alpha diversity was higher in PDAC than in HC; Genera of Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes, including Parabacteroides, Veillonella, Klebsiella, and Cardiobacterium were enriched and Megamonas and Prevotella were depleted in PDAC. |
| Ren et al. 2017 [43] | China HC (57) PC (85) | Gut 16S rRNA gene sequencing | Alpha diversity was lower in PC vs. HC; Beta diversity analysis distinguished PC and HC; 15 taxa including Prevotella, Veillonella, Klebsiella, Selenomonas, and Enterobacter were enriched, and 25 taxa including Bifidobacterium, Coprococcus, Clostridium IV, Blautia, Flavonifractor, and Anaerostipes were depleted in PC. |
| Petrick et al. 2022 [44] | USA (African-Americans) HC (354) PC (122) | Oral Metagenomics | No differences in alpha diversity and beta diversity between PC and HC; No individual microbial taxa were differentially abundant between PC and HC after multiple comparisons. |
| Sun et al. 2020 [45] | China HC (10) BPD (17) PC (10) | Oral 16S rRNA gene sequencing | Alpha diversity was higher in BPD and PC vs. HC; Beta diversity distinguished PC, BPD and HC; Fusobacterium, Megasphaera, Prevotella, Spirochaeta, and Treponema were enriched, and Leptotrichia and Neisseria were depleted in PC; Selenomonas was enriched in BPD. |
| Vogtmann et al. 2020 [46] | Iran HC (285) PDAC (273) | Oral 16S rRNA gene sequencing | No differences in alpha diversity between PDAC and HC; Beta diversity analysis distinguished PDAC and HC; Haemophilus abundance was associated with increased risk of PDAC; Presence of Enterobactericeae and Lachnospiraceae was associated with increased risk of PDAC. |
| Wei et al. 2020 [47] | China HC (69) PDAC (41) | Oral 16S rRNA gene sequencing | Higher number of species and species richness and lower microbial diversity in PDAC vs. HC; Streptococcus, Actinomyces, Rothia, Leptotrichia, Lactobacillus, Escherichia coli, and Enterobacteriales were enriched, and Selenomonas, Porphyromnas, Prevotella, Capnocytophaga, Alloprevotella, Tannerella, and Neisseria were depleted in PDAC. |
| Fan et al. 2018 [48] | USA HC (371) PC (361) | Oral 16S rRNA gene sequencing | No differences in beta diversity; Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans were associated with higher PC risk, and Fusobacteria and Leptotrichia were associated with lower PC risk. |
| Olson et al. 2017 [49] | USA HC (58) IPMN (39) PDAC (34) | Oral 16S rRNA gene sequencing | No differences in alpha and beta diversity between HC, IPMN and PDAC; Firmicutes was enriched in PDAC, and Haemophilus and Neisseria enriched in IPMN and HC. |
| Torres et al. 2015 [50] | USA HC (22) OD (78) PC (8) | Oral 16S rRNA gene sequencing; qPCR Leptotrichia | No differences in alpha and beta-diversity between HC, OD, and PC; Significantly higher ratio of Leptotrichia to Porphyromonas in PC vs. HC or OD. |
| Farrell et al. 2012 [51] | USA Discovery: HC (10) PC (10) Validation: HC (28) PC (28) | Oral Discovery: Oligonucleotide HOMIM array Validation: 16S rRNA gene qPCR | Discovery: 16 species were significantly different between PC and HC, and 6 were validated by qPCR (Atopobium parvulum, Granulicatella adiacens, Neisseria elongata, Prevotella nigrescens, and Streptococcus australis, and Streptococcus mitis) Validation: N. elongata and S. mitis decreased in PC vs. HC; G. adiacens increased in PC vs. all non-cancer subjects. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bastos, A.R.; Pereira-Marques, J.; Ferreira, R.M.; Figueiredo, C. Harnessing the Microbiome to Reduce Pancreatic Cancer Burden. Cancers 2023, 15, 2629. https://doi.org/10.3390/cancers15092629
Bastos AR, Pereira-Marques J, Ferreira RM, Figueiredo C. Harnessing the Microbiome to Reduce Pancreatic Cancer Burden. Cancers. 2023; 15(9):2629. https://doi.org/10.3390/cancers15092629
Chicago/Turabian StyleBastos, Ana Raquel, Joana Pereira-Marques, Rui Manuel Ferreira, and Ceu Figueiredo. 2023. "Harnessing the Microbiome to Reduce Pancreatic Cancer Burden" Cancers 15, no. 9: 2629. https://doi.org/10.3390/cancers15092629
APA StyleBastos, A. R., Pereira-Marques, J., Ferreira, R. M., & Figueiredo, C. (2023). Harnessing the Microbiome to Reduce Pancreatic Cancer Burden. Cancers, 15(9), 2629. https://doi.org/10.3390/cancers15092629






