Sensitivity and Specificity of Different Prognostic Systems in Guiding Surveillance for Metastases in Uveal Melanoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
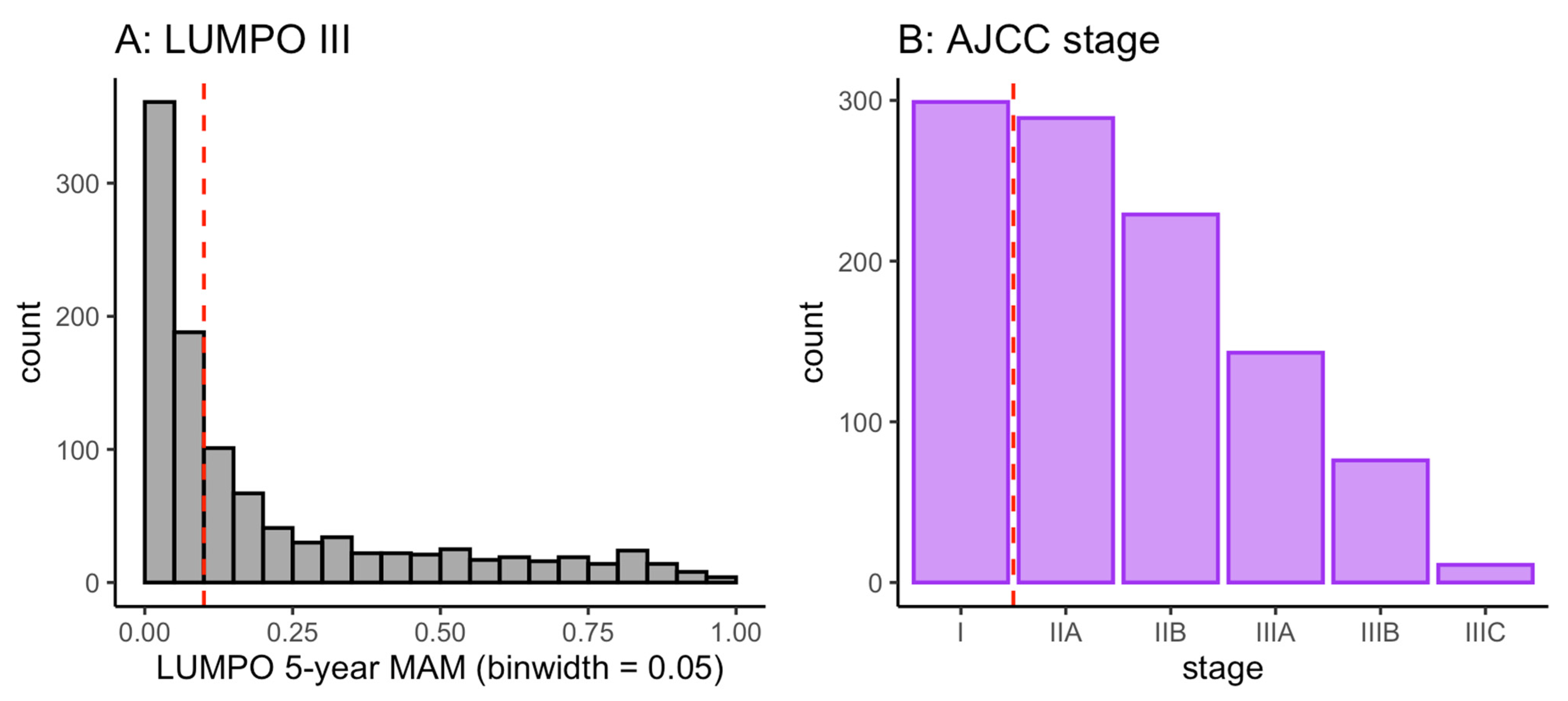
2.2. Distribution of Risk Scores and Classifications in the Patient Population
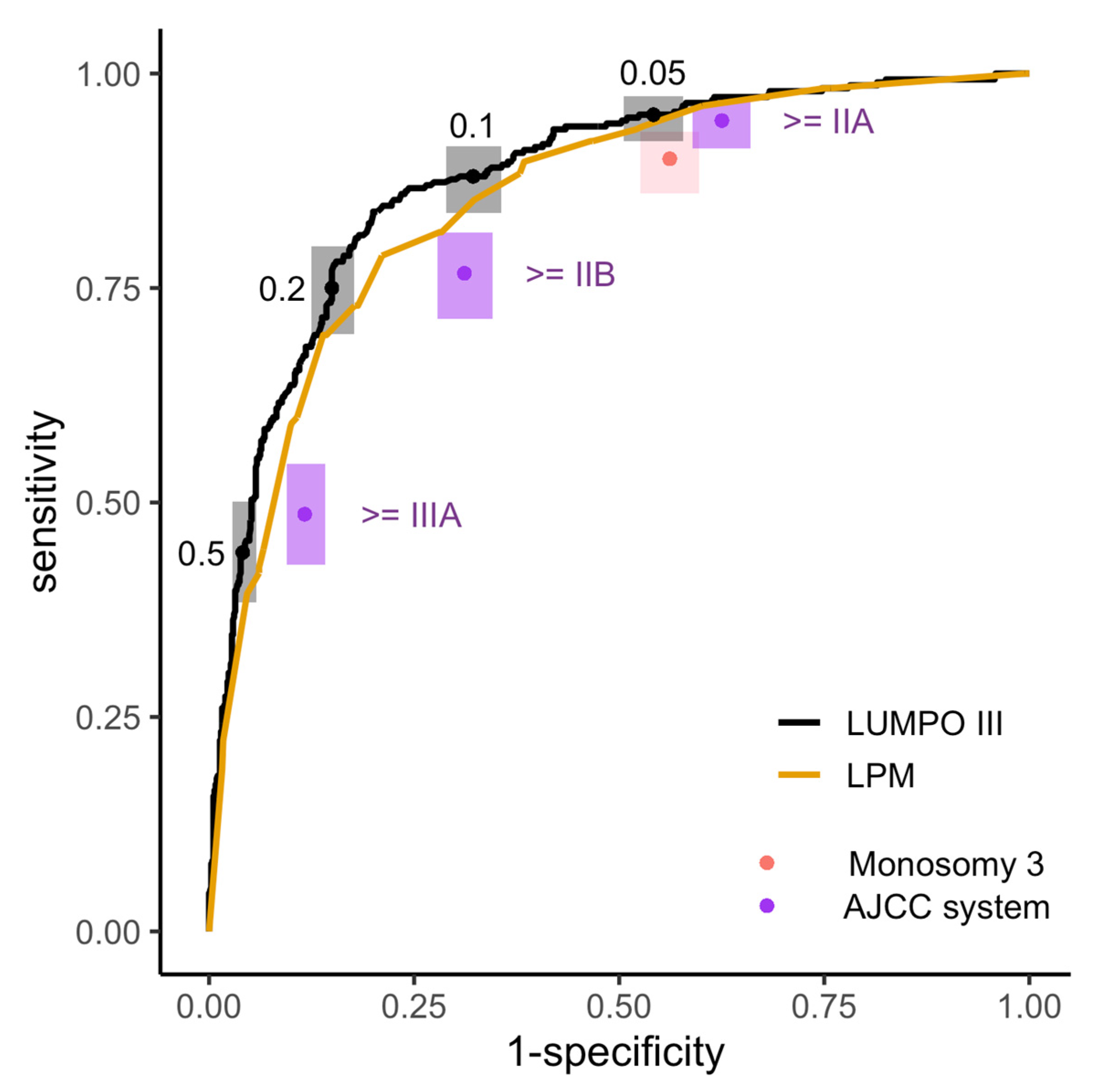
2.3. Sensitivity and Specificity of the Four Prognostic Systems for Risk Stratification at LOOC
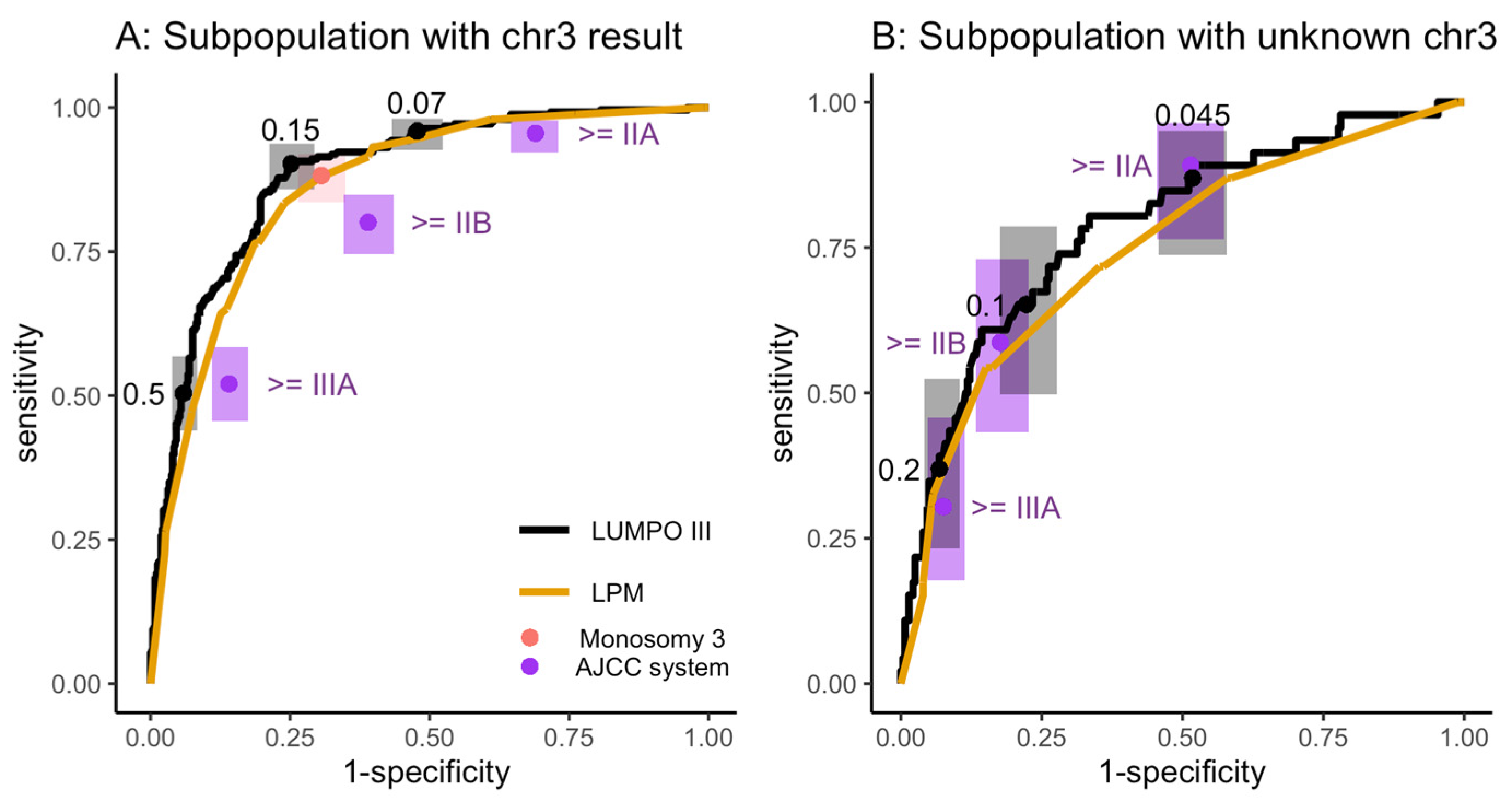
2.3.1. Subgroup Analysis of LOOC Patients with and without a Chromosome 3 Result
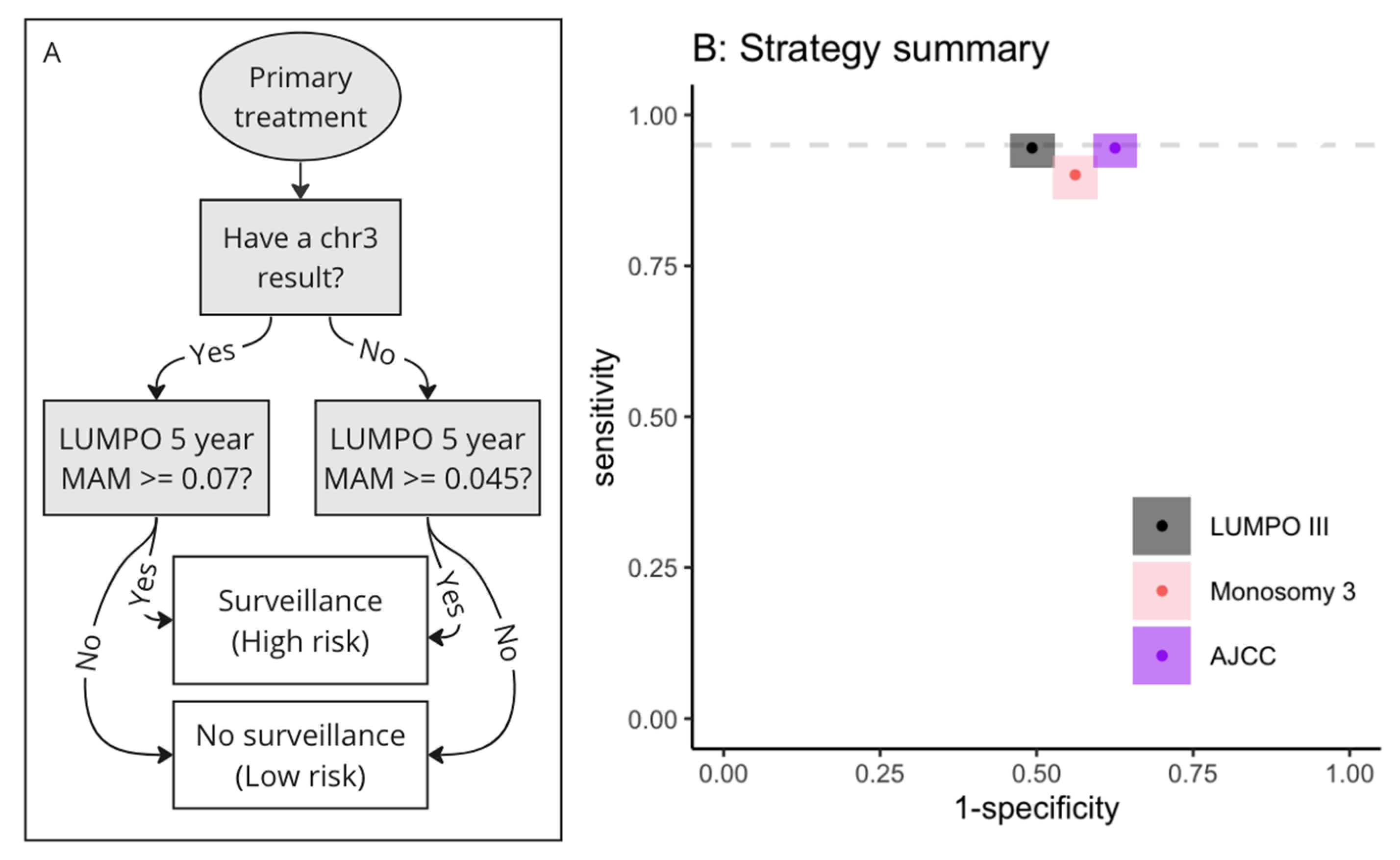
2.3.2. Decision Algorithm for Implementing Strategies Incorporating Two Thresholds
2.3.3. Comparison of Health Economic Impact of Using Different Strategies
2.4. Sensitivity and Specificity of Prognostic Systems for Risk Stratification at a Centre That Does Not Offer Genetic Testing
3. Results
3.1. Description of the Dataset Characteristics
3.2. Distribution of Risk Scores and Classifications in the Patient Population
3.3. Sensitivity and Specificity of the Four Prognostic Systems for Risk Stratification at LOOC
3.3.1. Subgroup Analysis of LOOC Patients with and without a Chromosome 3 Result
3.3.2. Decision Algorithm for Implementing Strategies Incorporating Two Thresholds
3.3.3. Comparison of Health Economic Impact of Using Different Strategies
3.4. Sensitivity and Specificity of Prognostic Systems for Risk Stratification at a Centre which Does Not Offer Genetic Testing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nathan, P.; Cohen, V.; Coupland, S.; Curtis, K.; Damato, B.; Evans, J.; Fenwick, S.; Kirkpatrick, L.; Li, O.; Marshall, E.; et al. Uveal Melanoma UK National Guidelines. Eur. J. Cancer 2015, 51, 2404–2412. [Google Scholar] [CrossRef] [PubMed]
- Damato, B.; Heimann, H. Personalized Treatment of Uveal Melanoma. Eye 2013, 27, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Jager, M.J.; Shields, C.L.; Cebulla, C.M.; Abdel-Rahman, M.H.; Grossniklaus, H.E.; Stern, M.-H.; Carvajal, R.D.; Belfort, R.N.; Jia, R.; Shields, J.A. Uveal Melanoma. Nat. Rev. Dis. Prim. 2020, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Eskelin, S.; Pyrhönen, S.; Summanen, P.; Prause, J.U.; Kivelä, T. Screening for Metastatic Malignant Melanoma of the Uvea Revisited. Cancer 1999, 85, 1151–1159. [Google Scholar] [CrossRef]
- Marshall, E.; Romaniuk, C.; Ghaneh, P.; Wong, H.; McKay, M.; Chopra, M.; Coupland, S.E.; Damato, B.E. MRI in the Detection of Hepatic Metastases from High-Risk Uveal Melanoma: A Prospective Study in 188 Patients. Br. J. Ophthalmol. 2013, 97, 159–163. [Google Scholar] [CrossRef]
- Choudhary, M.M.; Gupta, A.; Bena, J.; Emch, T.; Singh, A.D. Hepatic Ultrasonography for Surveillance in Patients With Uveal Melanoma. JAMA Ophthalmol. 2016, 134, 174–180. [Google Scholar] [CrossRef]
- Carvajal, R.D.; Sacco, J.J.; Jager, M.J.; Eschelman, D.J.; Olofsson Bagge, R.; Harbour, J.W.; Chieng, N.D.; Patel, S.P.; Joshua, A.M.; Piperno-Neumann, S. Advances in the Clinical Management of Uveal Melanoma. Nat. Rev. Clin. Oncol. 2023, 20, 99–115. [Google Scholar] [CrossRef]
- Gomez, D.; Wetherill, C.; Cheong, J.; Jones, L.; Marshall, E.; Damato, B.; Coupland, S.E.; Ghaneh, P.; Poston, G.J.; Malik, H.Z.; et al. The Liverpool Uveal Melanoma Liver Metastases Pathway: Outcome Following Liver Resection: Surgery for Uveal Melanoma Metastases. J. Surg. Oncol. 2014, 109, 542–547. [Google Scholar] [CrossRef]
- Piperno-Neumann, S.; Servois, V.; Mariani, P.; Plancher, C.; Lévy-Gabriel, C.; Lumbroso-Le Rouic, L.; Couturier, J.; Asselain, B.; Desjardins, L.; Cassoux, N. Prospective Study of Surveillance Testing for Metastasis in 100 High-Risk Uveal Melanoma Patients. J. Français Ophtalmol. 2015, 38, 526–534. [Google Scholar] [CrossRef]
- Ramtohul, T.; Abdul-Baki, M.; Rodrigues, M.; Cassoux, N.; Gardrat, S.; Ait Rais, K.; Pierron, G.; Bouhadiba, T.; Servois, V.; Mariani, P. Tumor Growth Rate as a Predictive Marker for Recurrence and Survival After Liver Resection in Patients with Liver Metastases of Uveal Melanoma. Ann. Surg. Oncol. 2022, 29, 8480–8491. [Google Scholar] [CrossRef]
- Zager, J.S.; Orloff, M.M.; Ferrucci, P.F.; Glazer, E.S.; Ejaz, A.; Richtig, E.; Ochsenreither, S.; Lowe, M.C.; Reddy, S.A.; Beasley, G.; et al. FOCUS Phase 3 Trial Results: Percutaneous Hepatic Perfusion (PHP) with Melphalan for Patients with Ocular Melanoma Liver Metastases (PHP-OCM-301/301A). JCO 2022, 40, 9510. [Google Scholar] [CrossRef]
- Mariani, P.; Almubarak, M.M.; Kollen, M.; Wagner, M.; Plancher, C.; Audollent, R.; Piperno-Neumann, S.; Cassoux, N.; Servois, V. Radiofrequency Ablation and Surgical Resection of Liver Metastases from Uveal Melanoma. Eur. J. Surg. Oncol. 2016, 42, 706–712. [Google Scholar] [CrossRef]
- Trivedi, D.B.; Aldulaimi, N.; Karydis, I.; Wheater, M.; Modi, S.; Stedman, B.; Karavias, D.; Primrose, J.; Pearce, N.; Takhar, A.S. Liver Resection for Metastatic Uveal Melanoma: Experience from a Supra-Regional Centre and Review of Literature. Melanoma Res. 2023, 33, 71–79. [Google Scholar] [CrossRef]
- Pelster, M.S.; Gruschkus, S.K.; Bassett, R.; Gombos, D.S.; Shephard, M.; Posada, L.; Glover, M.S.; Simien, R.; Diab, A.; Hwu, P.; et al. Nivolumab and Ipilimumab in Metastatic Uveal Melanoma: Results From a Single-Arm Phase II Study. J. Clin. Oncol. 2021, 39, 599–607. [Google Scholar] [CrossRef]
- Piulats, J.M.; Espinosa, E.; de la Cruz Merino, L.; Varela, M.; Alonso Carrión, L.; Martín-Algarra, S.; López Castro, R.; Curiel, T.; Rodríguez-Abreu, D.; Redrado, M.; et al. Nivolumab Plus Ipilimumab for Treatment-Naïve Metastatic Uveal Melanoma: An Open-Label, Multicenter, Phase II Trial by the Spanish Multidisciplinary Melanoma Group (GEM-1402). ASCO 2021, 39, 586. [Google Scholar] [CrossRef]
- Nathan, P.; Hassel, J.C.; Rutkowski, P.; Baurain, J.-F.; Butler, M.O.; Schlaak, M.; Sullivan, R.J.; Ochsenreither, S.; Dummer, R.; Kirkwood, J.M.; et al. Overall Survival Benefit with Tebentafusp in Metastatic Uveal Melanoma. N. Engl. J. Med. 2021, 385, 1196–1206. [Google Scholar] [CrossRef]
- Howlett, S.; Carter, T.J.; Shaw, H.M.; Nathan, P.D. Tebentafusp: A First-in-Class Treatment for Metastatic Uveal Melanoma. Ther. Adv. Med. Oncol. 2023, 15, 17588359231160140. [Google Scholar] [CrossRef]
- Dogrusöz, M.; Jager, M.J. Genetic Prognostication in Uveal Melanoma. Acta Ophthalmol. 2018, 96, 331–347. [Google Scholar] [CrossRef]
- Eleuteri, A.; Damato, B.; Coupland, S.E.; Taktak, A.F.G. Enhancing Survival Prognostication in Patients with Choroidal Melanoma by Integrating Pathologic, Clinical and Genetic Predictors of Metastasis. IJBET 2012, 8, 18. [Google Scholar] [CrossRef]
- Eleuteri, A.; Taktak, A.F.G.; Coupland, S.E.; Heimann, H.; Kalirai, H.; Damato, B. Prognostication of Metastatic Death in Uveal Melanoma Patients: A Markov Multi-State Model. Comput. Biol. Med. 2018, 102, 151–156. [Google Scholar] [CrossRef]
- Rola, A.C.; Taktak, A.; Eleuteri, A.; Kalirai, H.; Heimann, H.; Hussain, R.; Bonnett, L.J.; Hill, C.J.; Traynor, M.; Jager, M.J.; et al. Multicenter External Validation of the Liverpool Uveal Melanoma Prognosticator Online: An OOG Collaborative Study. Cancers 2020, 12, 477. [Google Scholar] [CrossRef] [PubMed]
- Royal Liverpool Universty Hospital LUMPO III. Available online: https://mpcetoolsforhealth.liverpool.ac.uk/matsoap/lumpo3cr.htm (accessed on 20 December 2022).
- Damato, B.; Eleuteri, A.; Hussain, R.; Kalirai, H.; Thornton, S.; Taktak, A.; Heimann, H.; Coupland, S.E. Parsimonious Models for Predicting Mortality from Choroidal Melanoma. Investig. Ophthalmol. Vis. Sci. 2020, 61, 35. [Google Scholar] [CrossRef] [PubMed]
- Baron, E.D.; Nicola, M.D.; Shields, C.L. Updated AJCC Classification for Posterior Uveal Melanoma. Retina Today 2018, 5, 30–34. [Google Scholar]
- Kivela, T.; Simpson, R.; Grossniklaus, H. Uveal melanoma. In AJCC Cancer Staging Manual; Springer: New York, NY, USA, 2016; pp. 805–817. [Google Scholar]
- Bornfeld, N.; Prescher, G.; Becher, R.; Hirche, H.; Jöckel, K.-H.; Horsthemke, B. Prognostic Implications of Monosomy 3 in Uveal Melanoma. Lancet 1996, 347, 1222–1225. [Google Scholar] [CrossRef] [PubMed]
- Damato, B.; Dopierala, J.A.; Coupland, S.E. Genotypic Profiling of 452 Choroidal Melanomas with Multiplex Ligation-Dependent Probe Amplification. Clin. Cancer Res. 2010, 16, 6083–6092. [Google Scholar] [CrossRef]
- Harbour, J.W.; Onken, M.D.; Roberson, E.D.O.; Duan, S.; Cao, L.; Worley, L.A.; Council, M.L.; Matatall, K.A.; Helms, C.; Bowcock, A.M. Frequent Mutation of BAP1 in Metastasizing Uveal Melanomas. Science 2010, 330, 1410–1413. [Google Scholar] [CrossRef]
- Thomas, S.; Pütter, C.; Weber, S.; Bornfeld, N.; Lohmann, D.R.; Zeschnigk, M. Prognostic Significance of Chromosome 3 Alterations Determined by Microsatellite Analysis in Uveal Melanoma: A Long-Term Follow-up Study. Br. J. Cancer 2012, 106, 1171–1176. [Google Scholar] [CrossRef]
- Angi, M.; Kalirai, H.; Taktak, A.; Hussain, R.; Groenewald, C.; Damato, B.E.; Heimann, H.; Coupland, S.E. Prognostic Biopsy of Choroidal Melanoma: An Optimised Surgical and Laboratory Approach. Br. J. Ophthalmol. 2017, 101, 1143–1146. [Google Scholar] [CrossRef]
- Damato, B.; Duke, C.; Coupland, S.E.; Hiscott, P.; Smith, P.A.; Campbell, I.; Douglas, A.; Howard, P. Cytogenetics of Uveal Melanoma: A 7-Year Clinical Experience. Ophthalmology 2007, 114, 1925–1931.e1. [Google Scholar] [CrossRef]
- Clopper, C.J.; Pearson, E.S. The Use of Confidence or Fiducial Limits Illustrated in the Case of the Binomial. Biometrika 1934, 26, 404–413. [Google Scholar] [CrossRef]
- Sachs, M.C. PlotROC: A Tool for Plotting ROC Curves. J. Stat. Softw. 2017, 79, 2. [Google Scholar] [CrossRef]
- Carter, J.V.; Pan, J.; Rai, S.N.; Galandiuk, S. ROC-Ing along: Evaluation and Interpretation of Receiver Operating Characteristic Curves. Surgery 2016, 159, 1638–1645. [Google Scholar] [CrossRef]
- Djulbegovic, B.; Hozo, I.; Dale, W. Transforming Clinical Practice Guidelines and Clinical Pathways into Fast-and-Frugal Decision Trees to Improve Clinical Care Strategies. J. Eval. Clin. Pract. 2018, 24, 1247–1254. [Google Scholar] [CrossRef]
- NHS England 2020/21 National Cost Collection Data Publication. Available online: https://www.england.nhs.uk/publication/2020-21-national-cost-collection-data-publication/ (accessed on 5 January 2023).
- Diener-West, M.; Reynolds, S.M.; Agugliaro, D.J.; Caldwell, R.; Cumming, K.; Earle, J.D.; Hawkins, B.S.; Hayman, J.A.; Jaiyesimi, I.; Jampol, L.M.; et al. Development of Metastatic Disease after Enrollment in the COMS Trials for Treatment of Choroidal Melanoma: Collaborative Ocular Melanoma Study Group Report No. 26. Arch. Ophthalmol. 2005, 123, 1639–1643. [Google Scholar] [CrossRef]
- Kujala, E.; Mäkitie, T.; Kivelä, T. Very Long-Term Prognosis of Patients with Malignant Uveal Melanoma. Investig. Ophthalmol. Vis. Sci. 2003, 44, 4651–4659. [Google Scholar] [CrossRef]
- Eleuteri, A.; Rola, A.C.; Kalirai, H.; Hussain, R.; Sacco, J.; Damato, B.E.; Heimann, H.; Coupland, S.E.; Taktak, A.F.G. Cost-Utility Analysis of a Decade of Liver Screening for Metastases Using the Liverpool Uveal Melanoma Prognosticator Online (LUMPO). Comput. Biol. Med. 2021, 130, 104221. [Google Scholar] [CrossRef]
- Robertson, A.G.; Shih, J.; Yau, C.; Gibb, E.A.; Oba, J.; Mungall, K.L.; Hess, J.M.; Uzunangelov, V.; Walter, V.; Danilova, L.; et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell 2017, 32, 204–220.e15. [Google Scholar] [CrossRef]
- NICE NICE Guidance: Ipilimumab for Previously Untreated Advanced (Unresectable or Metastatic) Melanoma. Available online: https://www.nice.org.uk/guidance/ta319 (accessed on 22 February 2023).
- Payne, K.; McAllister, M.; Davies, L.M. Valuing the Economic Benefits of Complex Interventions: When Maximising Health Is Not Sufficient. Health Econ. 2013, 22, 258–271. [Google Scholar] [CrossRef]


| System | Description | Inputs | Outputs |
|---|---|---|---|
| Liverpool Uveal Melanoma Prognosticator Online (LUMPOIII) | LUMPO III is a semiparametric Markov multi-state model developed using a large dataset of UK patients [19,20]. It has been externally validated on datasets from different centres and is available to clinicians via a website [21,22]. | Age, sex, tumour diameter, tumour height, ciliary body involvement, extraocular extension, presence of epithelioid cells, presence of closed Periodic Acid Schiff (PAS)-positive connective tissue loops, mitotic count (per 40 high power field [HPF]), monosomy 3, chromosome 8q gain | Probability (0–1) of death from metastasis (metastatic associated mortality [MAM]) and probability of death from other causes for each year up to 10 years after primary treatment |
| Liverpool Parsimonious Model (LPM) | LPM was developed from the same dataset as the LUMPOIII model [23]. Due to its relative simplicity, prognostication can be conducted using just a reference table, improving accessibility. | Age, tumour diameter, monosomy 3 | Probability (0–100) of death from metastasis for 2, 5 and 10 years after primary treatment |
| The American Joint Committee on Cancer (AJCC) staging system; 8th Edition | The AJCC system provides a universal staging system which has been adapted for use for cancer at any anatomical site [24,25]. | Tumour diameter, tumour height, ciliary body involvement, extraocular extension | Seven ordinal primary tumour stages (I, IIA, IIB, IIIA, IIIB, IIIC) |
| Monosomy 3 only system (Figure S1) | Monosomy 3 (and underlying loss of function of the tumour suppressor gene BAP1) is a strong independent prognostic factor present in ~50% of choroidal melanomas [26,27,28,29]. | Monosomy 3 | Patients with monosomy 3 classified as ‘high risk’; patients with disomy 3 (normal) status classified as ‘low risk’; patients without a chromosome 3 result classified as ‘unknown’ risk (and also recommended surveillance) |
| Variable | Count | Median | Range | Number Missing |
|---|---|---|---|---|
| Endpoint | Endpoint: 292 | |||
| No endpoint: 755 | ||||
| Age | 1047 | 61 | 18–94 | - |
| Sex | F: 490 | - | - | - |
| M: 557 | ||||
| Largest tumour diameter (mm) | 1047 | 12.7 | 1.2–26 | - |
| Tumour height (mm) | 1047 | 4.5 | 0.5–18.3 | - |
| Ciliary body involvement | Present: 210 | - | - | - |
| Absent: 837 | ||||
| Extraocular extension | Present: 54 | - | - | - |
| Absent: 993 | ||||
| Epithelioid cell type | Present: 463 | - | - | 158 |
| Absent: 426 | ||||
| Presence of PAS+ closed loops | Present: 239 | - | - | 610 |
| Absent: 198 | ||||
| Mitotic count/40 HPF | 0–1: 53 | - | - | 602 |
| 2–3: 143 | ||||
| 4–7: 148 | ||||
| 7+: 101 | ||||
| Chromosome 3 loss | Present: 363 | - | - | 324 |
| Absent: 360 | ||||
| Chromosome 8q gain | Present: 300 | - | - | 480 |
| Absent: 267 | ||||
| Primary treatment | Enucleation: 371 | - | - | - |
| Plaque radiotherapy (RT): 343 | ||||
| Proton Beam RT: 231 | ||||
| Endoresection + Plaque RT: 49 | ||||
| Local resection + Plaque RT: 46 | ||||
| Photodynamic therapy: 7 |
| System | Threshold | Sensitivity | Specificity | PPV | NPV | Surveillance |
|---|---|---|---|---|---|---|
| LUMPOIII | MAM ≥ 0.05 | 95% (92–97) | 46% (42–49) | 40% | 96% | 66% |
| LUMPOIII | MAM ≥ 0.1 | 88% (83–92) | 68% (64–71) | 51% | 94% | 48% |
| LUMPOIII | MAM ≥ 0.2 | 75% (79–82) | 85% (82–87) | 66% | 90% | 32% |
| LPM | MAM ≥ 5 | 93% (90–96) | 48% (44–51) | 41% | 95% | 64% |
| LPM | MAM ≥ 10 | 90% (86–93) | 62% (58–65) | 47% | 94% | 53% |
| AJCC | Stage ≥ IIA | 95% (91–96) | 37% (34–41) | 37% | 95% | 71% |
| AJCC | Stage ≥ IIB | 77% (71–81) | 69% (65–72) | 49% | 88% | 44% |
| Monosomy 3 | NA | 90% (86–93) | 44% (40–47) | 38% | 92% | 66% |
| No. | System | Threshold | Sensitivity | Specificity | PPV | NPV | Surveillance | |
|---|---|---|---|---|---|---|---|---|
| Known Chr3 | Unknown Chr3 | |||||||
| 1 | LUMPOIII | MAM ≥ 0.07 | MAM ≥ 0.045 | 95% (91–97) | 51% (47–54) | 43% | 96% | 62% |
| 2 | LUMPOIII | MAM ≥ 0.15 | MAM ≥ 0.045 | 90% (86–93) | 65% (62–68) | 50% | 94% | 50% |
| 3 | LPM | MAM ≥ 11 | MAM ≥ 7 | 92% (88–95) | 54% (50–57) | 43% | 95% | 59% |
| 4 | AJCC | Stage ≥ IIA | Stage ≥ IIA | 95% (91–96) | 37% (34–41) | 37% | 95% | 71% |
| 5 | Monosomy 3 | M3 or no result included in surveillance group | 90% (86–93) | 44% (40–47) | 38% | 92% | 66% | |
| System | Sensitivity | Specificity | True Positives | False Negatives | False Positives | True Negatives | Total |
|---|---|---|---|---|---|---|---|
| LUMPOIII | 95% | 51% | 53 | 3 | 71 | 73 | 200 |
| AJCC | 95% | 38% | 53 | 3 | 89 | 55 | 200 |
| System | Threshold | Sensitivity | Specificity | PPV | NPV | Surveillance |
|---|---|---|---|---|---|---|
| LUMPOIII | MAM ≥ 0.05 | 95% (91–97) | 44% (40–47) | 39% | 95% | 67% |
| LUMPOIII | MAM ≥ 0.07 | 92% (88–94) | 55% (52–59) | 44% | 95% | 58% |
| LUMPOIII | MAM ≥ 0.1 | 85% (80–89) | 66% (62–69) | 49% | 92% | 48% |
| LPM | MAM ≥ 5 | 93% (89–95) | 34% (31–38) | 35% | 93% | 73% |
| AJCC | Stage ≥ IIA | 95% (91–97) | 37% (34–41) | 37% | 95% | 71% |
| AJCC | Stage ≥ IIB | 77% (71–81) | 69% (65–72) | 49% | 88% | 44% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robinson, H.; Eleuteri, A.; Sacco, J.J.; Hussain, R.; Heimann, H.; Taktak, A.F.G.; Damato, B.; Thompson, A.J.; Allen, T.; Kalirai, H.; et al. Sensitivity and Specificity of Different Prognostic Systems in Guiding Surveillance for Metastases in Uveal Melanoma. Cancers 2023, 15, 2610. https://doi.org/10.3390/cancers15092610
Robinson H, Eleuteri A, Sacco JJ, Hussain R, Heimann H, Taktak AFG, Damato B, Thompson AJ, Allen T, Kalirai H, et al. Sensitivity and Specificity of Different Prognostic Systems in Guiding Surveillance for Metastases in Uveal Melanoma. Cancers. 2023; 15(9):2610. https://doi.org/10.3390/cancers15092610
Chicago/Turabian StyleRobinson, Helena, Antonio Eleuteri, Joseph J. Sacco, Rumana Hussain, Heinrich Heimann, Azzam F. G. Taktak, Bertil Damato, Alexander J. Thompson, Thomas Allen, Helen Kalirai, and et al. 2023. "Sensitivity and Specificity of Different Prognostic Systems in Guiding Surveillance for Metastases in Uveal Melanoma" Cancers 15, no. 9: 2610. https://doi.org/10.3390/cancers15092610
APA StyleRobinson, H., Eleuteri, A., Sacco, J. J., Hussain, R., Heimann, H., Taktak, A. F. G., Damato, B., Thompson, A. J., Allen, T., Kalirai, H., & Coupland, S. E. (2023). Sensitivity and Specificity of Different Prognostic Systems in Guiding Surveillance for Metastases in Uveal Melanoma. Cancers, 15(9), 2610. https://doi.org/10.3390/cancers15092610








