Simple Summary
In surgical pathology, the assessment of the presence of lymph node metastases is a key aspect in terms of the staging and prognosis of cancer patients. This type of work is time-consuming and prone to error. Owing to digital pathology, artificial intelligence (AI) applied to whole slide images (WSIs) of lymph nodes can be exploited for the automatic detection of metastatic cells, so this task can be automated and standardized, increasing diagnostic quality. This manuscript aims to systematically review the published literature regarding the application of various artificial intelligence systems for the assessment of metastases in lymph nodes in whole slide images.
Abstract
One of the most relevant prognostic factors in cancer staging is the presence of lymph node (LN) metastasis. Evaluating lymph nodes for the presence of metastatic cancerous cells can be a lengthy, monotonous, and error-prone process. Owing to digital pathology, artificial intelligence (AI) applied to whole slide images (WSIs) of lymph nodes can be exploited for the automatic detection of metastatic tissue. The aim of this study was to review the literature regarding the implementation of AI as a tool for the detection of metastases in LNs in WSIs. A systematic literature search was conducted in PubMed and Embase databases. Studies involving the application of AI techniques to automatically analyze LN status were included. Of 4584 retrieved articles, 23 were included. Relevant articles were labeled into three categories based upon the accuracy of AI in evaluating LNs. Published data overall indicate that the application of AI in detecting LN metastases is promising and can be proficiently employed in daily pathology practice.
1. Introduction
The incidence of cancer has been increasing worldwide due to a growing and aging population coupled with the adoption of screening programs [1]. One of the most relevant prognostic factors for cancer patients is the presence of lymph node (LN) metastasis. Metastatic disease is an important feature that impacts patient clinical staging and treatment decisions [2]. However, having pathologists manually review LNs microscopically for the presence of metastatic tumor cells is a tedious, time-consuming, and potentially error-prone process. Additionally, many hospitals require intraoperative examinations of sentinel LNs on frozen sections for guiding surgical procedures (Figure 1) [3]. Currently, pathologists may be required to screen a large number of slides of lymph nodes, often including additional immunohistochemical (IHC) stains to conventional hematoxylin and eosin (H&E)-stained sections. As a result, this has increased the workload for surgical pathologists.
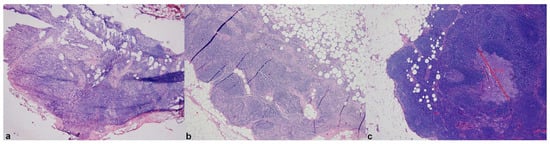
Figure 1.
Compared WSIs of frozen (a) and permanent sections (b,c) of a sentinel lymph node with a micrometastasis from breast carcinoma showing up only at deeper section examination of formalin-fixed paraffin-embedded material (c). Reproduced with permission from Girolami I et al. [3] (original magnification ×25 (a) and ×50 (b)).
Recently, whole slide imaging (WSI) has made digital pathology (DP) more useful for primary diagnosis [4,5] and non-clinical purposes (e.g., education and research) [6,7,8]. By transforming glass slides into WSIs, it has now become possible to use computerized digital image analysis systems including artificial intelligence (AI)-based deep learning (DL) algorithms to analyze digital slides [9]. DP has demonstrated a strong performance in various tasks in different fields, including pathology applications, some of which have already been approved by the Food and Drug Administration (FDA) [10]. In recent years, many AI-based algorithms have been created for the automatic detection of metastases in LNs in WSI. Such novel technology exhibits the potential to reduce pathologists’ workload and increase diagnostic accuracy. The aim of this manuscript is to systematically review the published literature regarding the application of various AI systems for the assessment of metastases in LNs in WSIs. In the next sections of the paper, the search strategy, along with the main characteristics of the retrieved studies, are, respectively, reported in the Material and Methods and the Results. Hence, the included papers are further analyzed in specific paragraphs of the Discussion section according to the following fields of application: (i) the value of AI in lymph node metastases of breast cancer, (ii) the results of public challenges employing and comparing different AI tools, (iii) the value of AI in lymph node metastases detection during intraoperative consultation, and (iv) the role of AI in identifying nodal metastases of tumors apart from breast cancer.
2. Materials and Methods
A systematic review of the literature was conducted, without language restrictions, according to the guideline for Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) [11] and Meta-Analysis of Observational Studies in Epidemiology (MOOSE) [12]. The databases, Pubmed and Embase, were systematically searched until August 2022 to identify any study regarding the application of image analysis and/or AI for the detection of lymph node metastases. The search strategy comprised a combination of terms including “image and analysis”, “artificial and intelligence”, “morphometry”, “histomorphometric”, “neural network”, “convolutional”, “computational”, “deep learning”, “automated”, “machine learning” “lymph node”, “metastasis”, “WSI”, and their spelling variations. The complete search strategy for these databases is detailed in Table S1. Two authors reviewed all article titles and abstracts with the aid of Rayyan QCRI reference manager web application [13]. Eligibility of published studies was determined independently by two reviewers with disagreement resolved through consensus. Full texts of the articles fulfilling the initial screening criteria were acquired and reviewed. Inclusion criteria encompassed the application of any kind of AI as a tool for automatic detection of metastasis in LNs. Only full texts of the articles fulfilling the initial screening criteria were acquired and reviewed. Any disagreement with respect to inclusion of a particular article was resolved by consensus.
Studies represented only by abstracts were excluded, as well as reviews and published letters to the editor with no original data. Two investigators independently extracted data from the included studies with a standardized form. Data extracted included author(s), publication year, country of origin for the research, type of metastatic cancer, type of AI employed, main results, and limitations of the study. As for the performances of the AI systems, three cut-offs were chosen based on whether the precision, accuracy, sensitivity, specificity, or area under the curve (AUC) of the receiver operating curves (ROC) was higher or lower than 95%. This threshold was chosen because it is equivalent to two standard deviations (σ). A shade of green was assigned according to the results achieved by the algorithm proposed by the authors. As the color gradation intensifies, the accuracy of AI for the identification of LN metastases increases as follows: (i) light green for every parameter <95%, (ii) medium colored green for at least one parameter >95%, and (iii) dark green: all parameters >95%.
3. Results
A flow diagram of the screening and exclusion of all the articles is shown in Figure 2.
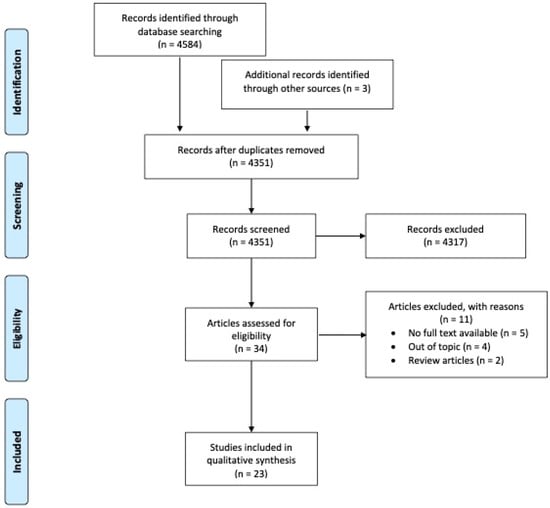
Figure 2.
Flow diagram of the screening and exclusion articles according to PRISMA’s guidelines [11].
A total of 4584 records were retrieved and screened, with only 23 suitable articles finally included in the analysis. An overview of the included studies is provided in Table 1 and Table 2. Publication dates range from 2000 to 2022 and were geographically distributed mainly in the USA (5/23, 22%) and Europe (5/23, 22%), followed by Taiwan (3/23, 13%), China (3/23, 13%), Canada (2/23, 9%), Japan (2/23, 9%), India (1/23, 4%), Bangladesh (1/23, 4%), and South Korea (1/23, 4%).
In more than half of the included studies (16/23, 70%), the investigated LNs were from breast cancer series. Nineteen (19/23, 83%) of the included studies relied only on H&E stains, while three (3/23, 13%) of them used only IHC for cytokeratin and one study coupled H&E with deep UV excitation fluorescence microscopy [14]. As for the employed AI technology, it is worth mentioning that three studies (3/23, 13%) employed a combination (ensemble) of DL algorithms, ranging from four to thirty-seven, for improving the evaluation of breast cancer metastases in LNs. For these reasons, the authors divided so-called ‘challenge’ studies in a different table (Table 2). DL-based computational pathology approaches required either manual annotation of regions of interest (ROI) on WSIs in fully supervised settings or large datasets with slide-level labels in a weakly supervised setting. Both methods required a training dataset. Nineteen (19/23, 87%) of the included studies used a fully supervised approach, while the remaining four (4/23, 13%) algorithms utilized weak supervision [15,16,17,18]. Most of the studies (21/23, 91%) had a WSI training set, whereas two (1/23, 4%) [19,20] studies did not employ a training set as the algorithm was directly able to recognize neoplastic cells stained by the IHC cytokeratin assay. A WSI validation (hold-out) data set was available in three of the papers (3/23 13%). The number of slides used for the training set ranged from 36 to 1963. In the weakly supervised setting, the number of slides of the training set was much higher than in algorithms that relied upon pixel-level annotations.
In terms of AI efficiency, 11 (11/20, 55%) algorithms achieved a performance of >95% across all parameters (precision, accuracy, sensitivity, specificity, or area under the curve of the receiver operating curves) with all four of the weekly supervised studies reaching this goal. As for the detection of isolated tumor cells (ITC), the 95% cut-off was almost never reached; rather, for these settings the AUC rates ranged from 0.575 [21] to 0.9228 [18]. Only one study reached 100% detection rate for ITC, but with the cost of 0% specificity [22]. Among the different employed AI systems, three (3/20, 15%) were programmed to recognize IHC cytokeratin stains, while the others analyzed only H&E slides. Twelve of the latter studies (12/20, 60%) used Convolution Neural Network (CNN) algorithms built on pre-existing platforms, including Googlenet, AlexNet-GRU, Resnet, Densenet, MobileNetV2, and LYNA, among others, and five (5/20, 25%) were ex novo designed algorithms. Finally, three of the included studies (3/20, 15%) simultaneously tested different AI algorithms utilizing the same dataset of H&E stained WSIs [14,23,24].

Table 1.
Characteristics of the included studies.
Table 1.
Characteristics of the included studies.
| Author, Year, Country | Organ | N. of WSI * | Stain | AI Employed | Scanner | Main Results | Limitations |
|---|---|---|---|---|---|---|---|
| Weaver, 2003, USA | BC LNS | NA 100 (20+, 80−) | CKAE1-AE3 | NA | ChromaVision Automated Cell Imaging and Medical System | AI-based identification of 19/20 micrometastatic cases | Use of IHC |
| Clarke, 2011, Canada | BC LNS | 36/ 102 (43+, 59−) | CK 8–18, CAM5.2 | CAD algorithm | Mirax slide scanner | Sensitivity detection of ITCs, micro-, and macrometastases of 57.5%, 89.5%, and 100% | Use of IHC |
| Litjens, 2016, Holland | BC LNS | 98, (48+, 50−) /(42+, 56−) | H&E | In house CNN | 3DHistech Pannoramic 250 Flash II slide scanner | Identification of 90% of all micro- and macrometastases | NA |
| Valkonen, 2017, Finland | BC LNS | 170 (70+, 100) /100 (40+, 60−) | H&E | In house CNN | # | Mean AUC 0.970-0.839 | NA |
| Holten-Rossing, 2017, Denmark | BC LNS | NA /900(139+, 761−) | CK7, CAM5.2 CKAE1-AE3 | Visiopharm APP 10104 | Hamamatsu NanoZoomer-XR | Sensitivity and specificity of 100% and 68.9% | NA |
| Campanella, 2019, USA | BC LNS | 9864/NA | H&E | Resnet 34 | Philips IntelliSite Ultra Fast | AUC of 0.966 | Big amount of data for the testing set |
| Liu, 2018, USA | BC LNS | 270(170−, 110+) /129(49+, 80−) | H&E | LYmph Node Assistant, or LYNA | 3DHISTECH Pannoramic 250 Flash II; Hamamatsu Aperio | AUC of 99.6%, no influence by artifacts (overfixation, poor staining, and air bubbles) | No ITC slides |
| Steiner, 2018, USA | BC LNS | 70 (24−, 46+) /NA | H&E | LYmph Node Assistant, or LYNA | Leica AT2 system at a resolution of 0.25 µm/pixel | Shorter turn-around times with AI for micrometastases and negative images | NA |
| Matsumoto, 2019, Japan | Gastric cancer | 56 (18−, 38+) 27 (26+, 1−) | H&E, UV excitation fluorescence microscopy | VGG16, INCEPTION V3, Inception ResNet V2 | Nanozoomer C9600-02, Hamamatsu Photonics | Mean accuracy in fluorescence patch classification of 97.4%, 98.2%, and 97.9%, respectively. | Technology not available in all laboratories |
| Pham, 2019, Japan | Lung cancer | 233, 10/106 | H&E | HALO-based AI (CNN VGG network) | Aperio Scanscope CS2 digital slide scanner | Sensitivity of 100% (macro–micro metastases, and ITC) | Limited setting parameters, low specificity |
| Pam, 2020, China | ESCC, lung cancer | 242(110−,132+) /795(222+,573−) | H&E | DeepLab model V3 with ResNet-5026 | NA | Accuracy of 94% and 90% in ESCC and lung cancer | No data about digital acquisition process |
| Jin, 2020, Canada | BC LNS | # | H&E | ConcatNet | # | AUC of 0.924 | NA |
| Hu, 2021, China | Gastric cancer | 594/327 | H&E | Xception, DenseNet-121, and fused networks | Leica Aperio Versa | Negative Predictive Value 97.99% in patients given neoadjuvant chemotherapy | Lot of work to classify at pixel level |
| Chuang, 2021, Taiwan | Colon rectal cancer | 1963, 219/1000 | H&E | ResNet 50 | NanoZoomer S360 with a 40× magnification | AUC of 0.99 and 0.99 with macro- and micrometastases | Worse performance with ITC (AUC 0.78) |
| Ming, 2021, USA | BC LNS | CAMELEON 16-17 dataset | H&E | Multiple instance learning | # | Average AUC of 0.953 ± 0.029 at ×40 magnification | Big amount of data for the training set |
| Shahab, 2022, Bangladesh | BC LNS | 270/54 | H&E | AlexNet-GRU | Kaggle dataset | Accuracy, precision, sensitivity, and specificity of 99.50%, 98.10%, 98.90%, and 97.50 | NA |
| Tang, 2022, China | HNSCC | 85 (38+,47−) /50 (21+, 29−) | H&E | GoogLeNet, MobileNet-v2, ResNet50, and ResNet101 | Pannoramic MIDI, 3DHISTECH Ltd. | Overall accuracy, sensitivity, and specificity of 86%, 100%, and 75.9% | NA |
| Vulli, 2022, India | BC LNS | CAMELEON 16 dataset | H&E | Fine-tuned DenseNet 169 | # | Overall accuracy >97.4% | No ITC slides |
| Khalil, 2022, Taiwan | BC LNS | 68 (54+, 18−)/26 (12+, 14−) | H&E, CK AE1-AE3 | Modified FCN based on Shelhamer model | 3DHISTECH Pannoramic | Including ITC overall precision 89.6%, recall 83.8%, F1-score 84.4%, and mIoU 74.9% | NA |
| Huang, 2022, Taiwan | Gastric cancer | 983, 110/201 | H&E | ResNet50 architecture | NanoZoomer S360 digital slide | AUC of 0.9936, improvement of ITC and micrometastasis identification in a shorter turn-around time (−31.5%, p < 0.001) | Big amount of data for the training set |
* testing/training. # data from references [25] and [26]. Abbreviations: AI: artificial intelligence, BC: breast cancer, NA: not available, LNS: sentinel lymph node, IHC: immunohistochemistry, ITC: isolated tumors cells, CNN: convolutional neural network, H&E: hematoxylin and eosin, AUC: Area under the curve mIoU: mean intersection over union, ESCC: esophageal squamous cell carcinoma, HNSCC: head and neck squamous cell carcinoma. Color legend: (i) light green: all parameters <95%, (ii) medium colored green: at least one parameter >95%, (iii) dark green: all parameters >95%.
4. Discussion
AI development is progressing rapidly, with the introduction of several models that can perform various tasks such as the detection and the segmentation of various malignancies such as breast, pharynx, and thyroid carcinoma [27,28,29,30], and non-tumoral specimens, among others [31,32].
LN metastases are one of the most important prognostic factors for staging malignancies [2]. The histological evaluation of nodal specimens must be performed with care and precision. However, this work, conducted manually by pathologists, is often a protracted, tedious, and possibly error-prone process that could benefit from the aid of digital pathology and AI-based algorithms designed to assist with screening LNs for metastatic disease. Indeed, the application of digital techniques to help detect LN metastases may allow pathologists to reduce turn-around times and increase their diagnostic accuracy. Several AI-based tools have been developed in the last decades for addressing this relevant issue, which are further discussed in the following sections, according to their fields of application.

Table 2.
Characteristics of the three studies testing several types of algorithms.
Table 2.
Characteristics of the three studies testing several types of algorithms.
| Author, Year, Country | Organ | N. of WSIs * | Stain | N. of Algorithms | Scanner | Limitations |
|---|---|---|---|---|---|---|
| Bejnordi, 2017, Holland | BC LNS | 160−, 110+/80−, 49+ | H&E | 32 | Pannoramic 250 Flash II,3DHISTECH, NanoZoomer-XR Digital slidescanner C12000-01 Hamamatsu Photonics | No ITC |
| Bandi, 2018, Holland | BC LNS | 899 (558 − 341+)/500 | H&E | 37 | 3D Histec P250; Philips IntelliSite Ultra Fast; Hamamatsu XR C12000 | NA |
| Kim, 2020 South Korea | BC LNS | 197/100 | H&E | 4 | Pannoramic 250 FLASH, 3DHISTECH Ltd. | TL |
* training/testing. Abbreviations: BC: breast cancer, LNS: sentinel lymph node, ITC: isolated tumors cells, H&E: hematoxylin and eosin, TL: transfer learning.
4.1. AI and Nodal Breast Cancer Metastases
The majority (70%) of the included studies in our systematic review focused on the evaluation of sentinel LNs in breast cancer patients. While localized breast cancer has a five-year survival rate of >95%, the presence of LN metastases drops the survival rate to 85% [33]. Based on the diameter of clusters of tumor cells, metastases can be divided into three categories: macrometastases, micrometastases, or isolated tumor cells (ITC), which reflect the “N” classification of breast cancer staging according to the eighth edition of the TNM staging criteria [2]. The biological significance of an ITC is debated and, according to the WHO, LNs just containing lTC are currently excluded from the total positive nodal count for the purposes of the N classification [33].
In 2003, Weaver et al. [19] were among the first investigators to use an automatic LN metastasis detection system on WSIs. Their system was based on a sensor capable of recognizing cells stained by IHC for cytokeratins. Those authors showed that their tool identified 19 of 20 (95.0%; 95% CI 75% to 100%) cases with micrometastases. In the only case where micrometastases were missed, the cancer cells were placed outside the physical limits of slide scanning for the instrument. It is also important to note that slides with excessive stain debris could not be analyzed by the system [19]. The use of IHC stained WSIs was reported by Clarke et al. [21] and Holten-Rossing et al. [20]. Clarke’s [21] models reached sensitivities of 57.5% for ITCs (<0.200 mm), 89.5% for micrometastases, and 100% for larger metastases, while Holten-Rossing and colleagues [20] achieved a sensitivity of 100% without any false negative. Despite the advantage of this IHC-based method, it requires longer stain times and is subject to increased complexity. However, this method could be particularly useful in specific settings such as patients that underwent neoadjuvant chemotherapy where nodal tissue may have drug-induced changes or an inflammatory/fibrotic response.
Undoubtedly, a turning point in the research of AI-based tools for detecting LN metastases was in 2016, represented by the CAMELYON16 challenge (CancerMetastases in Lymph Nodes Challenge) [25]. As a result, several improvements in computer programming resulted in AI capable of analyzing large WSI files. Several AI-based algorithms were developed for detecting metastases of various tumors. Among these was an interesting algorithm owned by Google called LYNA (LYmph Node Assistant), which highlights areas suspicious for the presence of metastatic cells. LYNA was reported in two different articles [34,35]. Steiner et al. [34] designed a fully crossed, intermodal, multi-reader study to evaluate the performance metrics for both assisted and unassisted reads. The pathologists in this study interpreted all the images in both modalities, with or without assistance, in two sessions separated by a wash-out period of at least four weeks. The results stated that all pathologists performed better than the algorithm alone with regard to both sensitivity and specificity; however, when they reviewed the images with AI assistance, the average time of review per image was significantly shorter, especially in negative and micrometastatic LNs. Of note in the study by Steiner et al. is that the researchers also examined WSI for ITC. Liu et al. [35] reached similar results with the algorithm that performed best in the CAMELEON16 challenge (slide level AUC 99.3 vs. 99.4). Relying on the LYNA, through an exhaustive screen for each slide at a high-power magnification, the authors propose the application of this AI in screening LNs highlighting ROIs. In a second phase, these areas could be evaluated by single pathologists, ignoring false positives, and interpreting only the true positive regions. Another algorithm proposed by Khalil et al. [36] took between 2.4 and 9.6 min per WSI to detect metastasis depending on the amount of the graphics processing unit (GPU) used.
As highlighted in Table 1, most of the algorithms in our review appeared to struggle with the task of detecting isolated tumor cells. In order to achieve high sensitivity for small ITCs, AI-based digital tools likely have to allow for a higher rate of false positive results. Indeed, examples of misdiagnosed tissue as micrometastasis could be hypertrophic lymphoid follicles, reactive venules or capillaries, and macrophages. Non-histological pollutants such as paraffin debris, bubbles, or stains can also be incorrectly interpreted as a metastasis by the algorithm. Such errors, however, can be addressed by incorporating oversight of the results by an experienced pathologist. For discrepant cases, additional H&E-stained sections or IHC can be performed. In the future, novel technology such as non-destructive 3D pathology may be used and coupled with the application of AI [37].
4.2. Public Challenges
One mechanism that has facilitated the development of AI algorithms has been through public challenges for specific tasks. Since the first challenges in 2007, the number of challenges per year has steadily been increasing [38]. As noted, a key turning point in this field occurred due to the CAMELYON16 challenge [25] where humans were compared with AI models to detect LN metastases by measuring the time required for reaching a correct diagnosis. The algorithms developed for this public challenge performed better than the 11 involved pathologists in identifying micrometastases. For example, when there was a time limit imposed for the detection of a tumor cell cluster of a diameter 0.2 to <2 mm, the AUC for the best algorithm was 0.994 (95% CI, 0.983–0.999) versus a mean AUC for the pathologists of 0.810 (range, 0.738–0.884; p < 0.001). Nevertheless, the AI models did not surpass the pathologists if no time restriction was applied (AUC = 0.943). A limitation of the CAMELYON16 competition was that WSI with ITCs were not provided. Further efforts were made with the CAMELYON17 challenge [26]. For this subsequent public challenge, the dataset of slides was divided into 100 artificial patients representing different pN stages for assessing the ability of the participating algorithms to perform automatic pN staging and getting closer to a real-world simulation. In general, the AI systems created were not only able to detect the presence of metastases, but were also able to measure their extent, including ITC, and hence better determine an accurate pN-stage.
However, even the best combination of algorithms only correctly classified 77% of patients at the slide level, where the best ranked team wrongly classified 13,4% of the slides in the test set. Overall, ten slides containing micrometastases and four slides containing macrometastases were missed [26], which would of course be unacceptable in clinical practice. One putative strategy to overcome such a problem would be to increase the sensitivity of these AI systems, even though this could increase the number of false positives. These early results imply that perhaps algorithms developed for research purposes to automatically detect LN metatastatic disease are not yet ready to be fully adopted in daily practice.
4.3. Intraoperative Consultation
Only a few studies investigated the implementation of AI to assist with frozen sections. The intraoperative evaluation of a sentinel LN is particularly demanding in this clinical setting, even for an experienced pathologist, because of artifacts such as tissue compression, nuclear ice crystals, sections with folds, and stain nuances which differ from formalin-fixed paraffin-embedded (FFPE) material, overall leading to inferior image quality [39]. Kim et al. [40,41] proposed a transfer learning to effectively train their CNN model for the identification of metastatic breast cancer cells on frozen tissue section digital slides. Transfer learning relies on the re-utilization of a model trained on one task to a second, related task by adding modifications. The authors exploited data of annotated WSIs from FFPE samples to train CNNs working on frozen sections. The best algorithms detected metastasis with an AUC of 0.805 and a processing time of 10.8 min. In conventional (human) frozen section examinations, the time between sample receipt and a rendered pathological diagnosis typically spans from 20 to 30 min, including gross specimen inspection, tissue freezing, sectioning, staining, and microscopic examination [42]. For a digital workflow, the time for scanning frozen sections may vary depending on the size, type of scanning machine, magnification, and focus z-stacking, but it generally ranges from 3 to 9 min [42,43]. Therefore, the application of an efficient AI system in the intraoperative consultation setting, despite increased diagnostic times, could be useful in particularly demanding cases, especially when a second opinion by a remotely located colleague is required. For these reasons, this technology is likely suitable for use in routine practice.
4.4. Tumors Other Than Breast Cancer
Our review identified seven studies where metastatic disease in LNs was from non-breast cancer series including gastric cancer (3/7), squamous carcinoma (2/7), colorectal carcinoma (1), and lung cancer. For gastric cancer metastatic LN series, Matsumoto et al. [14] combined H&E with deep UV excitation fluorescence microscopy and tested different AI models on both types of images acquired. Their results were excellent (AUC = 98.8) and these authors also demonstrated that automated analysis with fluorescence images achieved rates of detection of LN metastasis as accurately as that with H&E images. In the 222 patient cohort of Hu et al. [23], 51 were treated with neoadjuvant chemotherapy (NACT). In this particular study, these researchers demonstrated that their AI model was effective and can accordingly be confidently used for LN screening after NACT. Huang et al. obtained a slide-level [18] AUC curve of 0.9936 for gastric LN metastasis using a weakly supervised algorithm, based on ResnNet50 architecture. Moreover, their proposed method significantly enhanced the sensitivity of ITC recognition and micrometastases identification while shortening the review time per slide. In 2020, Pan et al. [44] employed an algorithm developed on a slide set of metastatic esophageal squamous cell carcinoma to screen LNs suspicious for metastases from the throat and lung. By relying on transfer learning, the applied AI tool reached an accuracy of 96.7% and 90%, respectively, for each type of cancer. Nodal metastases from head and neck squamous cell carcinoma cases were also tested by the algorithm developed by Tang et al. [24] in 2020, gaining ever higher sensitivity rates (100%), but with less specificity (75.9%). Chuang et al. [16], exploiting a ResNet50 model, built a weakly supervised algorithm that performed well in identifying both macro- and micrometastases from colorectal cancer with an AUC of 0.9993 and 0.9956, respectively, at the slide level. However, when focusing on ITC, these values dropped to 0.7828. Finally, in 2019, Pham et al. [22] employed an AI tool called HALO to recognize metastases of nearly all subtypes of lung carcinoma (excluding small cell lung cancer), achieving very high sensitivity rates but with lower specificity.
4.5. Limitations
One key limitation of all the herein studied AI systems is that they were designed to detect just one main pathological lesion (i.e., the detection of metastatic cells), so that they were unable to recognize other rare, but still relevant, key histological features such as co-occurring pathologies (e.g., lymphoma or infection) involving LNs. Secondly, the reported AI tools frequently faced difficulties when metastatic foci were particularly small in size (e.g., ITCs). Finally, DL-based algorithms were often expensive to develop and deploy, therefore hindering the widespread use of this technology.
5. Conclusions
The published data overall indicate that the application of AI in detecting LN metastasis, with due care, is feasible for routine clinical practice in the near future. Ideally, AI-based automated analysis of LNs would assist pathologists by screening these samples and thereby augment diagnostic pathology reporting and tumor staging. A high sensitivity rate is ideally required for these novel AI systems to reach this goal. Further studies are warranted to improve the performance and workflow of this promising technology, in order to validate their adoption in the routine workflow of pathology laboratories.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15092491/s1, Table S1: Search strategy for electronic databases.
Author Contributions
A.E. and S.M. were involved in the conceptualization of the study. N.C. and I.G. researched the literature. N.C., S.M. and I.G. wrote the first draft of the manuscript. N.C. and S.M. analyzed the data and created the figures. P.C.R., A.E., G.N.F., A.G.N., G.B., N.F., G.d., A.S. and L.P. reviewed and edited the manuscript for intellectual content. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the European Union—NextGenerationEU through the Italian Ministry of University and Research under PNRR—M4C2-I1.3 Project PE_00000019 “HEAL ITALIA” to Aldo Scarpa CUP: B33C22001030006.
Institutional Review Board Statement
Ethical review and approval were waived for this study as no ethical issue was raised by reviews.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Worldwide Cancer Data—World Cancer Research Fund. Available online: https://www.Wcrf.Org/Cancer-Trends/Worldwide-Cancer-Data/ (accessed on 24 March 2023).
- Amin, M.B.; Edge, S.; Greene, F.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C.; et al. (Eds.) AJCC Cancer Staging Manual, 8th ed.; Springer: Berlin/Heidelberg, Germany; American Joint Commission on Cancer: Chicago, IL, USA, 2017. [Google Scholar]
- Girolami, I.; Neri, S.; Eccher, A.; Brunelli, M.; Hanna, M.; Pantanowitz, L.; Hanspeter, E.; Mazzoleni, G. Frozen section telepathology service: Efficiency and benefits of an e-health policy in South Tyrol. Digit. Health 2022, 8, 20552076221116776. [Google Scholar] [CrossRef] [PubMed]
- Fraggetta, F.; Garozzo, S.; Zannoni, G.F.; Pantanowitz, L.; Rossi, E.D. Routine Digital Pathology Workflow: The Catania Experience. J. Pathol. Inform. 2017, 8, 51. [Google Scholar] [CrossRef] [PubMed]
- Pantanowitz, L.; Wiley, C.A.; Demetris, A.; Lesniak, A.; Ahmed, I.; Cable, W.; Contis, L.; Parwani, A.V. Experience with multimodality telepathology at the University of Pittsburgh Medical Center. J. Pathol. Inform. 2012, 3, 45. [Google Scholar] [CrossRef] [PubMed]
- Santonicco, N.; Marletta, S.; Pantanowitz, L.; Fadda, G.; Troncone, G.; Brunelli, M.; Ghimenton, C.; Antonini, P.; Paolino, G.; Girolami, I.; et al. Impact of mobile devices on cancer diagnosis in cytology. Diagn. Cytopathol. 2021, 50, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Eccher, A.; Fontanini, G.; Fusco, N.; Girolami, I.; Graziano, P.; Rocco, E.G.; Martini, M.; Morbini, P.; Pantanowitz, L.; Parwani, A.; et al. Digital Slides as an Effective Tool for Programmed Death Ligand 1 Combined Positive Score Assessment and Training: Lessons Learned from the “Programmed Death Ligand 1 Key Learning Program in Head-and-Neck Squamous Cell Carcinoma”. J. Pathol. Inform. 2021, 12. [Google Scholar] [CrossRef]
- Eccher, A.; Neil, D.; Ciangherotti, A.; Cima, L.; Boschiero, L.; Martignoni, G.; Ghimenton, C.; Chilosi, M.; Giobelli, L.; Zampicinini, L.; et al. Digital reporting of whole-slide images is safe and suitable for assessing organ quality in preimplantation renal biopsies. Hum. Pathol. 2016, 47, 115–120. [Google Scholar] [CrossRef]
- Tizhoosh, H.R.; Pantanowitz, L. Artificial Intelligence and Digital Pathology: Challenges and Opportunities. J. Pathol. Inform. 2018, 9, 38. [Google Scholar] [CrossRef]
- Joshi, G.; Jain, A.; Adhikari, S.; Garg, H.; Bhandari, M. FDA approved Artificial Intelligence and Machine Learning (AI/ML)-Enabled Medical Devices: An updated 2022 landscape. medRxiv 2022. preprint. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Stroup, D.F.; Berlin, J.A.; Morton, S.C.; Olkin, I.; Williamson, G.D.; Rennie, D.; Moher, D.; Becker, B.J.; Sipe, T.A.; Thacker, S.B. Meta-analysis of observational studies in epidemiology: A proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 2000, 283, 2008–2012. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Niioka, H.; Kumamoto, Y.; Sato, J.; Inamori, O.; Nakao, R.; Harada, Y.; Konishi, E.; Otsuji, E.; Tanaka, H.; et al. Deep-UV excitation fluorescence microscopy for detection of lymph node metastasis using deep neural network. Sci. Rep. 2019, 9, 16912. [Google Scholar] [CrossRef] [PubMed]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Silva, V.W.K.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Chuang, W.-Y.; Chen, C.-C.; Yu, W.-H.; Yeh, C.-J.; Chang, S.-H.; Ueng, S.-H.; Wang, T.-H.; Hsueh, C.; Kuo, C.-F.; Yeh, C.-Y. Identification of nodal micrometastasis in colorectal cancer using deep learning on annotation-free whole-slide images. Mod. Pathol. 2021, 34, 1901–1911. [Google Scholar] [CrossRef]
- Lu, M.Y.; Williamson, D.F.K.; Chen, T.Y.; Chen, R.J.; Barbieri, M.; Mahmood, F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 2021, 5, 555–570. [Google Scholar] [CrossRef]
- Huang, S.-C.; Chen, C.-C.; Lan, J.; Hsieh, T.-Y.; Chuang, H.-C.; Chien, M.-Y.; Ou, T.-S.; Chen, K.-H.; Wu, R.-C.; Liu, Y.-J.; et al. Deep neural network trained on gigapixel images improves lymph node metastasis detection in clinical settings. Nat. Commun. 2022, 13, 3347. [Google Scholar] [CrossRef]
- Weaver, D.L.; Krag, D.N.; Manna, E.A.; Ashikaga, T.; Harlow, S.P.; Bauer, K.D. Comparison of Pathologist-Detected and Automated Computer-Assisted Image Analysis Detected Sentinel Lymph Node Micrometastases in Breast Cancer. Mod. Pathol. 2003, 16, 1159–1163. [Google Scholar] [CrossRef]
- Holten-Rossing, H.; Talman, M.M.; Jylling, A.M.B.; Lænkholm, A.; Kristensson, M.; Vainer, B. Application of automated image analysis reduces the workload of manual screening of sentinel lymph node biopsies in breast cancer. Histopathology 2017, 71, 866–873. [Google Scholar] [CrossRef]
- Clarke, G.M.; Peressotti, C.; Holloway, C.M.B.; Zubovits, J.T.; Liu, K.; Yaffe, M.J. Development and evaluation of a robust algorithm for computer-assisted detection of sentinel lymph node micrometastases. Histopathology 2011, 59, 116–128. [Google Scholar] [CrossRef]
- Pham, H.H.N.; Futakuchi, M.; Bychkov, A.; Furukawa, T.; Kuroda, K.; Fukuoka, J. Detection of Lung Cancer Lymph Node Metastases from Whole-Slide Histopathologic Images Using a Two-Step Deep Learning Approach. Am. J. Pathol. 2019, 189, 2428–2439. [Google Scholar] [CrossRef]
- Hu, Y.; Su, F.; Dong, K.; Wang, X.; Zhao, X.; Jiang, Y.; Li, J.; Ji, J.; Sun, Y. Deep learning system for lymph node quantification and metastatic cancer identification from whole-slide pathology images. Gastric Cancer 2021, 24, 868–877. [Google Scholar] [CrossRef]
- Tang, H.; Li, G.; Liu, C.; Huang, D.; Zhang, X.; Qiu, Y.; Liu, Y. Diagnosis of lymph node metastasis in head and neck squamous cell carcinoma using deep learning. Laryngoscope Investig. Otolaryngol. 2022, 7, 161–169. [Google Scholar] [CrossRef]
- Bejnordi, B.E.; Veta, M.; Van Diest, P.J.; Van Ginneken, B.; Karssemeijer, N.; Litjens, G.; Van Der Laak, J.A.W.M.; Hermsen, M.; Manson, Q.F.; Balkenhol, M.; et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women with Breast Cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef]
- Bandi, P.; Geessink, O.; Manson, Q.; Van Dijk, M.; Balkenhol, M.; Hermsen, M.; Bejnordi, B.E.; Lee, B.; Paeng, K.; Zhong, A.; et al. From Detection of Individual Metastases to Classification of Lymph Node Status at the Patient Level: The CAMELYON17 Challenge. IEEE Trans. Med. Imaging 2018, 38, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Cireşan, D.C.; Giusti, A.; Gambardella, L.M.; Schmidhuber, J. Mitosis Detection in Breast Cancer Histology Images with Deep Neural Networks. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Nagoya, Japan, 22–26 September 2013; Springer: Berlin/Heidelberg, Germany, 2013; pp. 411–418. [Google Scholar]
- dos Santos, D.F.; de Faria, P.R.; Travençolo, B.A.; Nascimento, M.Z.D. Automated detection of tumor regions from oral histological whole slide images using fully convolutional neural networks. Biomed. Signal Process. Control. 2021, 69, 102921. [Google Scholar] [CrossRef]
- Pedersen, N.J.; Jensen, D.H.; Lelkaitis, G.; Kiss, K.; Charabi, B.; Specht, L.; von Buchwald, C. Construction of a pathological risk model of occult lymph node metastases for prognostication by semi-automated image analysis of tumor budding in early-stage oral squamous cell carcinoma. Oncotarget 2017, 8, 18227–18237. [Google Scholar] [CrossRef]
- Girolami, I.; Marletta, S.; Pantanowitz, L.; Torresani, E.; Ghimenton, C.; Barbareschis, M.; Scarpa, A.; Brunelli, M.; Barresi, V.; Trimboli, P.; et al. Impact of image analysis and artificial intelligence in thyroid pathology, with particular reference to cytological aspects. Cytopathology 2020, 31, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Marletta, S.; Pantanowitz, L.; Santonicco, N.; Caputo, A.; Bragantini, E.; Brunelli, M.; Girolami, I.; Eccher, A. Application of Digital Imaging and Artificial Intelligence to Pathology of the Placenta. Pediatr. Dev. Pathol. 2022, 26, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Girolami, I.; Pantanowitz, L.; Marletta, S.; Hermsen, M.; van der Laak, J.; Munari, E.; Furian, L.; Vistoli, F.; Zaza, G.; Cardillo, M.; et al. Artificial intelligence applications for pre-implantation kidney biopsy pathology practice: A systematic review. J. Nephrol. 2022, 35, 1801–1808. [Google Scholar] [CrossRef] [PubMed]
- WHO Classification of Tumours Editorial Board. Breast Tumours, 5th ed.; WHO Classification of Tumours Series; International Agency for Research on Cancer: Lyon, France, 2019; Volume 2, Available online: https://Publications.Iarc.Fr/581 (accessed on 24 January 2023).
- Steiner, D.F.; MacDonald, R.; Liu, Y.; Truszkowski, P.; Hipp, J.D.; Gammage, C.; Thng, F.; Peng, L.; Stumpe, M.C. Impact of Deep Learning Assistance on the Histopathologic Review of Lymph Nodes for Metastatic Breast Cancer. Am. J. Surg. Pathol. 2018, 42, 1636–1646. [Google Scholar] [CrossRef]
- Liu, Y.; Kohlberger, T.; Norouzi, M.; Dahl, G.E.; Smith, J.L.; Mohtashamian, A.; Olson, N.; Peng, L.H.; Hipp, J.D.; Stumpe, M.C.; et al. Artificial Intelligence–Based Breast Cancer Nodal Metastasis Detection: Insights into the Black Box for Pathologists. Arch. Pathol. Lab. Med. 2018, 143, 859–868. [Google Scholar] [CrossRef]
- Khalil, M.-A.; Lee, Y.-C.; Lien, H.-C.; Jeng, Y.-M.; Wang, C.-W. Fast Segmentation of Metastatic Foci in H&E Whole-Slide Images for Breast Cancer Diagnosis. Diagnostics 2022, 12, 990. [Google Scholar] [CrossRef]
- Liu, J.T.C.; Glaser, A.K.; Bera, K.; True, L.D.; Reder, N.P.; Eliceiri, K.W.; Madabhushi, A. Harnessing non-destructive 3D pathology. Nat. Biomed. Eng. 2021, 5, 203–218. [Google Scholar] [CrossRef]
- Hartman, D.J.; Van Der Laak, J.A.W.M.; Gurcan, M.N.; Pantanowitz, L. Value of Public Challenges for the Development of Pathology Deep Learning Algorithms. J. Pathol. Informatics 2020, 11, 7. [Google Scholar] [CrossRef]
- Chen, Y.; Anderson, K.R.; Xu, J.; Goldsmith, J.D.; Heher, Y.K. Frozen-Section Checklist Implementation Improves Quality and Patient Safety. Am. J. Clin. Pathol. 2019, 151, 607–612. [Google Scholar] [CrossRef]
- Kim, Y.-G.; Kim, S.; Cho, C.E.; Song, I.H.; Lee, H.J.; Ahn, S.; Park, S.Y.; Gong, G.; Kim, N. Effectiveness of transfer learning for enhancing tumor classification with a convolutional neural network on frozen sections. Sci. Rep. 2020, 10, 21899. [Google Scholar] [CrossRef]
- Kim, Y.-G.; Song, I.H.; Lee, H.; Kim, S.; Yang, D.H.; Kim, N.; Shin, D.; Yoo, Y.; Lee, K.; Kim, D.; et al. Challenge for Diagnostic Assessment of Deep Learning Algorithm for Metastases Classification in Sentinel Lymph Nodes on Frozen Tissue Section Digital Slides in Women with Breast Cancer. Cancer Res. Treat. 2020, 52, 1103–1111. [Google Scholar] [CrossRef]
- Laurent-Bellue, A.; Poullier, E.; Pomerol, J.-F.; Adnet, E.; Redon, M.-J.; Posseme, K.; Trassard, O.; Cherqui, D.; Zarca, K.; Guettier, C. Four-Year Experience of Digital Slide Telepathology for Intraoperative Frozen Section Consultations in a Two-Site French Academic Department of Pathology. Am. J. Clin. Pathol. 2020, 154, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Menter, T.; Nicolet, S.; Baumhoer, D.; Tolnay, M.; Tzankov, A. Intraoperative frozen section consultation by remote whole-slide imaging analysis–validation and comparison to robotic remote microscopy. J. Clin. Pathol. 2019, 73, 350–352. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Sun, Z.; Wang, W.; Yang, Z.; Jia, J.; Feng, X.; Wang, Y.; Fang, Q.; Li, J.; Dai, H.; et al. Automatic detection of squamous cell carcinoma metastasis in esophageal lymph nodes using semantic segmentation. Clin. Transl. Med. 2020, 10, e129. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).