Multi-View Radiomics Feature Fusion Reveals Distinct Immuno-Oncological Characteristics and Clinical Prognoses in Hepatocellular Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
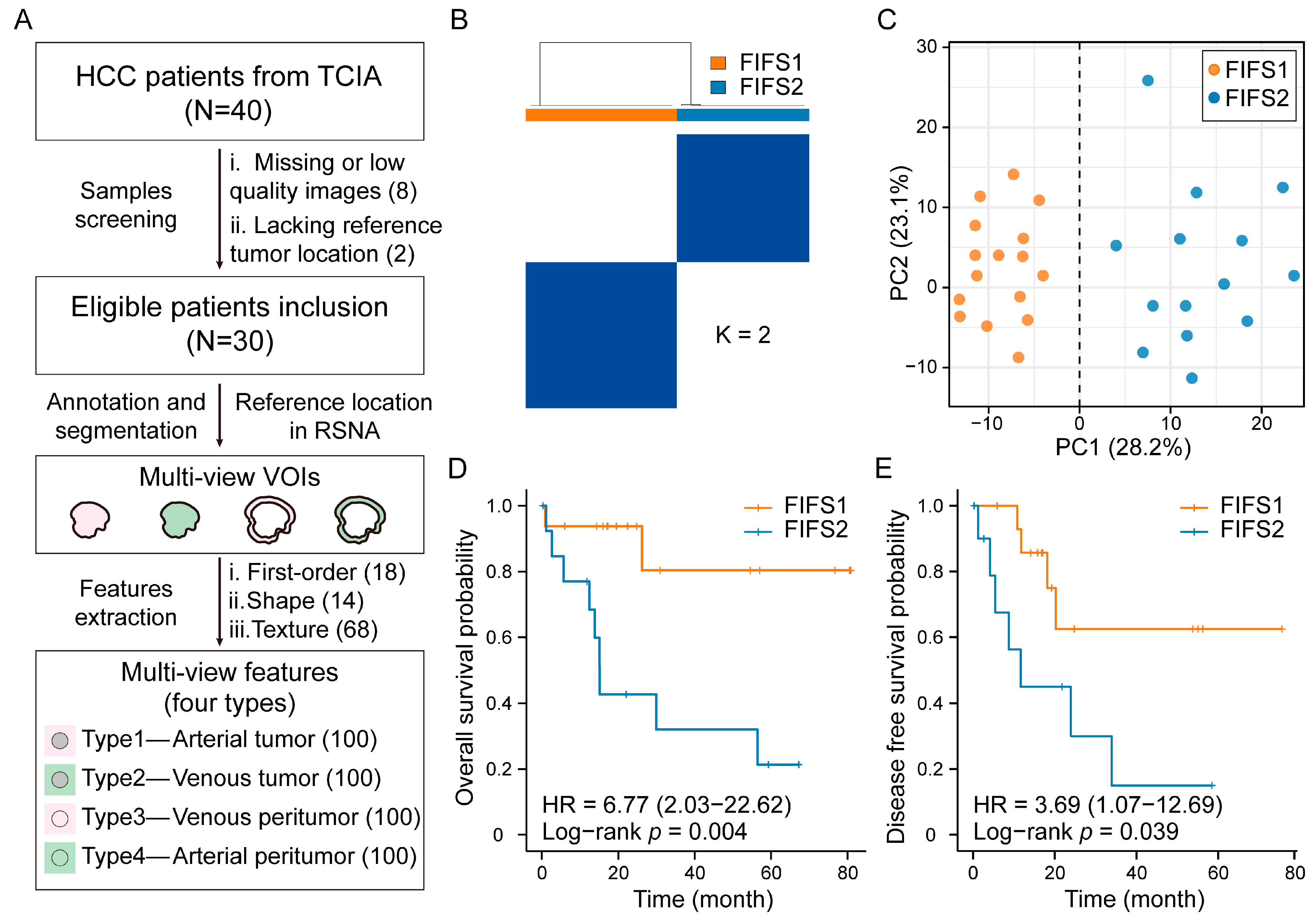
2.1. Data Collection
2.2. Volume of Interest Segmentation and Radiomics Feature Extraction
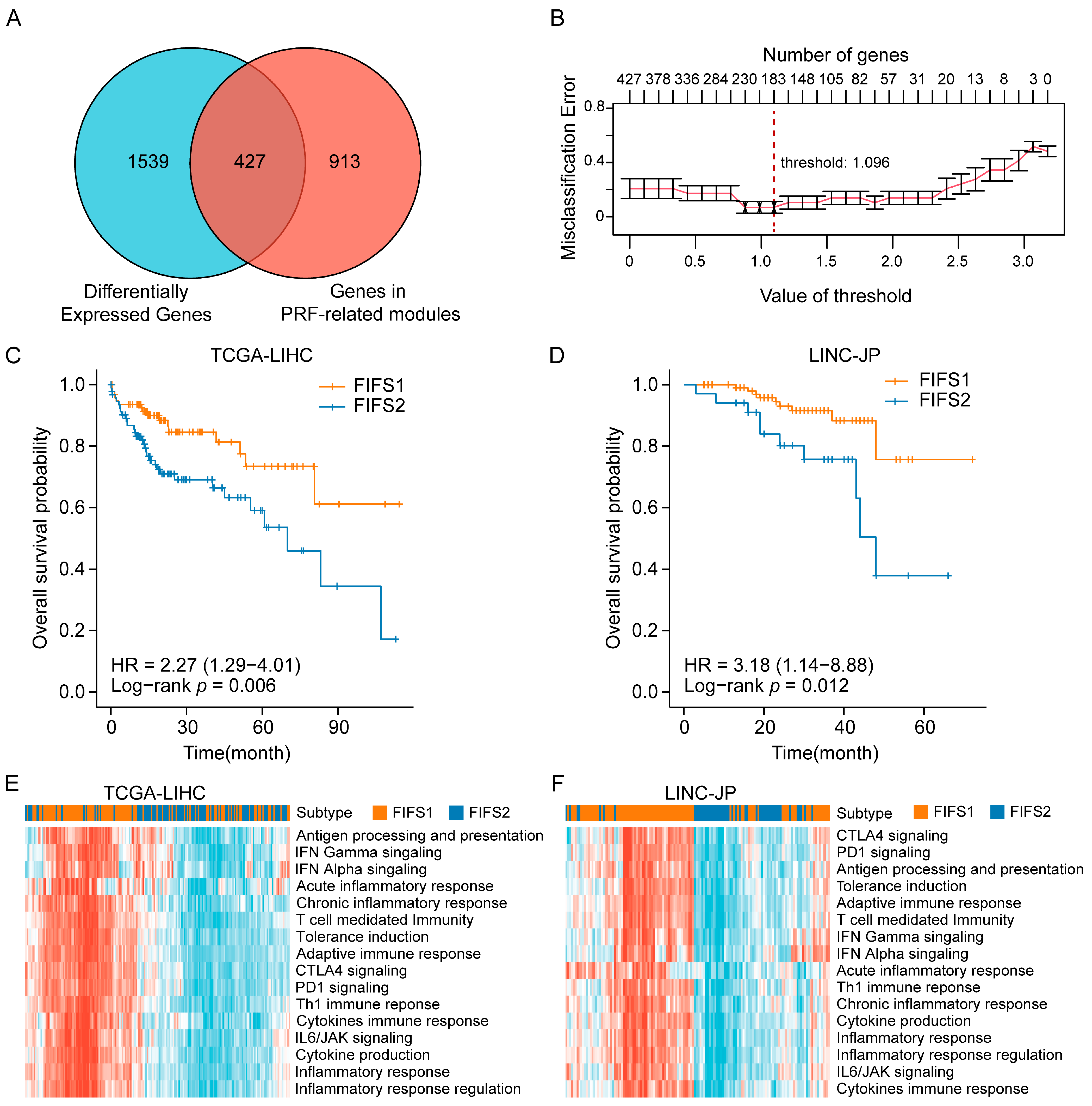
2.3. Radiomics Feature Fusion and Subtype Identification
2.4. Functional Enrichment Analysis and TIME Comparison between FIFS Subtypes
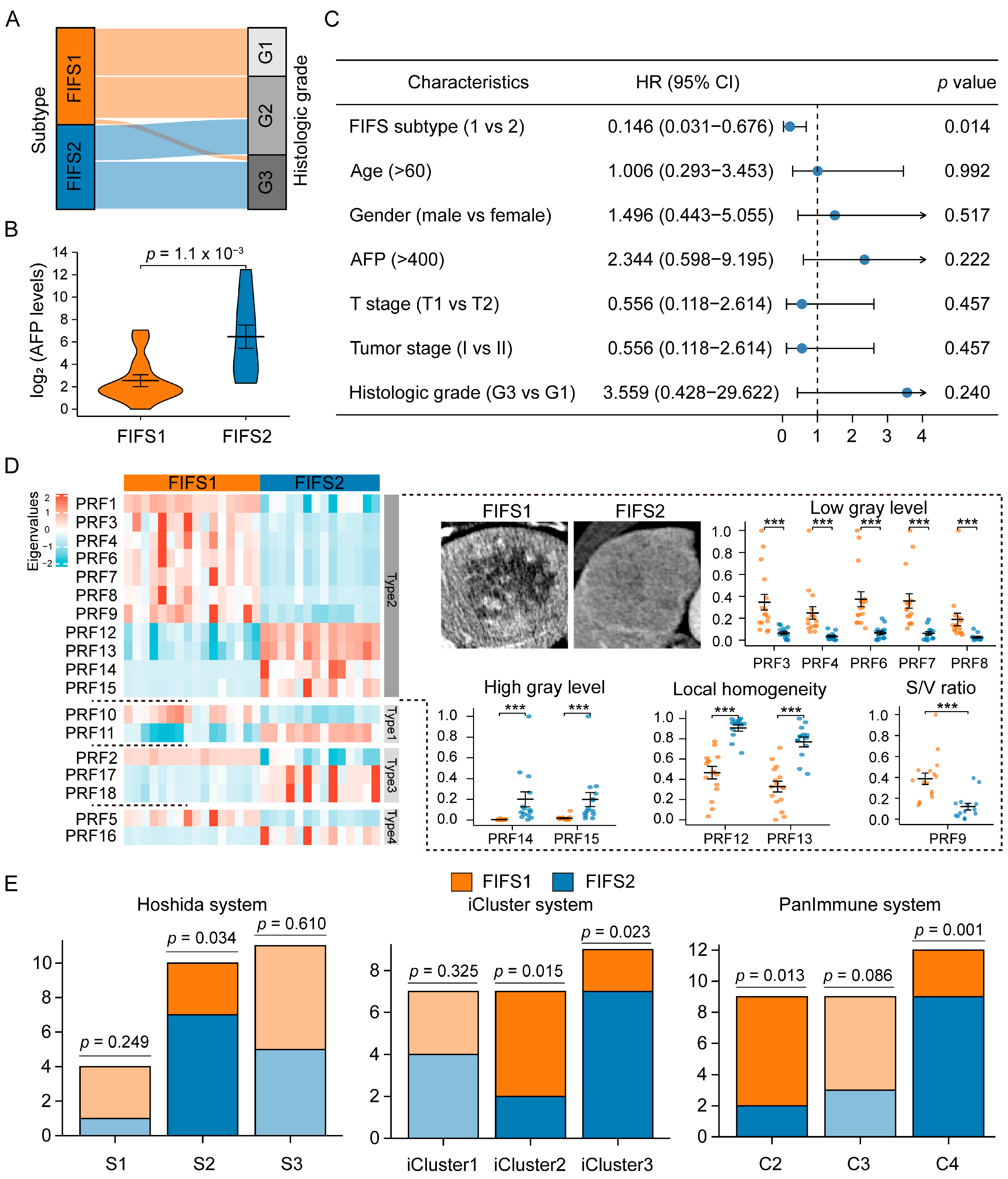
2.5. Comparative Analysis with Previous Molecular Subtypes
2.6. Radiogenomics Association Identification and Validation
2.7. Quantification and Statistical Analysis
3. Results
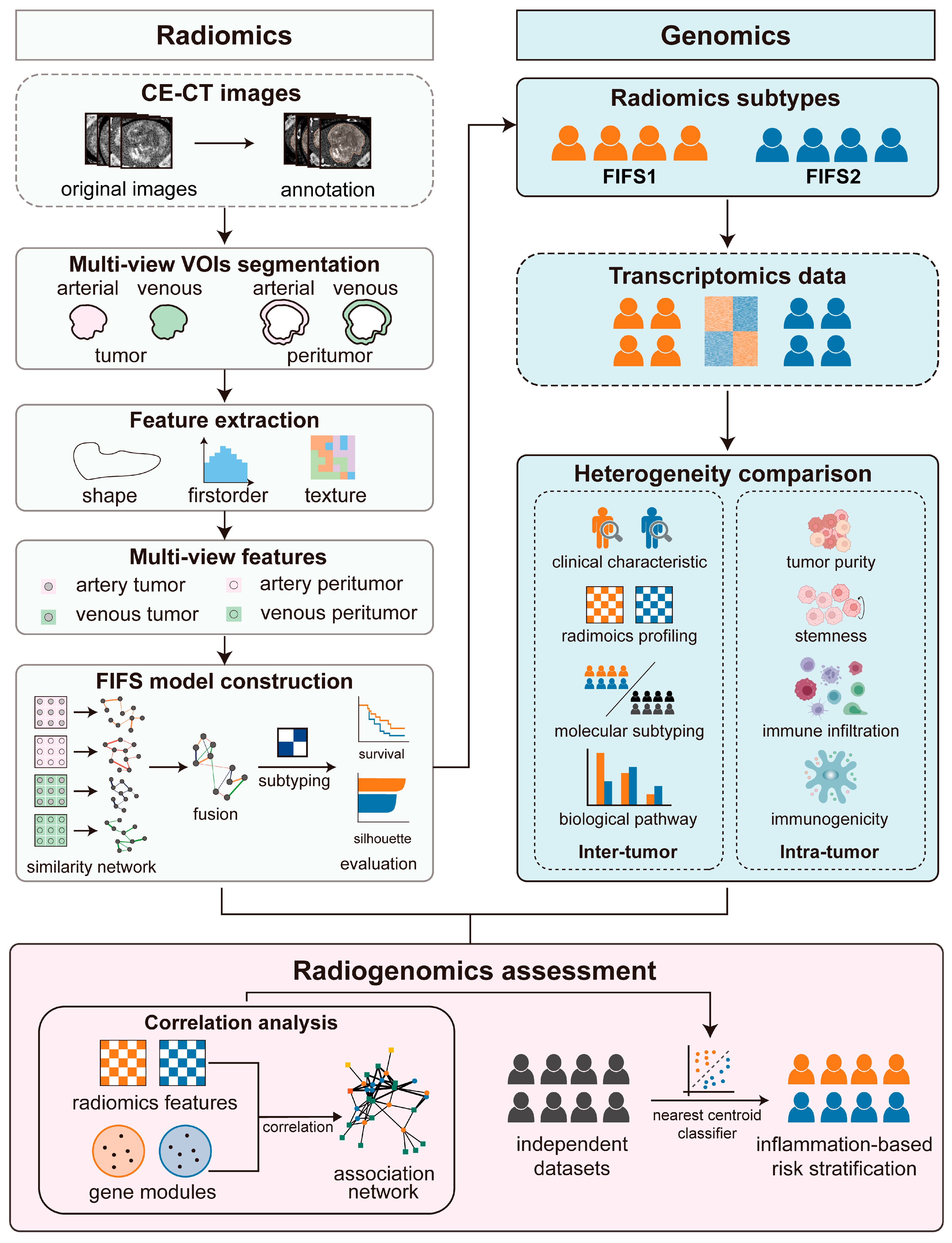
3.1. Study Design
3.2. Identifying HCC Imaging Subtypes Based on Multi-View Radiomics Feature Fusion
3.3. Radiomics Subtypes Describe Distinct Texture-Dominated Imaging Profiles
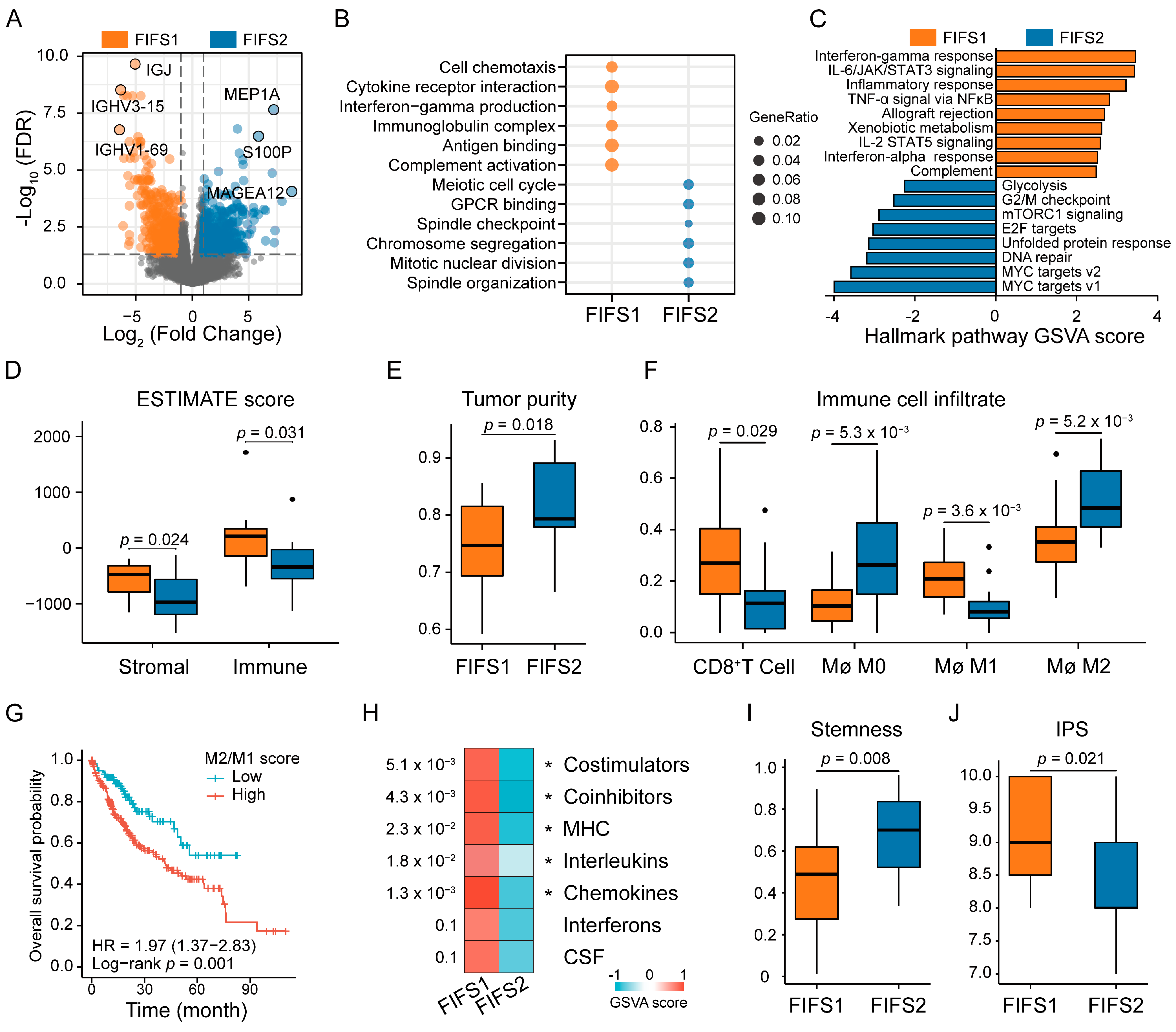
3.4. Distinct Biological Significance and Proinflammatory TIME Status of the FIFS Subtypes
3.5. Close Radiogenomics Association between Imaging Features and Immune Response as Well as a Cell Cycle Modulating Function
3.6. Independent Validation for the Immunocompetent Status and Prognostic Relevance Based on the FIFS System
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- European Association for the Study of the Liver. Easl Clinical Practice Guidelines: Management of Hepatocellular Carcinoma. J. Hepatol. 2018, 69, 182–236.
- Sugawara, Y.; Hibi, T. Surgical treatment of hepatocellular carcinoma. Biosci. Trends 2021, 15, 138–141. [Google Scholar] [CrossRef]
- Chen, S.; Cao, Q.; Wen, W.; Wang, H. Targeted therapy for hepatocellular carcinoma: Challenges and opportunities. Cancer Lett. 2019, 460, 1–9. [Google Scholar] [CrossRef]
- Liu, J.; Dang, H.; Wang, X.W. The significance of intertumor and intratumor heterogeneity in liver cancer. Exp. Mol. Med. 2018, 50, e416. [Google Scholar] [CrossRef]
- Vitale, I.; Shema, E.; Loi, S.; Galluzzi, L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat. Med. 2021, 27, 212–224. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network, Cancer Genome Atlas Research. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341.e1323. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e814. [Google Scholar] [CrossRef]
- Marrero, J.A.; Kulik, L.M.; Sirlin, C.B.; Zhu, A.X.; Finn, R.S.; Abecassis, M.M.; Roberts, L.R.; Heimbach, J.K. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018, 68, 723–750. [Google Scholar] [CrossRef]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Ming, W.; Li, F.; Zhu, Y.; Bai, Y.; Gu, W.; Liu, Y.; Sun, X.; Liu, X.; Liu, H. Predicting hormone receptors and PAM50 subtypes of breast cancer from multi-scale lesion images of DCE-MRI with transfer learning technique. Comput. Biol. Med. 2022, 150, 106147. [Google Scholar] [CrossRef] [PubMed]
- Devkota, L.; Starosolski, Z.; Rivas, C.H.; Stupin, I.; Annapragada, A.; Ghaghada, K.B.; Parihar, R. Detection of response to tumor microenvironment-targeted cellular immunotherapy using nano-radiomics. Sci. Adv. 2020, 6, eaba6156. [Google Scholar] [CrossRef] [PubMed]
- Harding-Theobald, E.; Louissaint, J.; Maraj, B.; Cuaresma, E.; Townsend, W.; Mendiratta-Lala, M.; Singal, A.G.; Su, G.L.; Lok, A.S.; Parikh, N.D. Systematic review: Radiomics for the diagnosis and prognosis of hepatocellular carcinoma. Aliment. Pharmacol. Ther. 2021, 54, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Asayama, Y.; Nishie, A.; Ishigami, K.; Ushijima, Y.; Takayama, Y.; Okamoto, D.; Fujita, N.; Morita, K.; Obara, M.; Honda, H. Heterogeneity of non-cancerous liver parenchyma on gadoxetic acid-enhanced MRI: An imaging biomarker for hepatocellular carcinoma development in chronic liver disease. Clin. Radiol. 2016, 71, 432–437. [Google Scholar] [CrossRef]
- Brenet Defour, L.; Mulé, S.; Tenenhaus, A.; Piardi, T.; Sommacale, D.; Hoeffel, C.; Thiéfin, G. Hepatocellular carcinoma: CT texture analysis as a predictor of survival after surgical resection. Eur. Radiol. 2019, 29, 1231–1239. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, H.L.; Liu, Q.P.; Sun, S.W.; Zhang, J.; Zhu, F.P.; Yang, G.; Yan, X.; Zhang, Y.D.; Liu, X.S. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J. Hepatol. 2019, 70, 1133–1144. [Google Scholar] [CrossRef]
- Chen, S.; Feng, S.; Wei, J.; Liu, F.; Li, B.; Li, X.; Hou, Y.; Gu, D.; Tang, M.; Xiao, H.; et al. Pretreatment prediction of immunoscore in hepatocellular cancer: A radiomics-based clinical model based on Gd-EOB-DTPA-enhanced MRI imaging. Eur. Radiol. 2019, 29, 4177–4187. [Google Scholar] [CrossRef]
- Kim, J.; Choi, S.J.; Lee, S.H.; Lee, H.Y.; Park, H. Predicting Survival Using Pretreatment CT for Patients With Hepatocellular Carcinoma Treated With Transarterial Chemoembolization: Comparison of Models Using Radiomics. AJR Am. J. Roentgenol. 2018, 211, 1026–1034. [Google Scholar] [CrossRef]
- Zinn, P.O.; Singh, S.K.; Kotrotsou, A.; Hassan, I.; Thomas, G.; Luedi, M.M.; Elakkad, A.; Elshafeey, N.; Idris, T.; Mosley, J.; et al. A Coclinical Radiogenomic Validation Study: Conserved Magnetic Resonance Radiomic Appearance of Periostin-Expressing Glioblastoma in Patients and Xenograft Models. Clin. Cancer Res. 2018, 24, 6288–6299. [Google Scholar] [CrossRef]
- Taouli, B.; Hoshida, Y.; Kakite, S.; Chen, X.; Tan, P.S.; Sun, X.; Kihira, S.; Kojima, K.; Toffanin, S.; Fiel, M.I.; et al. Imaging-based surrogate markers of transcriptome subclasses and signatures in hepatocellular carcinoma: Preliminary results. Eur. Radiol. 2017, 27, 4472–4481. [Google Scholar] [CrossRef]
- An, J.; Oh, M.; Kim, S.Y.; Oh, Y.J.; Oh, B.; Oh, J.H.; Kim, W.; Jung, J.H.; Kim, H.I.; Kim, J.S.; et al. PET-Based Radiogenomics Supports mTOR Pathway Targeting for Hepatocellular Carcinoma. Clin. Cancer Res. 2022, 28, 1821–1831. [Google Scholar] [CrossRef] [PubMed]
- Hectors, S.J.; Lewis, S.; Besa, C.; King, M.J.; Said, D.; Putra, J.; Ward, S.; Higashi, T.; Thung, S.; Yao, S.; et al. MRI radiomics features predict immuno-oncological characteristics of hepatocellular carcinoma. Eur. Radiol. 2020, 30, 3759–3769. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Bajari, R.; Andric, D.; Gerthoffert, F.; Lepsa, A.; Nahal-Bose, H.; Stein, L.D.; Ferretti, V. The International Cancer Genome Consortium Data Portal. Nat. Biotechnol. 2019, 37, 367–369. [Google Scholar] [CrossRef] [PubMed]
- Kalpathy-Cramer, J.; Beers, A.; Mamonov, A. Crowds Cure Cancer: Crowdsourced data collected at the RSNA 2017 annual meeting [Data set]. Cancer Imaging Arch. 2019. [Google Scholar] [CrossRef]
- Meng, X.P.; Wang, Y.C.; Ju, S.; Lu, C.Q.; Zhong, B.Y.; Ni, C.F.; Zhang, Q.; Yu, Q.; Xu, J.; Ji, J.; et al. Radiomics Analysis on Multiphase Contrast-Enhanced CT: A Survival Prediction Tool in Patients With Hepatocellular Carcinoma Undergoing Transarterial Chemoembolization. Front. Oncol. 2020, 10, 1196. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Wang, B.; Mezlini, A.M.; Demir, F.; Fiume, M.; Tu, Z.; Brudno, M.; Haibe-Kains, B.; Goldenberg, A. Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods 2014, 11, 333–337. [Google Scholar] [CrossRef]
- Wilkerson, M.D.; Hayes, D.N. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010, 26, 1572–1573. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.O. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Aran, D.; Sirota, M.; Butte, A.J. Systematic pan-cancer analysis of tumour purity. Nat. Commun. 2015, 6, 8971. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Su, G.H.; Xiao, Y.; Jiang, L.; Zheng, R.C.; Wang, H.; Chen, Y.; Gu, Y.J.; You, C.; Shao, Z.M. Radiomics features for assessing tumor-infiltrating lymphocytes correlate with molecular traits of triple-negative breast cancer. J. Transl. Med. 2022, 20, 471. [Google Scholar] [CrossRef]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Malta, T.M.; Sokolov, A.; Gentles, A.J.; Burzykowski, T.; Poisson, L.; Weinstein, J.N.; Kamińska, B.; Huelsken, J.; Omberg, L.; Gevaert, O.; et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 2018, 173, 338–354.e315. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, Y.; Nijman, S.M.; Kobayashi, M.; Chan, J.A.; Brunet, J.P.; Chiang, D.Y.; Villanueva, A.; Newell, P.; Ikeda, K.; Hashimoto, M.; et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009, 69, 7385–7392. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R.; Hastie, T.; Narasimhan, B.; Chu, G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. USA 2002, 99, 6567–6572. [Google Scholar] [CrossRef]
- Wu, J.; Cui, Y.; Sun, X.; Cao, G.; Li, B.; Ikeda, D.M.; Kurian, A.W.; Li, R. Unsupervised Clustering of Quantitative Image Phenotypes Reveals Breast Cancer Subtypes with Distinct Prognoses and Molecular Pathways. Clin. Cancer Res. 2017, 23, 3334–3342. [Google Scholar] [CrossRef]
- García-Mulero, S.; Alonso, M.H.; Pardo, J.; Santos, C.; Sanjuan, X.; Salazar, R.; Moreno, V.; Piulats, J.M.; Sanz-Pamplona, R. Lung metastases share common immune features regardless of primary tumor origin. J. Immunother. Cancer 2020, 8, e000491. [Google Scholar] [CrossRef]
- Dmitrieva-Posocco, O.; Dzutsev, A.; Posocco, D.F.; Hou, V.; Yuan, W.; Thovarai, V.; Mufazalov, I.A.; Gunzer, M.; Shilovskiy, I.P.; Khaitov, M.R.; et al. Cell-Type-Specific Responses to Interleukin-1 Control Microbial Invasion and Tumor-Elicited Inflammation in Colorectal Cancer. Immunity 2019, 50, 166–180.e167. [Google Scholar] [CrossRef]
- Teh, J.L.F.; Aplin, A.E. Arrested Developments: CDK4/6 Inhibitor Resistance and Alterations in the Tumor Immune Microenvironment. Clin. Cancer Res. 2019, 25, 921–927. [Google Scholar] [CrossRef]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef]
- Gabrielson, A.; Wu, Y.; Wang, H.; Jiang, J.; Kallakury, B.; Gatalica, Z.; Reddy, S.; Kleiner, D.; Fishbein, T.; Johnson, L.; et al. Intratumoral CD3 and CD8 T-cell Densities Associated with Relapse-Free Survival in HCC. Cancer Immunol. Res. 2016, 4, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Watkins-Schulz, R.; Tiet, P.; Gallovic, M.D.; Junkins, R.D.; Batty, C.; Bachelder, E.M.; Ainslie, K.M.; Ting, J.P.Y. A microparticle platform for STING-targeted immunotherapy enhances natural killer cell- and CD8(+) T cell-mediated anti-tumor immunity. Biomaterials 2019, 205, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Dragovich, M.A.; Adam, K.; Strazza, M.; Tocheva, A.S.; Peled, M.; Mor, A. SLAMF6 clustering is required to augment T cell activation. PLoS ONE 2019, 14, e0218109. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, M.; Rauen, T.; Kis-Toth, K.; Kyttaris, V.C.; Hedrich, C.M.; Terhorst, C.; Tsokos, G.C. Increased expression of SLAM receptors SLAMF3 and SLAMF6 in systemic lupus erythematosus T lymphocytes promotes Th17 differentiation. J. Immunol. 2012, 188, 1206–1212. [Google Scholar] [CrossRef]
- Alfieri, C.; Zhang, S.; Barford, D. Visualizing the complex functions and mechanisms of the anaphase promoting complex/cyclosome (APC/C). Open Biol. 2017, 7, 170204. [Google Scholar] [CrossRef] [PubMed]
- Gui, L.; Zhang, S.; Xu, Y.; Zhang, H.; Zhu, Y.; Kong, L. UBE2S promotes cell chemoresistance through PTEN-AKT signaling in hepatocellular carcinoma. Cell Death Discov. 2021, 7, 357. [Google Scholar] [CrossRef]
- Yao, J.; Shi, Y.; Cao, K.; Lu, L.; Lu, J.; Song, Q.; Jin, G.; Xiao, J.; Hou, Y.; Zhang, L. DeepPrognosis: Preoperative prediction of pancreatic cancer survival and surgical margin via comprehensive understanding of dynamic contrast-enhanced CT imaging and tumor-vascular contact parsing. Med. Image Anal. 2021, 73, 102150. [Google Scholar] [CrossRef]
- Markello, R.D.; Shafiei, G.; Tremblay, C.; Postuma, R.B.; Dagher, A.; Misic, B. Multimodal phenotypic axes of Parkinson’s disease. NPJ Park. Dis. 2021, 7, 6. [Google Scholar] [CrossRef]
- Han, S.; Xu, Y.; Guo, H.R.; Fang, K.; Wei, Y.; Liu, L.; Cheng, J.; Zhang, Y.; Cheng, J. Two distinct subtypes of obsessive compulsive disorder revealed by a framework integrating multimodal neuroimaging information. Hum. Brain Mapp. 2022, 43, 4254–4265. [Google Scholar] [CrossRef]
- Kim, S.H.; Kamaya, A.; Willmann, J.K. CT perfusion of the liver: Principles and applications in oncology. Radiology 2014, 272, 322–344. [Google Scholar] [CrossRef]
- Kelley, R.K.; Meyer, T.; Rimassa, L.; Merle, P.; Park, J.W.; Yau, T.; Chan, S.L.; Blanc, J.F.; Tam, V.C.; Tran, A.; et al. Serum Alpha-fetoprotein Levels and Clinical Outcomes in the Phase III CELESTIAL Study of Cabozantinib versus Placebo in Patients with Advanced Hepatocellular Carcinoma. Clin. Cancer Res. 2020, 26, 4795–4804. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, K.A.; Petrick, J.L.; El-Serag, H.B. Epidemiology of Hepatocellular Carcinoma. Hepatology 2021, 73 (Suppl. 1), 4–13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lin, J.B.; Jia, M.; Zhang, C.C.; Xu, R.; Guo, L.; Lin, X.J.; Wang, Q.S. Clinical and imaging features preoperative evaluation of histological grade and microvascular infiltration of hepatocellular carcinoma. BMC Gastroenterol. 2022, 22, 369. [Google Scholar] [CrossRef] [PubMed]
- Hasdemir, D.B.; Dávila, L.A.; Schweitzer, N.; Meyer, B.C.; Koch, A.; Vogel, A.; Wacker, F.; Rodt, T. Evaluation of CT vascularization patterns for survival prognosis in patients with hepatocellular carcinoma treated by conventional TACE. Diagn. Interv. Radiol. 2017, 23, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Ji, G.W.; Zhu, F.P.; Xu, Q.; Wang, K.; Wu, M.Y.; Tang, W.W.; Li, X.C.; Wang, X.H. Radiomic Features at Contrast-enhanced CT Predict Recurrence in Early Stage Hepatocellular Carcinoma: A Multi-Institutional Study. Radiology 2020, 294, 568–579. [Google Scholar] [CrossRef]
- Kalkavan, H.; Sharma, P.; Kasper, S.; Helfrich, I.; Pandyra, A.A.; Gassa, A.; Virchow, I.; Flatz, L.; Brandenburg, T.; Namineni, S.; et al. Spatiotemporally restricted arenavirus replication induces immune surveillance and type I interferon-dependent tumour regression. Nat. Commun. 2017, 8, 14447. [Google Scholar] [CrossRef]
- Kany, S.; Vollrath, J.T.; Relja, B. Cytokines in Inflammatory Disease. Int. J. Mol. Sci. 2019, 20, 6008. [Google Scholar] [CrossRef]
- West, E.E.; Kolev, M.; Kemper, C. Complement and the Regulation of T Cell Responses. Annu. Rev. Immunol. 2018, 36, 309–338. [Google Scholar] [CrossRef]
- Li, J.; Stanger, B.Z. Cell Cycle Regulation Meets Tumor Immunosuppression. Trends Immunol. 2020, 41, 859–863. [Google Scholar] [CrossRef]
- Casey, S.C.; Baylot, V.; Felsher, D.W. The MYC oncogene is a global regulator of the immune response. Blood 2018, 131, 2007–2015. [Google Scholar] [CrossRef]
- Shalapour, S.; Karin, M. Pas de Deux: Control of Anti-tumor Immunity by Cancer-Associated Inflammation. Immunity 2019, 51, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Pittet, M.J.; Michielin, O.; Migliorini, D. Clinical relevance of tumour-associated macrophages. Nat. Rev. Clin. Oncol. 2022, 19, 402–421. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Allavena, P.; Marchesi, F.; Garlanda, C. Macrophages as tools and targets in cancer therapy. Nat. Rev. Drug Discov. 2022, 21, 799–820. [Google Scholar] [CrossRef] [PubMed]
- Kurebayashi, Y.; Ojima, H.; Tsujikawa, H.; Kubota, N.; Maehara, J.; Abe, Y.; Kitago, M.; Shinoda, M.; Kitagawa, Y.; Sakamoto, M. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology 2018, 68, 1025–1041. [Google Scholar] [CrossRef]
- Ming, W.; Zhu, Y.; Bai, Y.; Gu, W.; Li, F.; Hu, Z.; Xia, T.; Dai, Z.; Yu, X.; Li, H.; et al. Radiogenomics analysis reveals the associations of dynamic contrast-enhanced-MRI features with gene expression characteristics, PAM50 subtypes, and prognosis of breast cancer. Front. Oncol. 2022, 12, 943326. [Google Scholar] [CrossRef]
- Ming, W.; Li, F.; Zhu, Y.; Bai, Y.; Gu, W.; Liu, Y.; Liu, X.; Sun, X.; Liu, H. Unsupervised Analysis Based on DCE-MRI Radiomics Features Revealed Three Novel Breast Cancer Subtypes with Distinct Clinical Outcomes and Biological Characteristics. Cancers 2022, 14, 5507. [Google Scholar] [CrossRef]

| Characteristics | Discovery Cohort (n = 30) | Validation Cohort 1 TCGA-LIHC (n = 192) | Validation Cohort 2 LINC-JP (n = 142) | p |
|---|---|---|---|---|
| Age (year) | 66 (56, 68) | 59.5 (51, 69) | 69 (62, 75) | <0.001 |
| Gender | 0.649 | |||
| male | 20 | 139 | 97 | |
| female | 10 | 53 | 45 | |
| AFP (ng/mL) | 73.5 ± 168.2 | 186.6 ± 496.6 | NA | 0.765 |
| T stage | 0.920 | |||
| T1 | 23 | 148 | NA | |
| T2 | 7 | 43 | NA | |
| Tumor stage | <0.001 | |||
| stage I | 23 | 149 | 36 | |
| stage II | 7 | 43 | 106 | |
| Histology grade | 0.006 | |||
| G1 | 6 | 21 | 17 | |
| G2 | 14 | 93 | 83 | |
| G3 | 10 | 65 | 31 | |
| G4 | 0 | 11 | 0 | |
| Treatment methods | <0.001 | |||
| segmentectomy | 19 | 103 | 2 | |
| lobectomy | 8 | 70 | 0 | |
| extended lobectomy | 3 | 7 | 0 | |
| total hepatectomy with transplant | 0 | 1 | 0 | |
| TACE | 0 | 0 | 25 | |
| chemotherapy | 0 | 0 | 1 | |
| Follow-up duration (day) | 552.0 (383.5, 1459.0) | 631.5 (381.2, 1289.0) | 870.0 (570.5, 1132.5) | 0.899 |
| Feature | Feature Name | Subtype | Type | Phase | Region | Class | HR (95% CI) | p |
|---|---|---|---|---|---|---|---|---|
| PRF1 | Minimum | FIFS1 | Type2 | venous | tumor | first-order | 0.17 (0.05–0.55) | 0.008 |
| PRF2 | LargeAreaHighGrayLevelEmphasis | FIFS1 | Type3 | venous | margin | texture-glszm | 0.07 (0.02–0.24) | 0.001 |
| PRF3 | LargeDependenceLowGrayLevelEmphasis | FIFS1 | Type2 | venous | tumor | texture-gldm | 0.28 (0.08–0.92) | 0.043 |
| PRF4 | LowGrayLevelZoneEmphasis | FIFS1 | Type2 | venous | tumor | texture-glszm | 0.19 (0.06–0.61) | 0.016 |
| PRF5 | ShortRunHighGrayLevelEmphasis | FIFS1 | Type4 | artery | peritumor | texture-glrlm | 0.09 (0.03–0.28) | 0.003 |
| PRF6 | LowGrayLevelEmphasis | FIFS1 | Type2 | venous | tumor | texture-gldm | 0.20 (0.06–0.66) | 0.021 |
| PRF7 | LowGrayLevelRunEmphasis | FIFS1 | Type2 | venous | tumor | texture-glrlm | 0.19 (0.06–0.61) | 0.016 |
| PRF8 | SmallAreaLowGrayLevelEmphasis | FIFS1 | Type2 | venous | tumor | texture-glszm | 0.23 (0.07–0.76) | 0.039 |
| PRF9 | SurfaceVolumeRatio | FIFS1 | Type2 | venous | tumor | shape | 0.06 (0.02–0.22) | <0.001 |
| PRF10 | SurfaceVolumeRatio | FIFS1 | Type1 | artery | tumor | shape | 0.06 (0.02–0.21) | <0.001 |
| PRF11 | Idmn | FIFS2 | Type1 | artery | tumor | texture-glcm | 3.74 (1.13–12.38) | 0.035 |
| PRF12 | Idmn | FIFS2 | Type2 | venous | tumor | texture-glcm | 14.84 (4.45–49.53) | 0.001 |
| PRF13 | Idn | FIFS2 | Type2 | venous | tumor | texture-glcm | 5.34 (1.63–17.50) | 0.016 |
| PRF14 | LargeAreaHighGrayLevelEmphasis | FIFS2 | Type2 | venous | tumor | texture-glszm | 15.61 (4.65–52.40) | <0.001 |
| PRF15 | LargeDependenceHighGrayLevelEmphasis | FIFS2 | Type2 | venous | tumor | texture-gldm | 6.28 (1.89–20.82) | 0.007 |
| PRF16 | LongRunLowGrayLevelEmphasis | FIFS2 | Type4 | artery | peritumor | texture-glrlm | 4.88 (1.49–15.92) | 0.024 |
| PRF17 | DifferenceVariance | FIFS2 | Type3 | venous | peritumor | texture-glcm | 4.73 (1.45–15.41) | 0.028 |
| PRF18 | Contrast | FIFS2 | Type3 | venous | peritumor | texture-glcm | 5.34 (1.63–17.48) | 0.016 |
| PRF-Related Gene | Imaging Feature | Subtype Specific | Pathway | Module | Correlation Coefficient | p |
|---|---|---|---|---|---|---|
| IRS1 | PRF1 | FIFS1 | Positive Regulation of Cellular Carbohydrate Metabolic Process | Darkorange | 0.534 | 0.003 |
| TBX21 | PRF2 | FIFS1 | T Cell Differentiation Involved in Immune Response | Green | 0.592 | 0.001 |
| CCR7 | PRF3 | FIFS1 | Regulation of JNK Cascade | Green | 0.679 | <0.001 |
| SLAMF6 | PRF4 | FIFS1 | CD4 Positive or CD8 Positive Alpha Beta T Cell Lineage Commitment | Green | 0.611 | 0.001 |
| IL6ST | PRF5 | FIFS1 | JAK-SKAT Signaling Pathway | Green | 0.556 | 0.002 |
| SLAMF6 | PRF6 | FIFS1 | T Helper 17 Cell Differentiation | Green | 0.595 | 0.001 |
| NFIL3 | PRF7 | FIFS1 | Natural Killer Cell Differentiation | Green | 0.577 | 0.001 |
| SPN | PRF8 | FIFS1 | CD4 Positive or CD8 Positive Alpha Beta T Cell Lineage Commitment | Green | 0.518 | 0.004 |
| PRKCQ | PRF9 | FIFS1 | Positive Regulation of Interleukin 17 Production | Green | 0.581 | 0.001 |
| LY9 | PRF10 | FIFS1 | Positive Regulation of Interleukin 17 Production | Green | 0.54 | 0.003 |
| CDC26 | PRF11 | FIFS2 | Anaphase Promoting Complex | Yellow | 0.578 | 0.001 |
| UBE2S | PRF12 | FIFS2 | Regulation of Ubiquitin Protein Ligase Activity | Yellow | 0.662 | <0.001 |
| BAG2 | PRF13 | FIFS2 | Regulation of Ubiquitin Protein Ligase Activity | Yellow | 0.601 | 0.001 |
| UBE2S | PRF14 | FIFS2 | Positive Regulation of Ubiquitin Protein Transferase Activity | Yellow | 0.527 | 0.004 |
| UBE2S | PRF15 | FIFS2 | Regulation of Ubiquitin Protein Ligase Activity | Yellow | 0.623 | <0.001 |
| MAD2L1BP | PRF16 | FIFS2 | Regulation of Mitotic Cell Cycle Spindle Assembly Checkpoint | Yellow | 0.539 | 0.003 |
| SIRT2 | PRF17 | FIFS2 | Positive Regulation of Meiotic Cell Cycle | Yellow | 0.486 | 0.008 |
| SIRT2 | PRF18 | FIFS2 | Regulation of Meiotic Nuclear Division | Yellow | 0.584 | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, Y.; Huang, H.; Tong, Q.; Cao, M.; Ming, W.; Zhang, R.; Zhu, W.; Wang, Y.; Sun, X. Multi-View Radiomics Feature Fusion Reveals Distinct Immuno-Oncological Characteristics and Clinical Prognoses in Hepatocellular Carcinoma. Cancers 2023, 15, 2338. https://doi.org/10.3390/cancers15082338
Gu Y, Huang H, Tong Q, Cao M, Ming W, Zhang R, Zhu W, Wang Y, Sun X. Multi-View Radiomics Feature Fusion Reveals Distinct Immuno-Oncological Characteristics and Clinical Prognoses in Hepatocellular Carcinoma. Cancers. 2023; 15(8):2338. https://doi.org/10.3390/cancers15082338
Chicago/Turabian StyleGu, Yu, Hao Huang, Qi Tong, Meng Cao, Wenlong Ming, Rongxin Zhang, Wenyong Zhu, Yuqi Wang, and Xiao Sun. 2023. "Multi-View Radiomics Feature Fusion Reveals Distinct Immuno-Oncological Characteristics and Clinical Prognoses in Hepatocellular Carcinoma" Cancers 15, no. 8: 2338. https://doi.org/10.3390/cancers15082338
APA StyleGu, Y., Huang, H., Tong, Q., Cao, M., Ming, W., Zhang, R., Zhu, W., Wang, Y., & Sun, X. (2023). Multi-View Radiomics Feature Fusion Reveals Distinct Immuno-Oncological Characteristics and Clinical Prognoses in Hepatocellular Carcinoma. Cancers, 15(8), 2338. https://doi.org/10.3390/cancers15082338





