Platelet-Based Liquid Biopsies through the Lens of Machine Learning
Abstract
Simple Summary
Abstract
1. Introduction
- We present a comparative study of various machine learning algorithms (convolutional neural networks (CNNs), boosting) on the task of liquid biopsy cancer classification using a recently introduced novel feature vector extraction.
- We show that using knowledge from the KEGG database works as efficient feature preselection.
- We study the robustness of the presented algorithms when presented with the samples collected at hospital locations that were not used in the training process.
- We identify the most important features for classification.
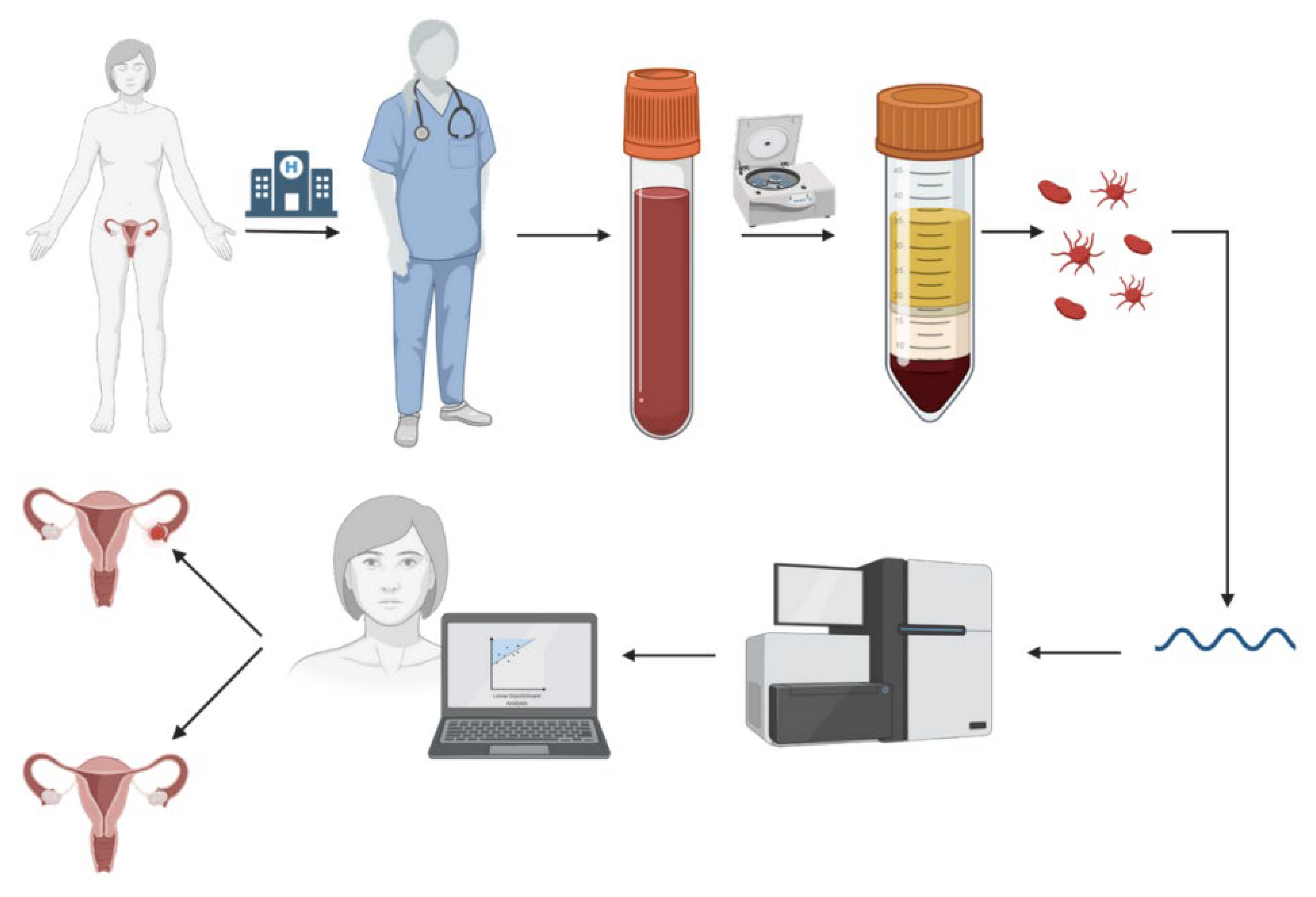
2. Data
2.1. Datasets
2.2. Data Preparation
2.3. Dataset Split by Location
3. Models
3.1. imPlatelet
3.2. Standard CNN
3.3. Gradient Boosting
4. Experiments
4.1. Model Comparison
4.1.1. Use of KEGG Expert Knowledge
4.1.2. Robustness Test
4.2. Feature Importance
4.3. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alabi, R.O.; Hietanen, P.; Elmusrati, M.; Youssef, O.; Almangush, A.; Mäkitie, A.A. Mitigating burnout in an oncological unit: A scoping review. Front. Public Health 2021, 9, 677915. [Google Scholar] [CrossRef] [PubMed]
- Vobugari, N.; Raja, V.; Sethi, U.; Gandhi, K.; Raja, K.; Surani, S.R. Advancements in oncology with artificial intelligence—A review article. Cancers 2022, 14, 1349. [Google Scholar] [CrossRef] [PubMed]
- Pastuszak, K.; Supernat, A.; Best, M.G.; In’t Veld, S.G.; Łapińska-Szumczyk, S.; Łojkowska, A.; Różański, R.; Żaczek, A.J.; Jassem, J.; Würdinger, T.; et al. imPlatelet classifier: Image converted RNA biomarker profiles enable blood based cancer diagnostics. Mol. Oncol. 2021, 15, 2688–2701. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, R.; Chiu, Y.-C.; Zhang, S.; Ramirez, J.; Chen, Y.; Huang, Y.; Jin, Y.-F. Prediction and interpretation of cancer survival using graph convolution neural networks. Methods 2021, 192, 120–130. [Google Scholar] [CrossRef]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2018, 20, 71–88. [Google Scholar] [CrossRef]
- Mader, S.; Pantel, K. Liquid Biopsy: Current Status and Future Perspectives. Oncol. Res. Treat. 2017, 40, 404–408. [Google Scholar] [CrossRef]
- Alix-Panabières, C. The future of liquid biopsy. Nature 2020, 579, S9. [Google Scholar] [CrossRef]
- Sol, N.; Leurs, C.E.; In’t Veld, S.G.; Strijbis, E.M.; Vancura, A.; Schweiger, M.W.; Teunissen, C.E.; Mateen, F.J.; Tannous, B.A.; Best, M.G.; et al. Blood platelet RNA enables the detection of multiple sclerosis. Mult. Scler. J. Exp. Transl. Clin. 2020, 6, 205521732094678. [Google Scholar] [CrossRef]
- Cygert, S.; Górski, F.; Juszczyk, P.; Lewalski, S.; Pastuszak, K.; Czyżewski, A.; Supernat, A. Towards cancer patients classification using liquid biopsy. In Proceedings of the Predictive Intelligence in Medicine: 4th International Workshop, PRIME 2021, Held in Conjunction with MICCAI 2021, Strasbourg, France, 1 October 2021; Springer International Publishing: Cham, Switzerland, 2021; pp. 221–230. [Google Scholar] [CrossRef]
- Gerratana, L.; Pierga, J.-Y.; Reuben, J.M.; Davis, A.A.; Wehbe, F.H.; Dirix, L.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; et al. Modeling the prognostic impact of circulating tumor cells enumeration in metastatic breast cancer for clinical trial design simulation. Oncologist 2022, 27, e561–e570. [Google Scholar] [CrossRef]
- Da Col, G.; Del Ben, F.; Bulfoni, M.; Turetta, M.; Gerratana, L.; Bertozzi, S.; Beltrami, A.P.; Cesselli, D. Image analysis of circulating tumor cells and leukocytes predicts survival and metastatic pattern in breast cancer patients. Front. Oncol. 2022, 12, 217. [Google Scholar] [CrossRef]
- Suzuki, K.; Igata, H.; Abe, M.; Yamamoto, Y.; Iwanaga, T.; Kanzaki, H.; Kato, N.; Tanaka, N.; Kawasaki, K.; Matsushita, K.; et al. Multiple cancer type classification by small RNA expression profiles with plasma samples from multiple facilities. Cancer Sci. 2022, 113, 2144–2166. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhao, J.; Wang, X.; Yan, S.; Chu, H.; Gao, M.; Zhang, X. Integrated pipeline of rapid isolation and analysis of human plasma exosomes for cancer discrimination based on deep learning of MALDI-TOF MS fingerprints. Anal. Chem. 2022, 94, 1831–1839. [Google Scholar] [CrossRef]
- Best, M.G.; Sol, N.; Kooi, I.; Tannous, J.; Westerman, B.A.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Koster, J.; et al. RNA-seq of tumor-educated platelets enables blood-based pancancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell 2015, 28, 666–676. [Google Scholar] [CrossRef] [PubMed]
- Best, M.G.; Sol, N.; In’t Veld, S.G.; Vancura, A.; Muller, M.; Niemeijer, A.-L.N.; Fejes, A.V.; Fat, L.-A.T.K.; In’t Veld, A.E.H.; Leurs, C.; et al. Swarm intelligence-enhanced detection of non-small-cell lung cancer using tumor-educated platelets. Cancer Cell 2017, 32, 238–252.e9. [Google Scholar] [CrossRef]
- Best, M.G.; Sol, N.; Tannous, B.A.; Wesseling, P.; Wurdinger, T. Re: A word of caution on new and revolutionary diagnostic tests. Cancer Cell 2016, 29, 143–144. [Google Scholar] [CrossRef]
- Zech, J.R.; Badgeley, M.A.; Liu, M.; Costa, A.B.; Titano, J.J.; Oermann, E.K. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS Med. 2018, 15, e1002683. [Google Scholar] [CrossRef]
- Esteva, A.; Chou, K.; Yeung, S.; Naik, N.; Madani, A.; Mottaghi, A.; Liu, Y.; Topol, E.; Dean, J.; Socher, R. Deep learning-enabled medical computer vision. npj Digit. Med. 2021, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Geirhos, R.; Jacobsen, J.-H.; Michaelis, C.; Zemel, R.; Brendel, W.; Bethge, M.; Wichmann, F.A. Shortcut learning in deep neural networks. Nat. Mach. Intell. 2020, 2, 665–673. [Google Scholar] [CrossRef]
- Supernat, A.; Popęda, M.; Pastuszak, K.; Best, M.G.; Grešner, P.; In’t Veld, S.; Siek, B.; Bednarz-Knoll, N.; Rondina, M.T.; Stokowy, T.; et al. Transcriptomic landscape of blood platelets in healthy donors. Sci. Rep. 2021, 11, 15679. [Google Scholar] [CrossRef]
- Łukasiewicz, M.; Pastuszak, K.; Łapinska-Szumczyk, S.; Rózanski, R.; In’t Veld, S.; Bieńkowski, M.; Stokowy, T.; Ratajska, M.; Best, M.G.; Würdinger, T.; et al. Diagnostic accuracy of liquid biopsy in endometrial cancer. Cancers 2021, 13, 5731. [Google Scholar] [CrossRef]
- Zhao, X.-M.; Wu, F.-X. Deep networks and network representation in bioinformatics. Methods 2021, 192, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Best, M.G.; In’t Veld, S.G.J.G.; Sol, N.; Wurdinger, T. RNA sequencing and swarm intelligence–enhanced classification algorithm development for blood-based disease diagnostics using spliced blood platelet RNA. Nat. Protoc. 2019, 14, 1206–1234. [Google Scholar] [CrossRef]
- Sol, N.; In’t Veld, S.G.; Vancura, A.; Tjerkstra, M.; Leurs, C.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Zwaan, K.; et al. Tumor-educated platelet RNA for the detection and (pseudo)progression monitoring of glioblastoma. Cell Rep. Med. 2020, 1, 100101. [Google Scholar] [CrossRef] [PubMed]
- Heinhuis, K.M.; In’t Veld, S.G.J.G.; Dwarshuis, G.; van den Broek, D.; Sol, N.; Best, M.G.; Van Coevorden, F.; Haas, R.L.; Beijnen, J.H.; Van Houdt, W.J.; et al. RNA-Sequencing of Tumor-Educated Platelets, a Novel Biomarker for Blood-Based Sarcoma Diagnostics. Cancers 2020, 12, 1372. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Huber, W.; von Heydebreck, A.; Sültmann, H.; Poustka, A.; Vingron, M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 2002, 18, S96–S104. [Google Scholar] [CrossRef]
- Frankish, A.; Diekhans, M.; Ferreira, A.-M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Luo, W.; Friedman, M.S.; Shedden, K.; Hankenson, K.D.; Woolf, P.J. GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinform. 2009, 10, 161. [Google Scholar] [CrossRef]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. Tensorflow: A system for large-scale machine learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation, OSDI 2016, Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer VISION and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. Pytorch: An imperative style, high-performance deep learning library. In Proceedings of the Advances in Neural Information Processing Systems 32: Annual Conference on Neural Information Processing Systems 2019, NeurIPS 2019, Vancouver, BC, Canada, 8–14 December 2019; pp. 8024–8035. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Zhang, H.; Cissé, M.; Dauphin, Y.N.; Lopez-Paz, D. Mixup: Beyond empirical risk minimization. In Proceedings of the 6th International Conference on Learning Representations, ICLR, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Camilli, M.; Iannaccone, G.; La Vecchia, G.; Cappannoli, L.; Scacciavillani, R.; Minotti, G.; Massetti, M.; Crea, F.; Aspromonte, N. Platelets: The point of interconnection among cancer, inflammation and cardiovascular diseases. Expert Rev. Hematol. 2021, 14, 537–546. [Google Scholar] [CrossRef]
- Gnatenko, D.V.; Dunn, J.J.; McCorkle, S.R.; Weissmann, D.; Perrotta, P.L.; Bahou, W.F. Transcript profiling of human platelets using microarray and serial analysis of gene expression. Blood 2003, 101, 2285–2293. [Google Scholar] [CrossRef] [PubMed]
- Arias-Salgado, E.G.; Haj, F.; Dubois, C.; Moran, B.; Kasirer-Friede, A.; Furie, B.C.; Furie, B.; Neel, B.G.; Shattil, S.J. PTP-1B is an essential positive regulator of platelet integrin signaling. J. Cell Biol. 2005, 170, 837–845. [Google Scholar] [CrossRef]
- Xu, R.; Chen, L.; Wei, W.; Tang, Q.; Yu, Y.; Hu, Y.; Kadasah, S.; Xie, J.; Yu, H. Single-Cell Sequencing Analysis Based on Public Databases for Constructing a Metastasis-Related Prognostic Model for Gastric Cancer. Appl. Bionics Biomech. 2022, 2022, 7061263. [Google Scholar] [CrossRef]
- Ren, H.; Cao, K.; Wang, M. A Correlation Between Differentiation Phenotypes of Infused T Cells and Anti-Cancer Immunotherapy. Front. Immunol. 2021, 12, 745109. [Google Scholar] [CrossRef] [PubMed]
- Best, M.G.; Vancura, A.; Wurdinger, T. Platelet RNA as a circulating biomarker trove for cancer diagnostics. J. Thromb. Haemost. 2017, 15, 1295–1306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tseng, J.T.-C.; Lien, I.-C.; Li, F.; Wu, W.; Li, H. mRNAsi Index: Machine Learning in Mining Lung Adenocarcinoma Stem Cell Biomarkers. Genes 2020, 11, 257. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Caselli, E.; Pelliccia, C.; Teti, V.; Bellezza, G.; Mandarano, M.; Ferri, I.; Hartmann, K.; Laible, M.; Sahin, U.; Varga, Z.; et al. Looking for more reliable biomarkers in breast cancer: Comparison between routine methods and RT-qPCR. PLoS ONE 2021, 16, e0255580. [Google Scholar] [CrossRef]
- In’t Veld, S.G.J.G.; Wurdinger, T. Tumor-educated platelets. Blood 2019, 133, 2359–2364. [Google Scholar] [CrossRef]



| - | EC | OC | NSCLC | GBM | Brain Metastasis | Sarcoma | Asymptomatic Controls | Multiple Sclerosis |
|---|---|---|---|---|---|---|---|---|
| Num patients | 39 | 28 | 329 | 234 | 51 | 39 | 338 | 84 |
| - | EC | OC | NSCLC | GBM | Brain Metastasis | Sarcoma | Asymptomatic Controls | Multiple Sclerosis |
|---|---|---|---|---|---|---|---|---|
| Training | 39 | 28 | 142 | 215 | 25 | Not included | 260 | 65 |
| Test | 0 | 0 | 185 | 4 | 26 | Not included | 54 | 19 |
| Total | 39 | 28 | 327 | 219 | 51 | Not included | 314 | 84 |
| Model | Val Bal. Acc. | Test Bal. Acc. | Val AUC | Test AUC |
|---|---|---|---|---|
| imPlatelet | 0.902 | 0.891 | 0.970 | 0.966 |
| ResNet-18 | 0.898 | 0.883 | 0.957 | 0.950 |
| Boosting | 0.907 | 0.889 | 0.962 | 0.960 |
| Model | Val AUC | Test AUC |
|---|---|---|
| ResNet-18 | 0.957 | 0.950 |
| Permuted rows | 0.959 | 0.951 |
| Permuted columns | 0.955 | 0.945 |
| Model | Val Bal. Acc | Test Bal. Acc | Val AUC | Test AUC |
|---|---|---|---|---|
| imPlatelet | 0.898 | 0.854 | 0.970 | 0.966 |
| ResNet-18 | 0.913 | 0.857 | 0.965 | 0.958 |
| Boosting | 0.909 | 0.878 | 0.967 | 0.953 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cygert, S.; Pastuszak, K.; Górski, F.; Sieczczyński, M.; Juszczyk, P.; Rutkowski, A.; Lewalski, S.; Różański, R.; Jopek, M.A.; Jassem, J.; et al. Platelet-Based Liquid Biopsies through the Lens of Machine Learning. Cancers 2023, 15, 2336. https://doi.org/10.3390/cancers15082336
Cygert S, Pastuszak K, Górski F, Sieczczyński M, Juszczyk P, Rutkowski A, Lewalski S, Różański R, Jopek MA, Jassem J, et al. Platelet-Based Liquid Biopsies through the Lens of Machine Learning. Cancers. 2023; 15(8):2336. https://doi.org/10.3390/cancers15082336
Chicago/Turabian StyleCygert, Sebastian, Krzysztof Pastuszak, Franciszek Górski, Michał Sieczczyński, Piotr Juszczyk, Antoni Rutkowski, Sebastian Lewalski, Robert Różański, Maksym Albin Jopek, Jacek Jassem, and et al. 2023. "Platelet-Based Liquid Biopsies through the Lens of Machine Learning" Cancers 15, no. 8: 2336. https://doi.org/10.3390/cancers15082336
APA StyleCygert, S., Pastuszak, K., Górski, F., Sieczczyński, M., Juszczyk, P., Rutkowski, A., Lewalski, S., Różański, R., Jopek, M. A., Jassem, J., Czyżewski, A., Wurdinger, T., Best, M. G., Żaczek, A. J., & Supernat, A. (2023). Platelet-Based Liquid Biopsies through the Lens of Machine Learning. Cancers, 15(8), 2336. https://doi.org/10.3390/cancers15082336






