DSCC_Net: Multi-Classification Deep Learning Models for Diagnosing of Skin Cancer Using Dermoscopic Images
Abstract
Simple Summary
Abstract
1. Introduction
- The novel proposed DSCC_Net model is designed to identify four different types of skin cancer. The proposed model has the capability of extracting dominant features from dermoscopy images that can assist in the accurate identification of the disease.
- In this study, we reduce the complexity of the model by decreasing the number of trainable parameters to obtain a significant classifier.
- The CNN model’s accuracy is compromised as a result of the problem of class imbalance in medical datasets. We overcome this issue by using an up-sampling technique, SMOTE Tomek, to obtain concoction samples of the image at each class to gain enhanced accuracy.
- The Grad-CAM heat-map technique is utilized to illustrate the visible features of skin cancer disease classification approaches.
- The proposed model achieved superior results, as compared to six baseline classifiers, Vgg-19, ResNet-152, Vgg-16, MobileNet, Inception-V3, and EfficientNet-B0, in terms of many evaluation metrics, i.e., accuracy, area under the curve (AUC), precision, recall, loss, and F1 score.
- Additionally, the proposed model also produced significant results as compared to the recent state-of-the-art classifiers.
2. Literature Review
3. Materials and Methods
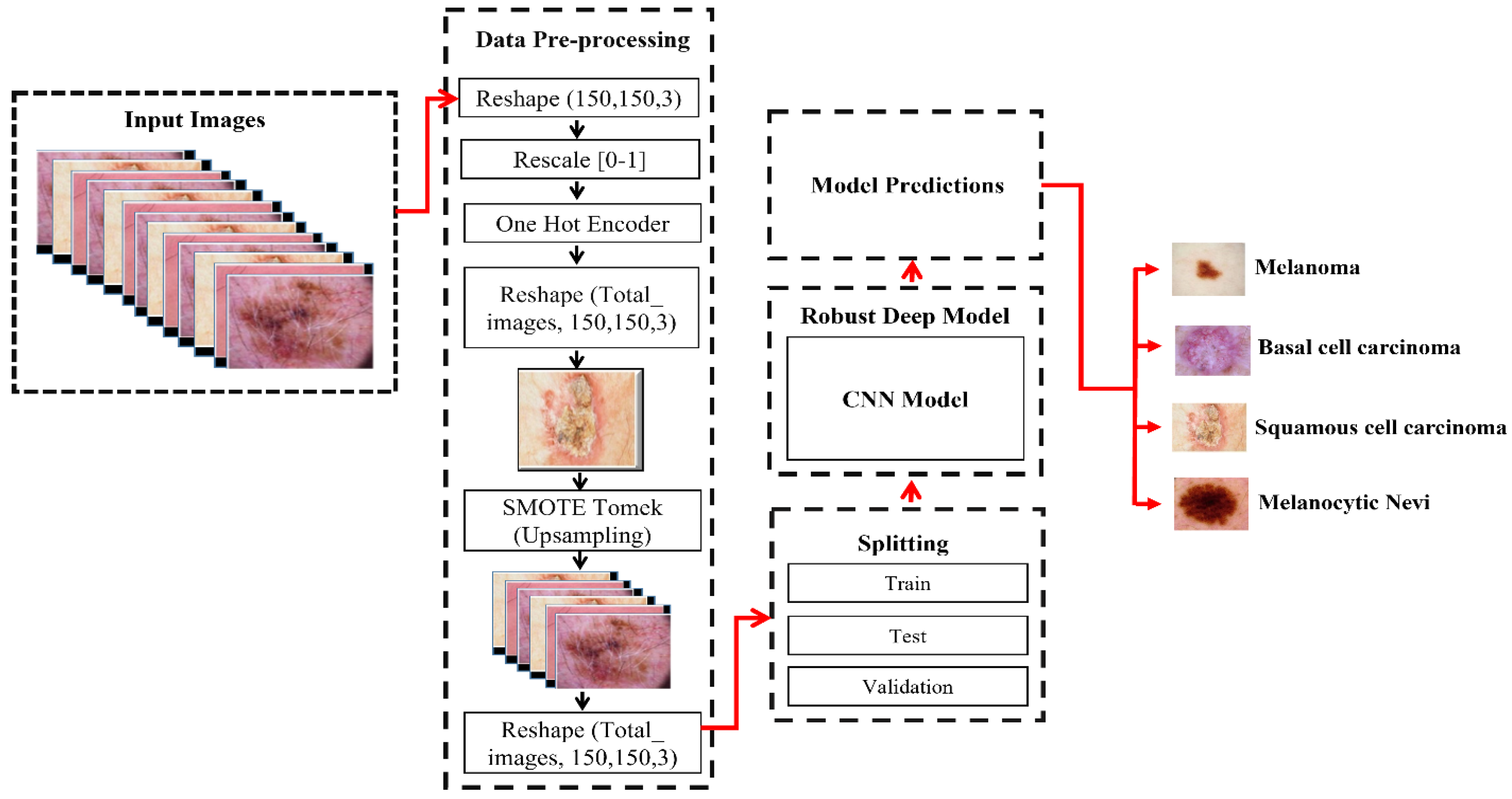
3.1. Proposed Study Flow for the Diagnosis of Skin Cancer
3.2. Dataset Description
3.3. Using SMOTE Tomek to Balance Dataset
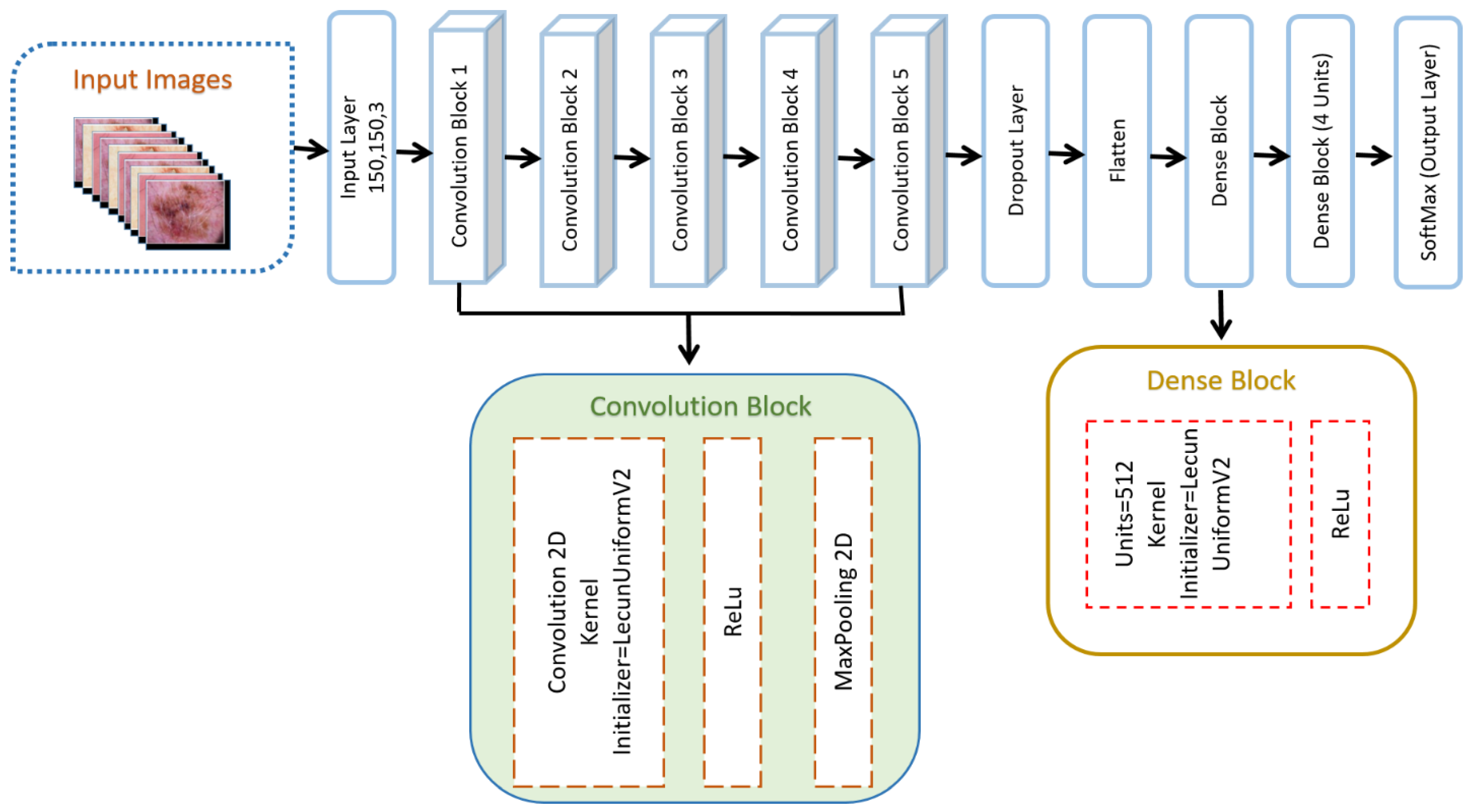
3.4. Proposed Model
3.4.1. Structure of the Proposed DSCC_Net
3.4.2. Convolutional Blocks of CNN Model
3.4.3. Flattened Layer
3.4.4. Dropout Layer
3.4.5. Dense Block of Proposed DSCC_Net
- ReLU Function
- Dense Layer
3.5. Model Evaluations
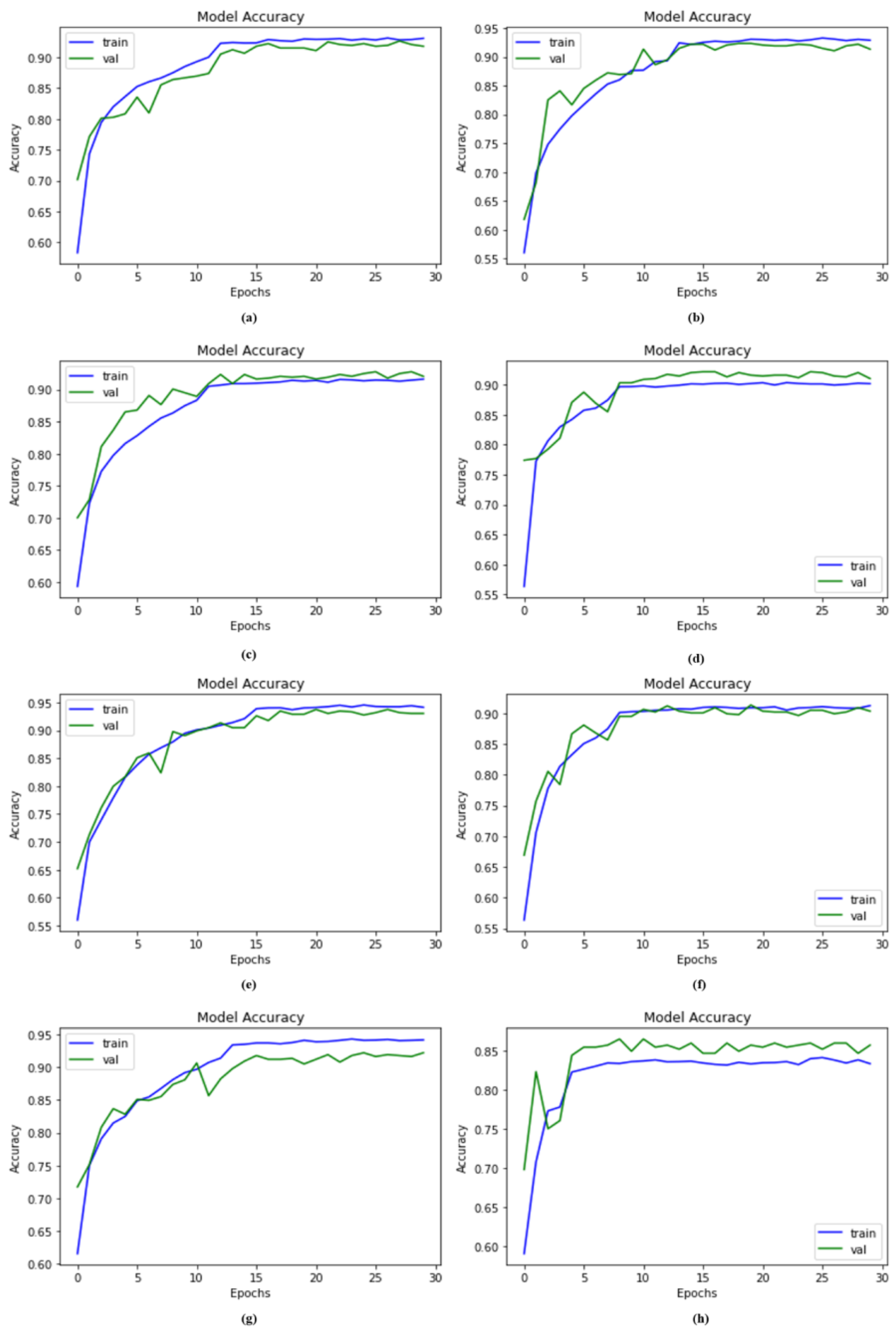
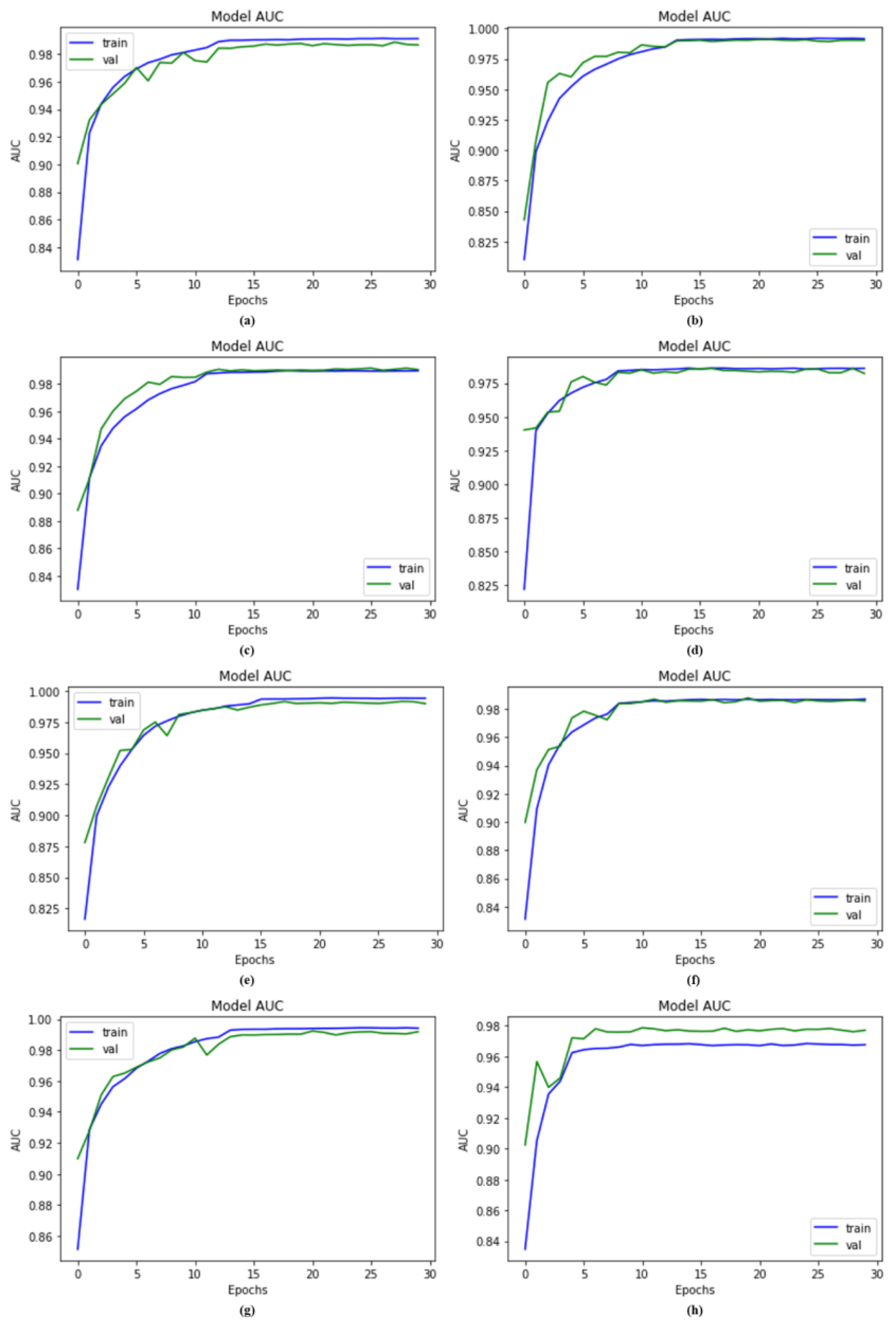
4. Results and Discussion
4.1. Experimental Setup
4.2. Accuracy Compared with Other Models
4.3. AUC Comparison with Other Models
4.4. Compared with Other Models Using Precision
4.5. Compared of DSCC_Net against Other Models Using Recall
4.6. F1-Score Comparison with Recent Deep Model
4.7. Comparison of Proposed Model with Other Models Using Loss
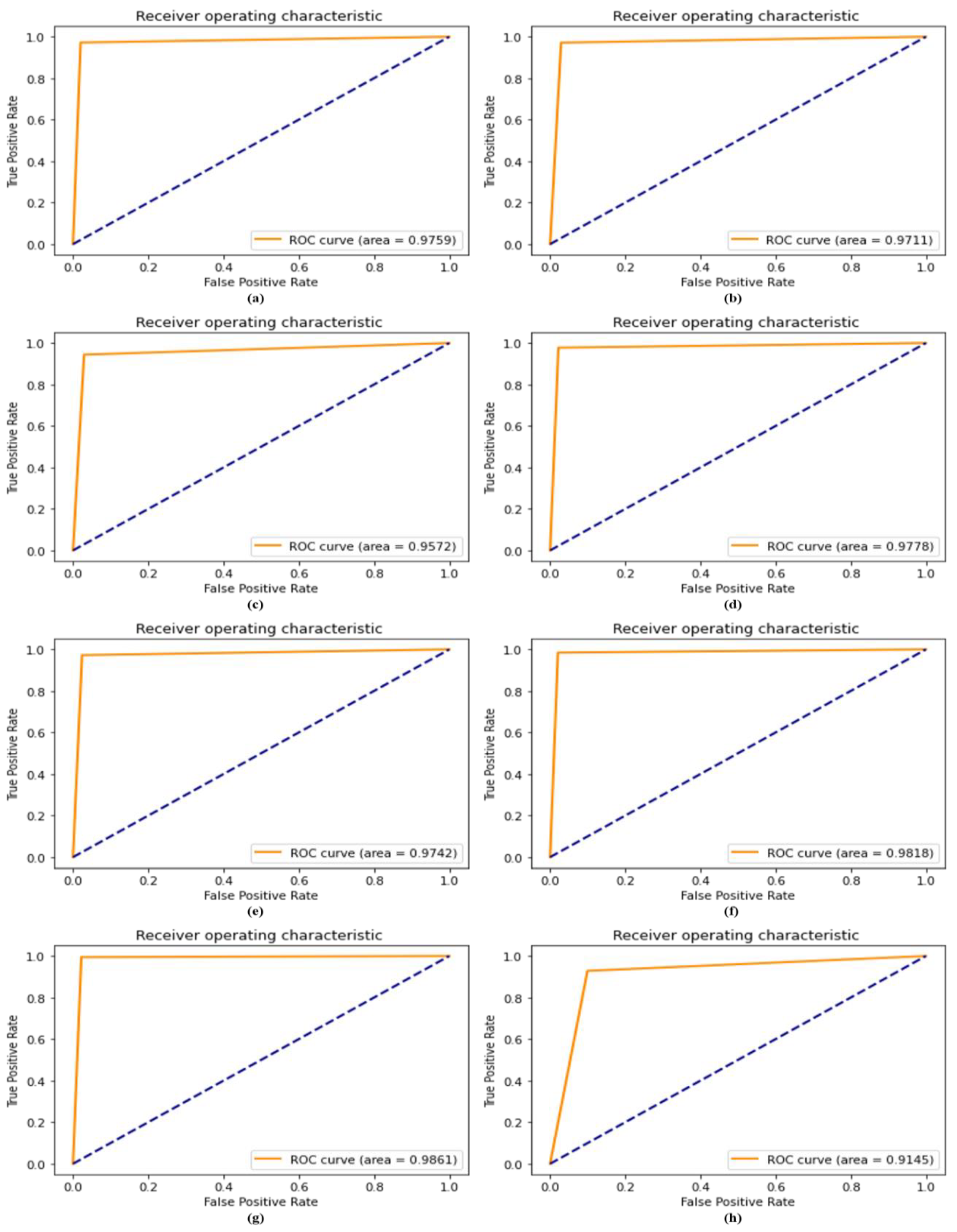
4.8. ROC Compared with Recent Model
4.9. AU(ROC) Extension for Multi-Class Comparison against Recent Models
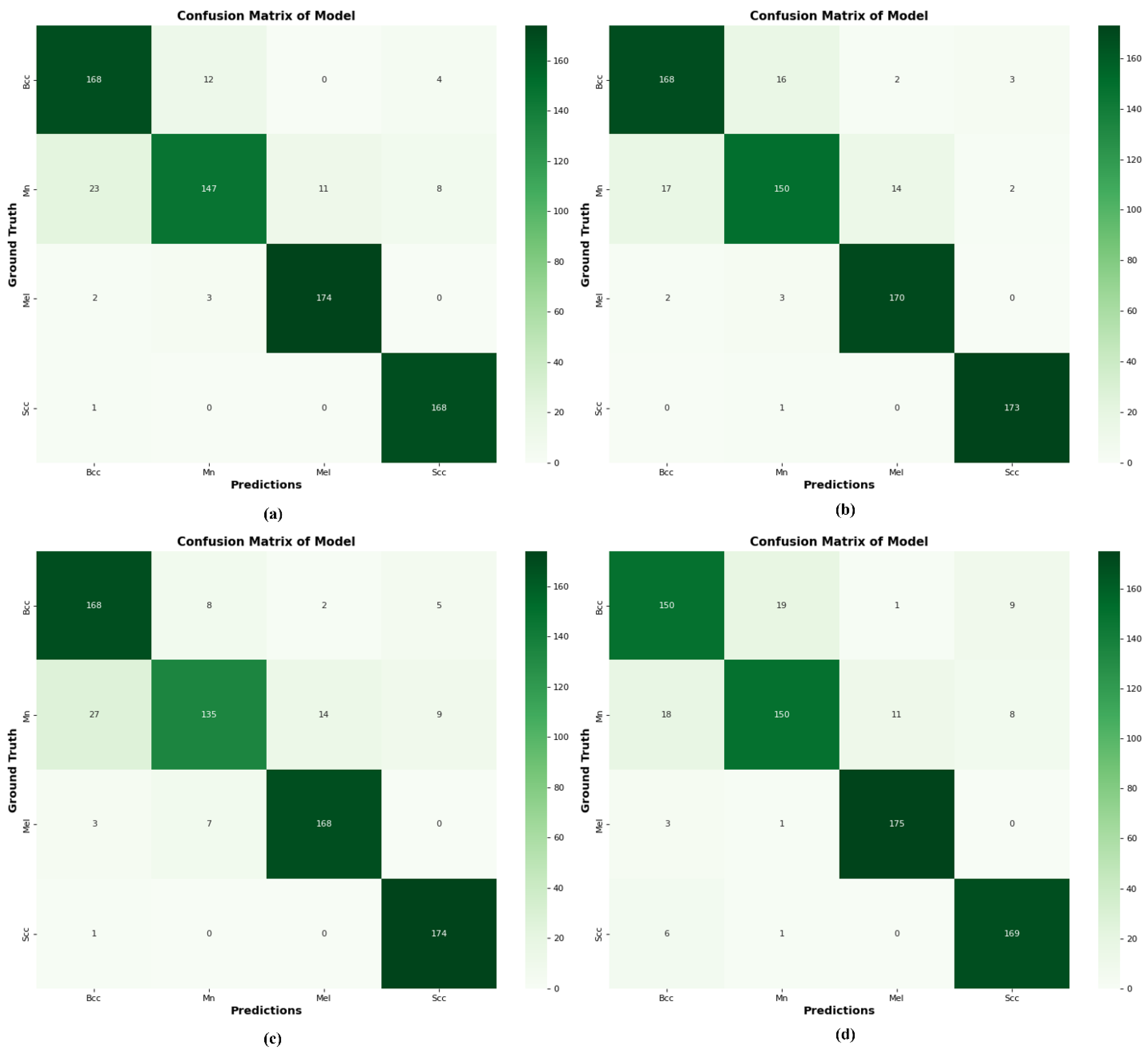
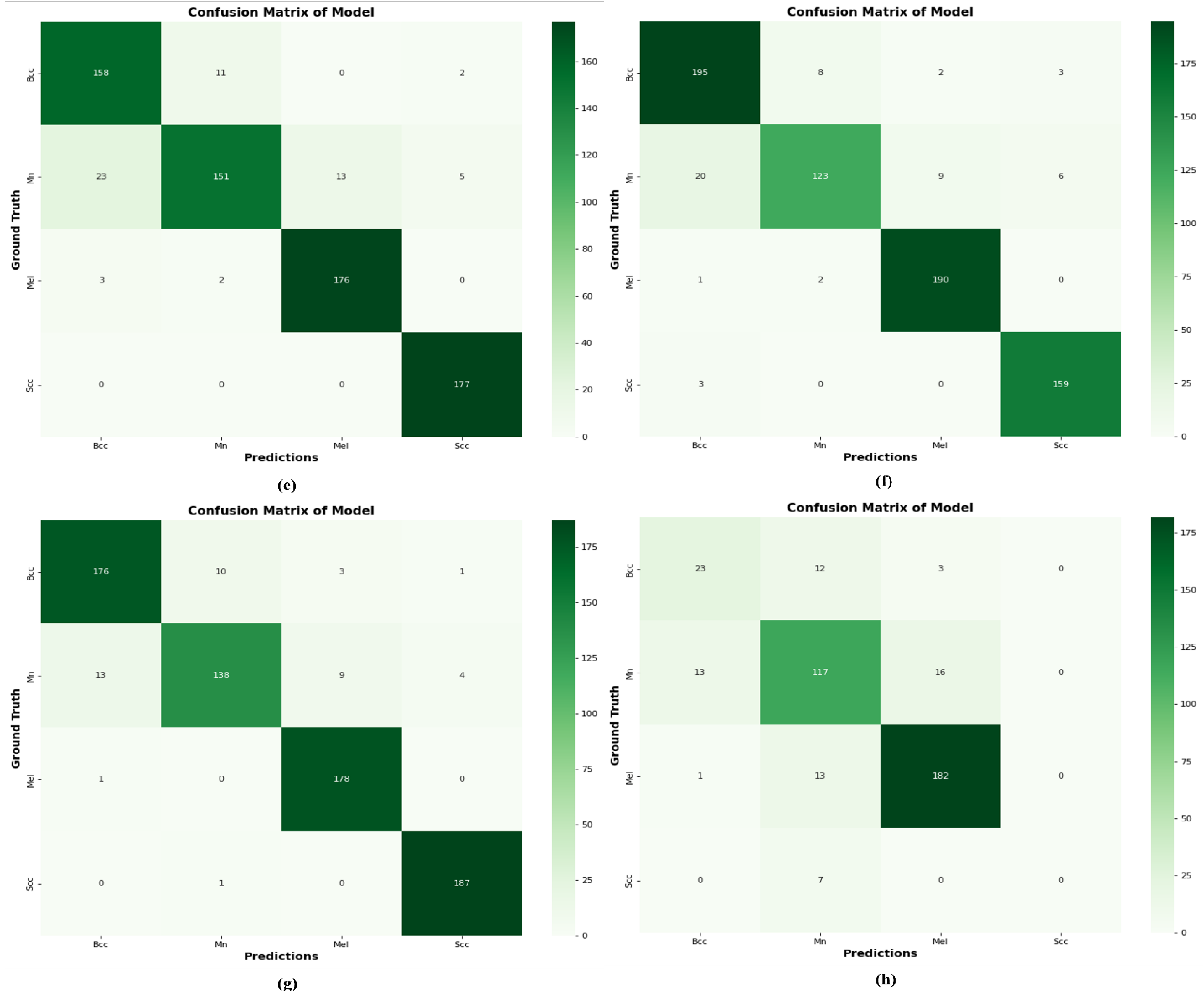
4.10. Comparison of DSCC_Net with Six Models Using a Confusion Matrix
4.11. Comparison of the Proposed Model with State-Of-The-Art
4.12. Discussions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization Radiation: Ultraviolet (UV) Radiation and Skin Cancer|How Common Is Skin Cancer. Available online: https://www.who.int/news-room/q-a-detail/radiation-ultraviolet-(uv)-radiation-and-skin-cancer# (accessed on 2 March 2023).
- Piccialli, F.; Di Somma, V.; Giampaolo, F.; Cuomo, S.; Fortino, G. A survey on deep learning in medicine: Why, how and when? Inf. Fusion 2021, 66, 111–137. [Google Scholar] [CrossRef]
- Navid, R.; Ashourian, M.; Karimifard, M.; Estrela, V.V.; Loschi, H.J.; Nascimento, D.D.; França, R.P.; Vishnevski, M. Computer-aided diagnosis of skin cancer: A review. Curr. Med. Imaging 2020, 16, 781–793. [Google Scholar]
- Ahmad, N.; Farooq, M.S.; Khelifi, A.; Abid, A. Malignant melanoma classification using deep learning: Datasets, performance measurements, challenges and opportunities. IEEE Access 2020, 8, 110575–110597. [Google Scholar]
- O’Sullivan, D.E.; Brenner, D.R.; Demers, P.A.; Villeneuve, P.J.; Friedenreich, C.M.; King, W.D. Indoor tanning and skin cancer in Canada: A meta-analysis and attributable burden estimation. Cancer Epidemiol. 2019, 59, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Walters-Davies, R. Skin cancer: Types, diagnosis and prevention. Evaluation 2020, 14, 34. [Google Scholar]
- Hodis, E. The Somatic Genetics of Human Melanoma. Ph.D. Thesis, Harvard University, Cambridge, MA, USA, 2018. [Google Scholar]
- Nathan, N.; Hubbard, M.; Nordmann, T.; Sperduto, P.W.; Clark, H.B.; Hunt, M.A. Effect of gamma knife radiosurgery and programmed cell death 1 receptor antagonists on metastatic melanoma. Cureus 2017, 9, e1943. [Google Scholar]
- Ahmad, N.; Anees, T.; Naqvi, R.A.; Loh, W.-K. A comprehensive analysis of recent deep and federated-learning-based methodologies for brain tumor diagnosis. J. Pers. Med. 2022, 12, 275. [Google Scholar]
- Rogers, H.W.; Weinstock, M.A.S.; Feldman, R.; Coldiron, B.M. Incidence estimate of non-melanoma skin cancer (keratinocyte carcinomas) in the US population 2012. JAMA Dermatol. 2015, 151, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Bomm, L.; Benez, M.D.V.; Maceira, J.M.P.; Succi, I.C.B.; Scotelaro, M.D.F.G. Biopsy guided by dermoscopy in cutaneous pigmented lesion-case report. An. Bras. Dermatol. 2013, 88, 125–127. [Google Scholar] [CrossRef] [PubMed]
- Kato, J.; Horimoto, K.; Sato, S.; Minowa, T.; Uhara, H. Dermoscopy of melanoma and non-melanoma skin cancers. Front. Med. 2019, 6, 180. [Google Scholar] [CrossRef]
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hadj, H.A.B.; Thomas, L.; Enk, A.; et al. Reader study level-I and level-II Groups, Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef]
- Ibrahim, H.; El-Taieb, M.; Ahmed, A.; Hamada, R.; Nada, E. Dermoscopy versus skin biopsy in diagnosis of suspicious skin lesions. Al-Azhar Assiut Med. J. 2017, 15, 203. [Google Scholar] [CrossRef]
- Duggani, K.; Nath, M.K. A technical review report on deep learning approach for skin cancer detection and segmentation. Data Anal. Manag. Proc. ICDAM 2021, 54, 87–99. [Google Scholar]
- Carli, P.; Quercioli, E.; Sestini, S.; Stante, M.; Ricci, L.; Brunasso, G.; DeGiorgi, V. Patternanalysis, notsimplifiedalgorithms, isthe most reliable method for teaching dermoscopy for melanoma diagnosis to residents in dermatology. Br. J. Dermatol. 2003, 148, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Carrera, C.; Marchetti, M.A.; Dusza, S.W.; Argenziano, G.; Braun, R.P.; Halpern, A.C.; Jaimes, N.; Kittler, H.J.; Malvehy, J.; Menzies, S.W.; et al. Validity and reliability of dermoscopic criteria used to differentiate nevi from melanoma: A web-based international dermoscopy society study. JAMA Dermatol. 2016, 152, 798–806. [Google Scholar] [CrossRef] [PubMed]
- Celebi, M.E.; Kingravi, H.A.; Uddin, B.; Iyatomi, H.; Aslandogan, Y.A.; Stoecker, W.V.; Moss, R.H. A methodological approach to the classification of dermoscopy images. Comput. Med. Imaging Graph. 2007, 31, 362–373. [Google Scholar] [CrossRef]
- Maglogiannis, I.; Doukas, C.N. Overview of advanced computer vision systems for skin lesions characterization. IEEE Trans. Inf. Technol. Biomed. 2009, 13, 721–733. [Google Scholar] [CrossRef]
- Celebi, M.E.; Iyatomi, H.; Stoecker, W.V.; Moss, R.H.; Rabinovitz, H.S.; Argenziano, G.; Soyer, H.P. Automatic detection of blue-white veil and related structures in dermoscopy images. Comput. Med. Imaging Graph. 2008, 32, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Hassaan, M.; Anees, T.; Din, M.; Ahmad, N. CDC_Net: Multi-classification convolutional neural network model for detection of COVID-19, pneumothorax, pneumonia, lung Cancer, and tuberculosis using chest X-rays. Multimed. Tools Appl. 2022, 82, 13855–13880. [Google Scholar]
- Lu, S.; Lu, Z.; Zhang, Y.D. Pathological brain detection based on AlexNet and transfer learning. J. Comput. Sci. 2019, 30, 41–47. [Google Scholar] [CrossRef]
- Ahmad, N.; Anees, T.; Ahmed, K.T.; Naqvi, R.A.; Ahmad, S.; Whangbo, T. Deep learned vectors’ formation using auto-correlation, scaling, and derivations with CNN for complex and huge image retrieval. Complex Intell. Syst. 2022, 4, 1–23. [Google Scholar]
- Sajjad, M.; Khan, S.; Muhammad, K.; Wu, W.; Ullah, A.; Baik, S.W. Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J. Comput. Sci. 2019, 30, 174–182. [Google Scholar] [CrossRef]
- Alom, M.Z.; Aspiras, T.; Taha, T.M.; Asari, V.K. Skin cancer segmentation and classification with improved deep convolutional neural network. In Medical Imaging 2020: Imaging Informatics for Healthcare, Research, and Applications; International Society for Optics and Photonics: Bellingham, WA, USA, 2020; Volume 11318, p. 1131814. [Google Scholar]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Polat, K.; Koc, K.O. Detection of skin diseases from dermoscopy image using the combination of convolutional neural network and one-versus-all. J. Artif. Intell. Syst. 2020, 2, 80–97. [Google Scholar] [CrossRef]
- Ratul, M.A.R.; Mozaffari, M.H.; Lee, W.S.; Parimbelli, E. Skin lesions classification using deep learning based on dilated convolution. BioRxiv 2020, 860700. [Google Scholar] [CrossRef]
- Ranpreet, K.; GholamHosseini, H.; Sinha, R.; Lindén, M. Melanoma classification using a novel deep convolutional neural network with dermoscopic images. Sensors 2022, 22, 1134. [Google Scholar]
- Javed, R.; Ishfaq, M.; Ali, G.; Saeed, M.R.; Hussain, M.; Alkhalifah, T.; Alturise, F.; Samand, N. Skin cancer disease detection using transfer learning technique. Appl. Sci. 2022, 12, 5714. [Google Scholar]
- Shahin, A.; Miah, S.; Haque, J.; Rahman, M.; Islam, K. An enhanced technique of skin cancer classification using deep convolutional neural network with transfer learning models. Mach. Learn. Appl. 2021, 5, 100036. [Google Scholar]
- Tanzila, S.; Khan, M.A.; Rehman, A.; Marie-Sainte, S.L. Region extraction and classification of skin cancer: A heterogeneous framework of deep CNN features fusion and reduction. J. Med. Syst. 2019, 43, 289. [Google Scholar]
- Hiam, A.; Qasmieh, I.A.; Alqudah, A.M.; Alhammouri, S.; Alawneh, E.; Abughazaleh, A.; Hasayen, F. The melanoma skin cancer detection and classification using support vector machine. In Proceedings of the 2017 IEEE Jordan Conference on Applied Electrical Engineering and Computing Technologies (AEECT), Amman, Jordania, 11–13 October 2017; pp. 1–5. [Google Scholar]
- Hardik, N.; Singh, S.P. Deep learning solutions for skin cancer detection and diagnosis. Mach. Learn. Health Care Perspect. Mach. Learn. Healthc. 2020, 13, 159–182. [Google Scholar]
- Duggani, K.; Venugopal, V.; Nath, M.K.; Mishra, M. Hybrid convolutional neural networks with SVM classifier for classification of skin cancer. Biomed. Eng. Adv. 2023, 5, 100069. [Google Scholar]
- Gilani, Q.; Syed, S.T.; Umair, M.; Marques, O. Skin Cancer Classification Using Deep Spiking Neural Network. J. Digit. Imaging 2023, 1–11. [Google Scholar]
- Ioannis, K.; Perikos, I.; Hatzilygeroudis, I.; Virvou, M. Deep learning methods for accurate skin cancer recognition and mobile application. Electronics 2022, 11, 1294. [Google Scholar]
- Ghadah, A.; Gouda, W.; Humayun, M.; Sama, N.U. Melanoma Detection Using Deep Learning-Based Classifications. Healthcare 2022, 10, 2481. [Google Scholar]
- Khalil, A.; Turki, T. Automatic Classification of Melanoma Skin Cancer with Deep Convolutional Neural Networks. AI 2022, 3, 512–525. [Google Scholar]
- Karar, A.; Shaikh, Z.A.; Khan, A.A.; Laghari, A.A. Multiclass skin cancer classification using EfficientNets—A first step towards preventing skin cancer. Neurosci. Inform. 2022, 2, 100034. [Google Scholar] [CrossRef]
- Naseer, B.M.; Muta, K.; Malik, M.I.; Siddiqui, S.A.; Braun, S.A.; Homey, B.; Dengel, A.; Ahmed, S. Computer-aided diagnosis of skin diseases using deep neural networks. Appl. Sci. 2020, 10, 2488. [Google Scholar]
- Adi, N.A.; Slamet, I.S. Skins cancer identification system of HAMl0000 skin cancer dataset using convolutional neural network. AIP Conf. Proc. 2019, 2202, 020039. [Google Scholar]
- Moldovan, D. Transfer learning based method for two-step skin cancer images classification. In Proceedings of the 2019 E-Health and Bioengineering Conference (EHB), Iasi, Romania, 21–23 November 2019; pp. 1–4. [Google Scholar]
- Le Duyen, N.T.; Hieu, X.L.; Lua, T.N.; Hoan, T.N. Transfer learning with class-weighted and focal loss function for automatic skin cancer classification. arXiv 2020, arXiv:2009.05977. [Google Scholar]
- Saksham, B.; Gomekar, A. Deep learning diagnosis of pigmented skin lesions. In Proceedings of the 2019 10th International Conference on Computing, Communication and Networking Technologies (ICCCNT), Kanpur, India, 6–8 July 2019; pp. 1–6. [Google Scholar]
- Emrah, Ç.; Zengin, K. Classification of skin lesions in dermatoscopic images with deep convolution network. Avrupa Bilim Ve Teknol. Derg. 2019, 6, 309–318. [Google Scholar]
- Hasan, M.; Barman, S.D.; Islam, S.; Reza, A.W. Skin cancer detection using convolutional neural network. In Proceedings of the 2019 5th International Conference on Computing and Artificial Intelligence, Bali, Indonesia, 19–22 April 2019; pp. 254–258. [Google Scholar]
- Tomáš, M.; Bajić, B.; Yildirim, S.; Hardeberg, J.Y.; Lindblad, J.; Sladoje, N. Ensemble of convolutional neural networks for dermoscopic images classification. arXiv 2018, arXiv:1808.05071. [Google Scholar]
- Lopez; Romero, A.; Giro-i-Nieto, X.; Burdick, J.; Marques, O. Skin lesion classification from dermoscopic images using deep learning techniques. In Proceedings of the 2017 13th IASTED International Conference on Biomedical Engineering (BioMed), Innsbruck, Austria, 20–21 February 2017; pp. 49–54. [Google Scholar]
- Jeremy, K.; BenTaieb, A.; Hamarneh, G. Deep features to classify skin lesions. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 1397–1400. [Google Scholar]
- Noel, C.; Cai, J.; Abedini, M.; Garnavi, R.; Halpern, A.; Smith, J.R. Deep learning, sparse coding, and SVM for melanoma recognition in dermoscopy images. In Proceedings of the Machine Learning in Medical Imaging: 6th International Workshop, MLMI 2015, Held in Conjunction with MICCAI 2015, Munich, Germany, 5 October 2015; pp. 118–126. [Google Scholar]
- Chadaga, K.; Prabhu, S.; Sampathila, N.; Chadaga, R.; Sengupta, S. Predicting cervical cancer biopsy results using demographic and epidemiological parameters: A custom stacked ensemble machine learning approach. Cogent Eng. 2022, 9, 2143040. [Google Scholar] [CrossRef]
- Sampathila, N.; Chadaga, K.; Goswami, N.; Chadaga, R.P.; Pandya, M.; Prabhu, S.; Bairy, M.G.; Katta, S.S.; Bhat, D.; Upadya, S.P. Customized Deep Learning Classifier for Detection of Acute Lymphoblastic Leukemia Using Blood Smear Images. Healthcare 2022, 10, 1812. [Google Scholar] [CrossRef] [PubMed]
- Krishnadas, P.; Chadaga, K.; Sampathila, N.; Rao, S.; Prabhu, S. Classification of Malaria Using Object Detection Models. Informatics 2022, 9, 76. [Google Scholar] [CrossRef]
- Acharya, V.; Dhiman, G.; Prakasha, K.; Bahadur, P.; Choraria, A.; Prabhu, S.; Chadaga, K.; Viriyasitavat, W.; Kautish, S. AI-assisted tuberculosis detection and classification from chest X-rays using a deep learning normalization-free network model. Comput. Intell. Neurosci. 2022, 2022, 2399428. [Google Scholar] [CrossRef]
- Khanna, V.V.; Chadaga, K.; Sampathila, N.; Prabhu, S.; Bhandage, V.; Hegde, G.K. A Distinctive Explainable Machine Learning Framework for Detection of Polycystic Ovary Syndrome. Appl. Syst. Innov. 2023, 6, 32. [Google Scholar] [CrossRef]
- Imran, I.; Younus, M.; Walayat, K.; Kakar, M.U.; Ma, J. Automated multi-class classification of skin lesions through deep convolutional neural network with dermoscopic images. Comput. Med. Imaging Graph. 2021, 88, 101843. [Google Scholar]
- WHO. Gastrointestinal Cancer. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 2 March 2023).
- Yogapriya, J.; Venkatesan Chandran, M.G.; Sumithra, P.; Anitha, P.; Jenopaul, C.; Dhas, S.G. Gastrointestinal tract disease classification from wireless endoscopy images using pretrained deep learning model. Comput. Math. Methods Med. 2021, 2021, 5940433. [Google Scholar] [CrossRef] [PubMed]
- Laith, A.; Fadhel, M.A.; Al-Shamma, O.; Zhang, J.; Santamaría, J.; Duan, Y.; Oleiwi, S.R. Towards a better understanding of transfer learning for medical imaging: A case study. Appl. Sci. 2020, 10, 4523. [Google Scholar]
- Yixuan, Y.; Li, B.; Meng, M.Q.-H. Bleeding frame and region detection in the wireless capsule endoscopy video. IEEE J. Biomed. Health Inform. 2015, 20, 624–630. [Google Scholar]
- Naveen, S.; Zverev, V.I.; Keller, H.; Pane, S.; Egolf, P.W.; Nelson, B.J.; Tishin, A.M. Magnetically guided capsule endoscopy. Med. Phys. 2017, 44, e91–e111. [Google Scholar]
- Benjamin, J.S.; Ferdinand, J.R.; Clatworthy, M.R. Using single-cell technologies to map the human immune system—Implications for nephrology. Nat. Rev. Nephrol. 2020, 16, 112–128. [Google Scholar]
- Hui, H.; Wang, W.-Y.; Mao, B.-H. Borderline-SMOTE: A new over-sampling method in imbalanced data sets learning. In Proceedings of the Advances in Intelligent Computing: International Conference on Intelligent Computing, ICIC 2005, Hefei, China, 23–26 August 2005; pp. 878–887. [Google Scholar]
- Vasileios, C.; Tsiligiri, A.; Hadjileontiadis, L.J.; Liatsos, C.N.; Mavrogiannis, C.C.; Sergiadis, G.D. Ulcer detection in wireless capsule endoscopy images using bidimensional nonlinear analysis. In Proceedings of the XII Mediterranean Conference on Medical and Biological Engineering and Computing 2010, Chalkidiki, Greece, 27–30 May 2010; pp. 236–239. [Google Scholar]
- Ayidzoe, A.; Mighty; Yu, Y.; Mensah, P.K.; Cai, J.; Adu, K.; Tang, Y. Gabor capsule network with preprocessing blocks for the recognition of complex images. Mach. Vis. Appl. 2021, 32, 91. [Google Scholar] [CrossRef]
- Mohapatra, S.; Nayak, J.; Mishra, M.; Pati, G.K.; Naik, B.; Swarnkar, T. Wavelet transform and deep convolutional neural network-based smart healthcare system for gastrointestinal disease detection. Interdiscip. Sci. Comput. Life Sci. 2021, 13, 212–228. [Google Scholar] [CrossRef] [PubMed]
- The ISIC 2020 Challenge Dataset. Available online: https://challenge2020.isic-archive.com/ (accessed on 2 March 2023).
- Philipp, T.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar]
- Dermtology Information System. Available online: http://www.dermis.net (accessed on 2 March 2023).
- Sen, W.; Xing, Y.; Zhang, L.; Gao, H.; Zhang, H. Deep convolutional neural network for ulcer recognition in wireless capsule endoscopy: Experimental feasibility and optimization. Comput. Math. Methods Med. 2019, 2019, 7546215. [Google Scholar]
- Nature. Olympus. The Endocapsule 10 System. Olympus Homepage. 2021. Available online: https://www.olympus-europa.com/medical/en/Products–and–Solutions/Products/Product/ENDOCAPSULE-10-System.html (accessed on 2 March 2023).
- Fushuan, W.; David, A.K. A genetic algorithm based method for bidding strategy coordination in energy and spinning reserve markets. Artif. Intell. Eng. 2001, 15, 71–79. [Google Scholar]
- Hassaan, M.; Farooq, M.S.; Khelifi, A.; Abid, A.; Qureshi, J.N.; Hussain, M. A comparison of transfer learning performance versus health experts in disease diagnosis from medical imaging. IEEE Access 2020, 8, 139367–139386. [Google Scholar]
- Ling, W.; Wang, X.; Fu, J.; Zhen, L. A Novel Probability Binary Particle Swarm Optimization Algorithm and its Application. J. Softw. 2008, 3, 28–35. [Google Scholar]
- Yufei, Z.; Koyuncu, C.; Lu, C.; Grobholz, R.; Katz, I.; Madabhushi, A.; Janowczyk, A. Multi-site cross-organ calibrated deep learning (MuSClD): Automated diagnosis of non-melanoma skin cancer. Med. Image Anal. 2023, 84, 102702. [Google Scholar]
- Alam, T.M.; Shaukat, K.; Khan, W.A.; Hameed, I.A.; Almuqren, L.A.; Raza, M.A.; Aslam, M.; Luo, S. An Efficient Deep Learning-Based Skin Cancer Classifier for an Imbalanced Dataset. Diagnostics 2022, 12, 2115. [Google Scholar] [CrossRef] [PubMed]
- Manash, E.B.K.; Suhasini, A.; Satyala, N. Intelligent skin cancer diagnosis using adaptive k-means segmentation and deep learning models. Concurr. Comput. Pract. Exp. 2023, 35, e7546. [Google Scholar]
- Mijwil, M.M. Skin cancer disease images classification using deep learning solutions. Multimed. Tools Appl. 2021, 80, 26255–26271. [Google Scholar] [CrossRef]
- Farhat, A.; Sharif, M.; Mittal, M.; Khan, M.A.; Hemanth, D.J. A hierarchical three-step superpixels and deep learning framework for skin lesion classification. Methods 2022, 202, 88–102. [Google Scholar]
- Khan, A.M.; Akram, T.; Zhang, Y.-D.; Sharif, M. Attributes based skin lesion detection and recognition: A mask RCNN and transfer learning-based deep learning framework. Pattern Recognit. Lett. 2021, 143, 58–66. [Google Scholar] [CrossRef]
- Naeem, A.; Anees, T.; Fiza, M.; Naqvi, R.A.; Lee, S.-W. SCDNet: A Deep Learning-Based Framework for the Multiclassification of Skin Cancer Using Dermoscopy Images. Sensors 2022, 22, 5652. [Google Scholar] [CrossRef]









| Ref | Model | Type | Limitations | Dataset | Accuracy |
|---|---|---|---|---|---|
| [35] | Two hybrid CNN Models | Benign vs. Melanoma. | The classification accuracy of the model may be enhanced by using more advanced sampling techniques and data preparation. | ISBI 2016 | 88.02% |
| [36] | Spiking Vgg-13 | Melanoma vs. non-Melanoma. | The model’s interpretability has to be improved. | ISIC 2019 | 89.57% |
| [37] | CNN, AlexNet, Vgg-16, Vgg-19 | MEL, BCC, AKIEC, NV, BKL, DF, and VASC. | There is a limited selection of lightweight networks and hyperparameters for evaluation. | HAM10000 | 92.25% |
| [29] | Deep CNN | Malignant vs. Benign. | The model’s segmentation performance is fragile to occlusions in skin pictures, and it struggles with low-contrast skin disease images. | ISIC 2016, ISIC 2017, ISIC 2020 | 90.42% |
| [38] | CNN and ResNet-50 | MEL, BCC, AKIEC, NV, BKL. | Different models and datasets call for various hyperparameter settings. | HAM10000 | 86% |
| [39] | DenseNet-201 | MEL & non-MEL. | To further enhance the model’s generality, a more clinical dataset of skin-cancer cases is required. | ISIC 2019 | 76.08% |
| [30] | MobileNet-V2 | Malignant & benign. | Overall accuracy drops when there is a large gap between the data domain and the target domain. | ISIC 2020 | 98.2% |
| [40] | EfficientNets B0-B7 | MEL, BCC, AKIEC, NV, BKL, DF, and VASC. | The proposed model was trained and tested on an imbalanced dataset of skin cancer, and it affects the model performance. | HAM10000 | 87.91% |
| [31] | DCNN | Benign & malignant. | Due to the small sample size of the datasets used in this study, local optimizations may have been achieved. | HAM10000 | 91.93% |
| [41] | ResNet-152, SE-ResNeXt-101, DenseNet-161 | MEL, BCC, AKIEC, NV, BKL, DF, and VASC. | The computational cost was significant, and the system did not take into account all possible skin cancers. | ISIC 2018 | 93% |
| [42] | CNN | MEL, BCC, AKIEC, NV, BKL, DF, and VASC. | Classification persists, however, because the model relies on a small quantity of training data and the hazy borders of skin disease pictures. | HAM10000 | 78% |
| [43] | DenseNet-121 | MEL, BCC, and AKIEC. | Due to the lack of adversarial training on other skin cancer datasets, the method’s model remains vulnerable. | HAM10000 | 85% |
| No. of Classes | Class Name | No. of Images |
|---|---|---|
| 0 | BCC | 510 |
| 1 | MEL | 1686 |
| 2 | MN | 2007 |
| 3 | SCC | 97 |
| No. of Classes | Class Name | No. of Images |
|---|---|---|
| 0 | BCC | 2035 |
| 1 | MEL | 1952 |
| 2 | MN | 2007 |
| 3 | SCC | 2018 |
| Layer Type | Output Shape | Parameters |
|---|---|---|
| Input Layer | (None, 150, 150, 3) | 0 |
| Block01 | (None, 150, 150, 8) | 224 |
| Block02 | (None, 75, 75, 16) | 1168 |
| Block03 | (None, 37, 37, 32) | 4640 |
| Block04 | (None, 18, 18, 64) | 18,496 |
| Block05 | (None, 9, 9, 128) | 73,856 |
| Dropout_1 | (None, 4, 4, 128) | 0 |
| Flatten | (None, 2048) | 0 |
| Dense_1 | (None, 512) | 1,049,088 |
| ReLu | (None, 512) | 0 |
| Dense_2 | (None, 4) | 2052 |
| Output: SoftMax | (None, 4) | 0 |
| Total Parameters: | 1,149,524 | |
| Trainable Parameters: | 1,149,524 | |
| Non-Trainable Parameters: | 0 | |
| Classifiers | Accuracy | Precision | Recall | F1-Score | AUC |
|---|---|---|---|---|---|
| Vgg-16 | 91.12% | 92.09% | 90.43% | 91.13% | 99.02% |
| Vgg-19 | 91.68% | 92.23% | 90.57% | 91.71% | 98.14% |
| MobileNet | 92.51% | 92.95% | 91.40% | 92.17% | 98.75% |
| ResNet-152 | 89.32% | 90.73% | 88.21% | 89.27% | 98.74% |
| EfficientNet-B0 | 89.46% | 90.21% | 88.21% | 89.31% | 98.43% |
| Inception-V3 | 91.82% | 92.28% | 91.12% | 91.76% | 99.06% |
| Proposed Model (With SMOTE Tomek) | 94.17% | 94.28% | 93.76% | 93.93% | 99.43% |
| Proposed Model (Without SMOTE Tomek) | 83.20% | 85.01% | 80.62% | 58.09% | 96.65% |
| Ref | Year | Model | Datasets | Accuracy | Recall | Precision | F1-Score |
|---|---|---|---|---|---|---|---|
| [70] | 2023 | CNN | ISIC-2017 | 92.00% | 91.90% | 91.65% | 91.99% |
| [71] | 2023 | Vgg-13 | ISIC-2019, Derm-IS | 89.57% | 90.70% | 89.66% | 89.65% |
| [72] | 2023 | Deep Belief Network | HAM-10000 | 93.00% | 92.91% | 92.45% | 92.65% |
| [73] | 2021 | ConvNet | ISIC-2018, Derm-IS | 86.90% | 86.14% | 87.47% | - |
| [74] | 2022 | 2D superpixels + RCNN | HAM-10000 | 85.50% | 83.40% | 84.50% | 85.30% |
| [75] | 2021 | ResNeXt101 | ISIC-2019 | 88.50% | 87.40% | 88.10% | 88.30% |
| [76] | 2022 | SCDNet | ISIC-2019 | 92.91% | 92.18% | 92.19% | 92.18% |
| Ours | - | DSCC_Net with SMOTE Tomek | ISIC-2020, Derm-IS, HAM-10000 | 94.17% | 94.28% | 93.76% | 93.93% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tahir, M.; Naeem, A.; Malik, H.; Tanveer, J.; Naqvi, R.A.; Lee, S.-W. DSCC_Net: Multi-Classification Deep Learning Models for Diagnosing of Skin Cancer Using Dermoscopic Images. Cancers 2023, 15, 2179. https://doi.org/10.3390/cancers15072179
Tahir M, Naeem A, Malik H, Tanveer J, Naqvi RA, Lee S-W. DSCC_Net: Multi-Classification Deep Learning Models for Diagnosing of Skin Cancer Using Dermoscopic Images. Cancers. 2023; 15(7):2179. https://doi.org/10.3390/cancers15072179
Chicago/Turabian StyleTahir, Maryam, Ahmad Naeem, Hassaan Malik, Jawad Tanveer, Rizwan Ali Naqvi, and Seung-Won Lee. 2023. "DSCC_Net: Multi-Classification Deep Learning Models for Diagnosing of Skin Cancer Using Dermoscopic Images" Cancers 15, no. 7: 2179. https://doi.org/10.3390/cancers15072179
APA StyleTahir, M., Naeem, A., Malik, H., Tanveer, J., Naqvi, R. A., & Lee, S.-W. (2023). DSCC_Net: Multi-Classification Deep Learning Models for Diagnosing of Skin Cancer Using Dermoscopic Images. Cancers, 15(7), 2179. https://doi.org/10.3390/cancers15072179








