Microvasculature Features Derived from Hybrid EPI MRI in Non-Enhancing Adult-Type Diffuse Glioma Subtypes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. MRI Acquisition
2.3. MR Image Pre-Processing
2.4. Histopathological Analysis
2.5. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. MRI-Based Measurements
3.3. Histology-Derived Measurements
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Lebelt, A.; Dziecioł, J.; Guzińska-Ustymowicz, K.; Lemancewicz, D.; Zimnoch, L.; Czykier, E. Angiogenesis in Gliomas. Folia Histochem. Cytobiol. 2008, 46, 69–72. [Google Scholar] [CrossRef]
- Plate, K.H.; Mennel, H.D. Vascular Morphology and Angiogenesis in Glial Tumors. Exp. Toxicol. Pathol. 1995, 47, 89–94. [Google Scholar] [CrossRef]
- Chen, W.; He, D.; Li, Z.; Zhang, X.; Pan, D.; Chen, G. Overexpression of Vascular Endothelial Growth Factor Indicates Poor Outcomes of Glioma: A Systematic Review and Meta-Analysis. Int. J. Clin. Exp. Med. 2015, 8, 8709–8719. [Google Scholar]
- Emblem, K.E.; Nedregaard, B.; Nome, T.; Due-Tonnessen, P.; Hald, J.K.; Scheie, D.; Borota, O.C.; Cvancarova, M.; Bjornerud, A. Glioma Grading by Using Histogram Analysis of Blood Volume Heterogeneity from MR-Derived Cerebral Blood Volume Maps. Radiology 2008, 247, 808–817. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. AI Neuropathologist: An Innovative Technology Enabling a Faultless Pathological Diagnosis? Neuro-Oncology 2021, 23, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Dubbink, H.J.; Atmodimedjo, P.N.; Kros, J.M.; French, P.J.; Sanson, M.; Idbaih, A.; Wesseling, P.; Enting, R.; Spliet, W.; Tijssen, C.; et al. Molecular Classification of Anaplastic Oligodendroglioma Using Next-Generation Sequencing: A Report of the Prospective Randomized EORTC Brain Tumor Group 26951 Phase III Trial. Neuro-Oncology 2016, 18, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Kang, H.; Tong, H.; Du, X.; Liu, H.; Tan, Y.; Yang, Y.; Wang, S.; Zhang, W. Microvascular Characteristics of Lower-Grade Diffuse Gliomas: Investigating Vessel Size Imaging for Differentiating Grades and Subtypes. Eur. Radiol. 2019, 29, 1893–1902. [Google Scholar] [CrossRef] [PubMed]
- Kickingereder, P.; Sahm, F.; Radbruch, A.; Wick, W.; Heiland, S.; von Deimling, A.; Bendszus, M.; Wiestler, B. IDH Mutation Status Is Associated with a Distinct Hypoxia/Angiogenesis Transcriptome Signature Which Is Non-Invasively Predictable with RCBV Imaging in Human Glioma. Sci. Rep. 2015, 5, 16238. [Google Scholar] [CrossRef]
- Cha, S.; Tihan, T.; Crawford, F.; Fischbein, N.J.; Chang, S.; Bollen, A.; Nelson, S.J.; Prados, M.; Berger, M.S.; Dillon, W.P. Differentiation of Low-Grade Oligodendrogliomas from Low-Grade Astrocytomas by Using Quantitative Blood-Volume Measurements Derived from Dynamic Susceptibility Contrast-Enhanced MR Imaging. AJNR Am. J. Neuroradiol. 2005, 26, 266–273. [Google Scholar]
- Kellner, E.; Breyer, T.; Gall, P.; Müller, K.; Trippel, M.; Staszewski, O.; Stein, F.; Saborowski, O.; Dyakova, O.; Urbach, H.; et al. MR Evaluation of Vessel Size Imaging of Human Gliomas: Validation by Histopathology. J. Magn. Reson. Imaging 2015, 42, 1117–1125. [Google Scholar] [CrossRef]
- Kiselev, V.G.; Strecker, R.; Ziyeh, S.; Speck, O.; Hennig, J. Vessel Size Imaging in Humans. Magn. Reson. Med. 2005, 53, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Shiroishi, M.S.; Castellazzi, G.; Boxerman, J.L.; D’Amore, F.; Essig, M.; Nguyen, T.B.; Provenzale, J.M.; Enterline, D.S.; Anzalone, N.; Dörfler, A.; et al. Principles of T2∗-Weighted Dynamic Susceptibility Contrast MRI Technique in Brain Tumor Imaging. J. Magn. Reson. Imaging 2015, 41, 296–313. [Google Scholar] [CrossRef]
- Chakhoyan, A.; Yao, J.; Leu, K.; Pope, W.B.; Salamon, N.; Yong, W.; Lai, A.; Nghiemphu, P.L.; Everson, R.G.; Prins, R.M.; et al. Validation of Vessel Size Imaging (VSI) in High-Grade Human Gliomas Using Magnetic Resonance Imaging, Image-Guided Biopsies, and Quantitative Immunohistochemistry. Sci. Rep. 2019, 9, 2846. [Google Scholar] [CrossRef] [PubMed]
- Pruis, I.J.; Koene, S.R.; van der Voort, S.R.; Incekara, F.; Vincent, A.J.P.E.; van den Bent, M.J.; Lycklama à Nijeholt, G.J.; Nandoe Tewarie, R.D.S.; Veldhuijzen van Zanten, S.E.M.; Smits, M. Noninvasive Differentiation of Molecular Subtypes of Adult Nonenhancing Glioma Using MRI Perfusion and Diffusion Parameters. Neuro-Oncol. Adv. 2022, 4, vdac023. [Google Scholar] [CrossRef] [PubMed]
- Leu, K.; Ott, G.A.; Lai, A.; Nghiemphu, P.L.; Pope, W.B.; Yong, W.H.; Liau, L.M.; Cloughesy, T.F.; Ellingson, B.M. Perfusion and Diffusion MRI Signatures in Histologic and Genetic Subtypes of WHO Grade II–III Diffuse Gliomas. J. Neuro-Oncol. 2017, 134, 177–188. [Google Scholar] [CrossRef]
- Arzanforoosh, F.; Croal, P.L.; van Garderen, K.A.; Smits, M.; Chappell, M.A.; Warnert, E.A.H. Effect of Applying Leakage Correction on RCBV Measurement Derived From DSC-MRI in Enhancing and Nonenhancing Glioma. Front. Oncol. 2021, 11, 648528. [Google Scholar] [CrossRef]
- Lemasson, B.; Chenevert, T.L.; Lawrence, T.S.; Tsien, C.; Sundgren, P.C.; Meyer, C.R.; Junck, L.; Boes, J.; Galbán, S.; Johnson, T.D.; et al. Impact of Perfusion Map Analysis on Early Survival Prediction Accuracy in Glioma Patients. Transl. Oncol. 2013, 6, 766–774. [Google Scholar] [CrossRef]
- Kang, H.Y.; Xiao, H.L.; Chen, J.H.; Tan, Y.; Chen, X.; Xie, T.; Fang, J.Q.; Wang, S.; Yang, Y.; Zhang, W.G. Comparison of the Effect of Vessel Size Imaging and Cerebral Blood Volume Derived from Perfusion MR Imaging on Glioma Grading. Am. J. Neuroradiol. 2016, 37, 51–57. [Google Scholar] [CrossRef]
- Ferreira, P.F.; Gatehouse, P.D.; Firmin, D.N. Myocardial First-Pass Perfusion Imaging with Hybrid-EPI: Frequency-Offsets and Potential Artefacts. J. Cardiovasc. Magn. Reson. 2012, 14, 44. [Google Scholar] [CrossRef]
- Jenkinson, M. Improved Optimization for the Robust and Accurate Linear Registration and Motion Correction of Brain Images. Neuroimage 2002, 17, 825–841. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, M.; Smith, S. A Global Optimisation Method for Robust Affine Registration of Brain Images. Med. Image Anal. 2001, 5, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Bell, L.C.; Stokes, A.M.; Quarles, C.C. Analysis of Postprocessing Steps for Residue Function Dependent Dynamic Susceptibility Contrast (DSC)-MRI Biomarkers and Their Clinical Impact on Glioma Grading for Both 1.5 and 3T. J. Magn. Reson. Imaging 2020, 51, 547–553. [Google Scholar] [CrossRef]
- Leenders, K.L.; Perani, D.; Lammertsma, A.A.; Heather, J.D.; Buckingham, P.; Jones, T.; Healy, M.J.R.; Gibbs, J.M.; Wise, R.J.S.; Hatazawa, J.; et al. Cerebral Blood Flow, Blood Volume And Oxygen Utilization. Brain 1990, 113, 27–47. [Google Scholar] [CrossRef] [PubMed]
- McKinley, R.; Rebsamen, M.; Dätwyler, K.; Meier, R.; Radojewski, P.; Wiest, R. Uncertainty-Driven Refinement of Tumor-Core Segmentation Using 3D-to-2D Networks with Label Uncertainty. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Springer: Cham, Switzerland, 2021; pp. 401–411. [Google Scholar]
- Luu, H.M.; Park, S.-H. Extending Nn-UNet for Brain Tumor Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Springer: Cham, Switzerland, 2022; pp. 173–186. [Google Scholar]
- Kickingereder, P.; Isensee, F.; Tursunova, I.; Petersen, J.; Neuberger, U.; Bonekamp, D.; Brugnara, G.; Schell, M.; Kessler, T.; Foltyn, M.; et al. Automated Quantitative Tumour Response Assessment of MRI in Neuro-Oncology with Artificial Neural Networks: A Multicentre, Retrospective Study. Lancet Oncol. 2019, 20, 728–740. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. NnU-Net: A Self-Configuring Method for Deep Learning-Based Biomedical Image Segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Zhang, Y.; Brady, M.; Smith, S. Segmentation of Brain MR Images through a Hidden Markov Random Field Model and the Expectation-Maximization Algorithm. IEEE Trans. Med. Imaging 2001, 20, 45–57. [Google Scholar] [CrossRef]
- Klein, S.; Staring, M.; Murphy, K.; Viergever, M.A.; Pluim, J. Elastix: A Toolbox for Intensity-Based Medical Image Registration. IEEE Trans. Med. Imaging 2010, 29, 196–205. [Google Scholar] [CrossRef]
- Reyes-Aldasoro, C.C.; Williams, L.J.; Akerman, S.; Kanthou, C.; Tozer, G.M. An Automatic Algorithm for the Segmentation and Morphological Analysis of Microvessels in Immunostained Histological Tumour Sections. J. Microsc. 2011, 242, 262–278. [Google Scholar] [CrossRef]
- Smits, M.; van den Bent, M.J. Imaging Correlates of Adult Glioma Genotypes. Radiology 2017, 284, 316–331. [Google Scholar] [CrossRef]
- Vanchinathan, V.; Mizramani, N.; Kantipudi, R.; Schwartz, E.J.; Sundram, U.N. The Vascular Marker CD31 Also Highlights Histiocytes and Histiocyte-Like Cells Within Cutaneous Tumors. Am. J. Clin. Pathol. 2015, 143, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Wesseling, P.; van der Laak, J.A.; Link, M.; Teepen, H.L.; Ruiter, D.J. Quantitative Analysis of Microvascular Changes in Diffuse Astrocytic Neoplasms with Increasing Grade of Malignancy. Hum. Pathol. 1998, 29, 352–358. [Google Scholar] [CrossRef]
- Wesseling, P.; van der Laak, J.A.; de Leeuw, H.; Ruiter, D.J.; Burger, P.C. Quantitative Immunohistological Analysis of the Microvasculature in Untreated Human Glioblastoma Multiforme. Computer-Assisted Image Analysis of Whole-Tumor Sections. J. Neurosurg. 1994, 81, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.A.; Farrell, C.L.; del Maestro, R.F. The Effect of Cellular Microenvironment on Vessels in the Brain. Part 1: Vessel Structure in Tumour, Peritumour and Brain from Humans with Malignant Glioma. Int. J. Radiat. Biol. 1991, 60, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Morgan, P.S.; Ashburner, J.; Smith, J.; Rorden, C. The First Step for Neuroimaging Data Analysis: DICOM to NIfTI Conversion. J. Neurosci. Methods 2016, 264, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Tustison, N.J.; Avants, B.B.; Cook, P.A.; Zheng, Y.; Egan, A.; Yushkevich, P.A.; Gee, J.C. N4ITK: Improved N3 Bias Correction. IEEE Trans. Med. Imaging 2010, 29, 1310–1320. [Google Scholar] [CrossRef]
- Isensee, F.; Schell, M.; Pflueger, I.; Brugnara, G.; Bonekamp, D.; Neuberger, U.; Wick, A.; Schlemmer, H.; Heiland, S.; Wick, W.; et al. Automated Brain Extraction of Multisequence MRI Using Artificial Neural Networks. Hum. Brain Mapp. 2019, 40, 4952–4964. [Google Scholar] [CrossRef]
- Smith, S.M. Fast Robust Automated Brain Extraction. Hum. Brain Mapp. 2002, 17, 143–155. [Google Scholar] [CrossRef]
- Fonov, V.; Evans, A.C.; Botteron, K.; Almli, C.R.; McKinstry, R.C.; Collins, D.L. Unbiased Average Age-Appropriate Atlases for Pediatric Studies. Neuroimage 2011, 54, 313–327. [Google Scholar] [CrossRef]
- Shamonin, D. Fast Parallel Image Registration on CPU and GPU for Diagnostic Classification of Alzheimer’s Disease. Front. Neuroinformatics 2013, 7, 50. [Google Scholar] [CrossRef]
- Rohlfing, T.; Zahr, N.M.; Sullivan, E.v.; Pfefferbaum, A. The SRI24 Multichannel Atlas of Normal Adult Human Brain Structure. Hum. Brain Mapp. 2009, 31, 798–819. [Google Scholar] [CrossRef] [PubMed]
- Rohlfing, T.; Russakoff, D.B.; Maurer, C.R. Performance-Based Classifier Combination in Atlas-Based Image Segmentation Using Expectation-Maximization Parameter Estimation. IEEE Trans. Med. Imaging 2004, 23, 983–994. [Google Scholar] [CrossRef] [PubMed]


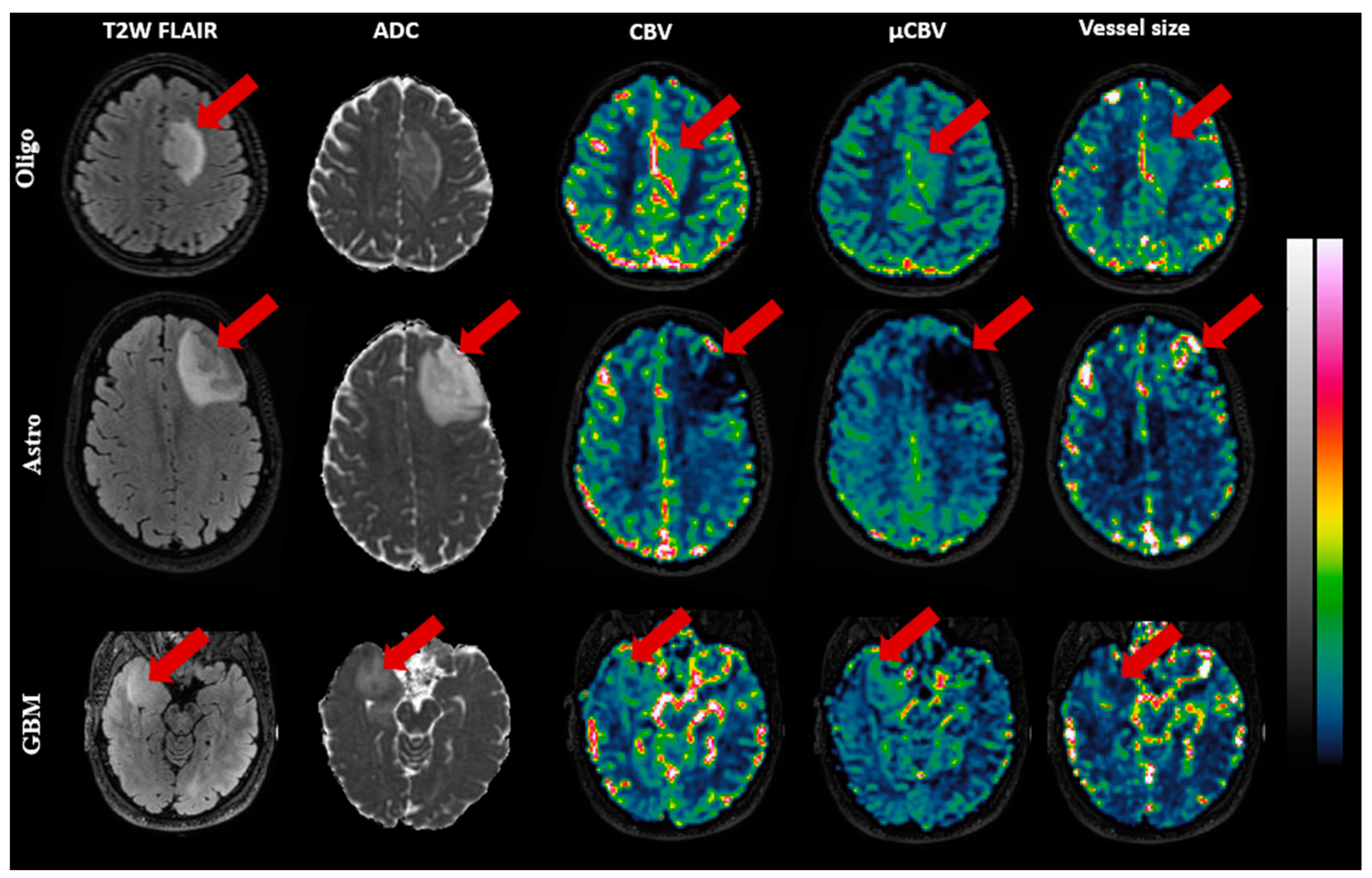
| Tumor Subtype | Grade 2 | Grade 3 | Grade 4 | Total | Sex | Age (Years) |
|---|---|---|---|---|---|---|
| Oligo (IDH-mut&1p/19q-codeleted) | 15 | 3 | - | 18 | 13M/5F | 40 ± 12 |
| (6) | (2) | - | (8) | (7M/1F) | (43 ± 13) | |
| Astro (IDH-mut) | 9 | 4 | - | 13 | 5M/8F | 36 ± 8 |
| (7) | (-) | (-) | (7) | (4M/3F) | (35 ± 6) | |
| GBM (IDH-wt) | - | - | 7 | 7 | 7M/0F | 60 ± 10 |
| (-) | (-) | (6) | (6) | (6M/0F) | (59 ± 8) |
| Oligo (IDH-mut&1p/19q-codeleted) | Astro (IDH-mut) | GBM (IDH-wt) | p-Value | ||
|---|---|---|---|---|---|
| MRI-derived parameters | |||||
| CBV (%) | Mean (std) | 1.43 (0.44) | 0.96 (0.36) | 1.20 (0.39) | 0.01 a |
| Median (std) | 1.17 (0.36) | 0.70 (0.18) | 0.92 (0.37) | <0.001 b | |
| Hot spot (std) | 3.31 (1.14) | 2.79 (1.71) | 3.34 (0.85) | 0.13 | |
| µCBV (%) | Mean (std) | 1.45 (0.29) | 0.86 (0.26) | 1.36 (0.71) | <0.001 b |
| Median (std) | 1.31 (0.25) | 0.74 (0.21) | 1.29 (0.73) | <0.001 b | |
| Hot spot (std) | 2.37 (0.60) | 1.62 (0.77) | 2.22 (0.83) | 0.02 a | |
|
Vessel size () | Mean (std) | 12.48 (2.61) | 15.55 (5.10) | 10.33 (3.62) | 0.01 a |
| Median (std) | 10.18 (2.02) | 10.91 (2.47) | 7.02 (2.32) | 0.002 a | |
| Hot spot (std) | 44.91 (15.55) | 78.85 (46.48) | 53.81 (34.96) | 0.06 a | |
| Histopathology-derived parameters | |||||
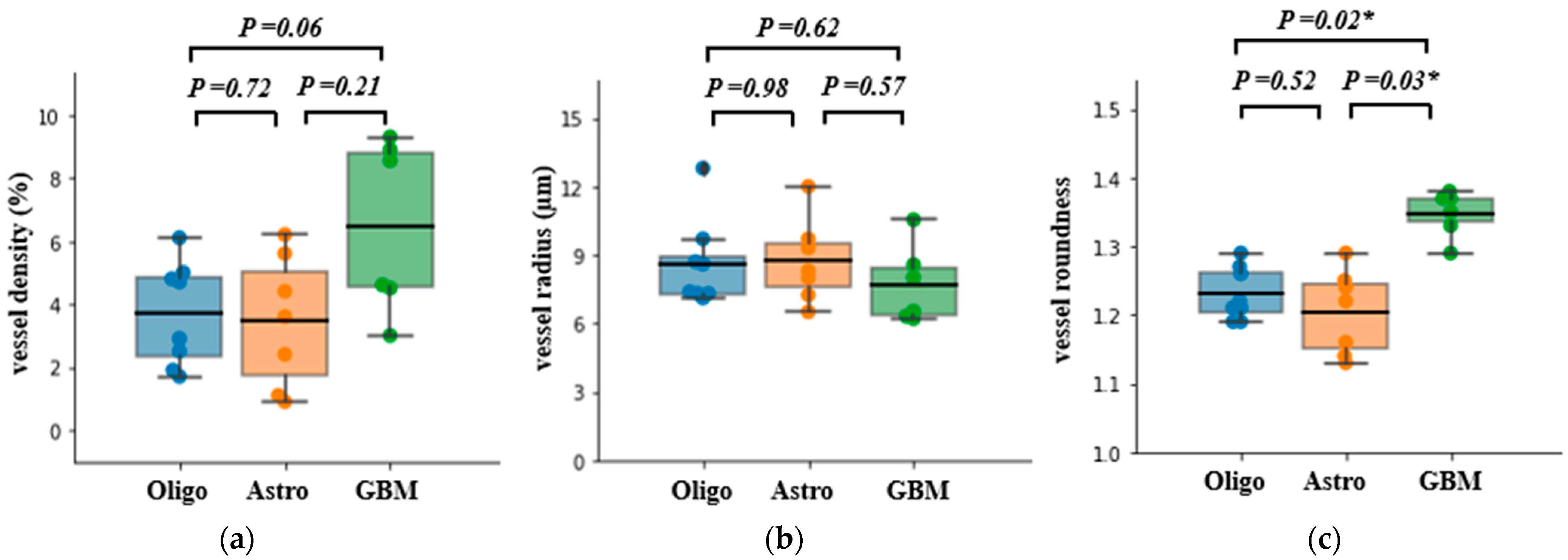
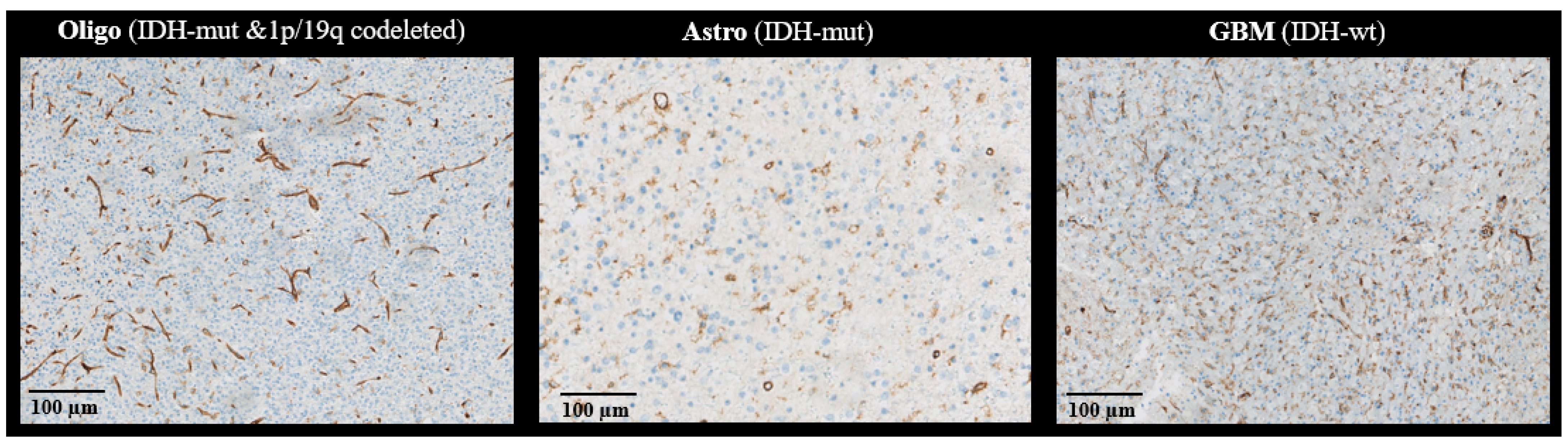
| Vessel density | Mean (std) | 3.69 (1.64) | 3.45 (2.09) | 6.47 (2.74) | 0.03 a |
| Vessel radius () | Mean (std) | 8.61 (1.92) | 8.72 (1.82) | 7.68 (1.70) | 0.55 b |
| Vessel roundness | Mean (std) | 1.23 (0.03) | 1.20 (0.06) | 1.34 (0.03) | <0.001 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arzanforoosh, F.; van der Voort, S.R.; Incekara, F.; Vincent, A.; Van den Bent, M.; Kros, J.M.; Smits, M.; Warnert, E.A.H. Microvasculature Features Derived from Hybrid EPI MRI in Non-Enhancing Adult-Type Diffuse Glioma Subtypes. Cancers 2023, 15, 2135. https://doi.org/10.3390/cancers15072135
Arzanforoosh F, van der Voort SR, Incekara F, Vincent A, Van den Bent M, Kros JM, Smits M, Warnert EAH. Microvasculature Features Derived from Hybrid EPI MRI in Non-Enhancing Adult-Type Diffuse Glioma Subtypes. Cancers. 2023; 15(7):2135. https://doi.org/10.3390/cancers15072135
Chicago/Turabian StyleArzanforoosh, Fatemeh, Sebastian R. van der Voort, Fatih Incekara, Arnaud Vincent, Martin Van den Bent, Johan M. Kros, Marion Smits, and Esther A. H. Warnert. 2023. "Microvasculature Features Derived from Hybrid EPI MRI in Non-Enhancing Adult-Type Diffuse Glioma Subtypes" Cancers 15, no. 7: 2135. https://doi.org/10.3390/cancers15072135
APA StyleArzanforoosh, F., van der Voort, S. R., Incekara, F., Vincent, A., Van den Bent, M., Kros, J. M., Smits, M., & Warnert, E. A. H. (2023). Microvasculature Features Derived from Hybrid EPI MRI in Non-Enhancing Adult-Type Diffuse Glioma Subtypes. Cancers, 15(7), 2135. https://doi.org/10.3390/cancers15072135







