Integrated Analysis of N1-Methyladenosine Methylation Regulators-Related lncRNAs in Hepatocellular Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
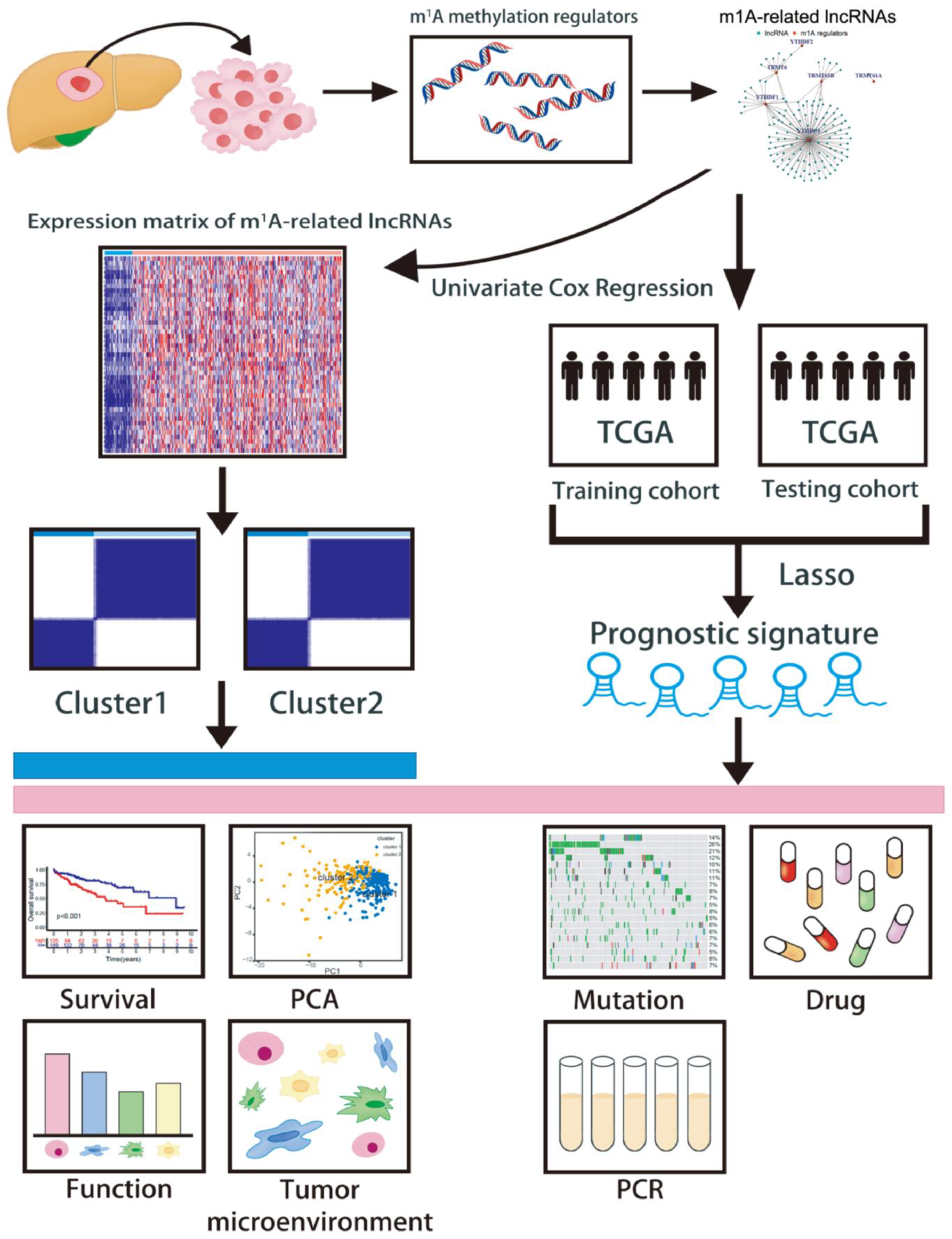
2. Materials and Methods
2.1. Data Collection and Processing
2.2. Unsupervised Clustering Analysis of m1A-Related lncRNAs
2.3. m1AScore Construction
2.4. Predictive Performance of the Prognostic Signature
2.5. Generating a Nomogram
2.6. Correlation between Single-Nucleotide Variants and the Prognostic Signature
2.7. Assessment of the Prognostic Signature in Immune Landscapes
2.8. Therapy Response Assessment of the Prognostic Signature
2.9. Specimen Collection
2.10. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.11. Statistical Analysis
3. Results
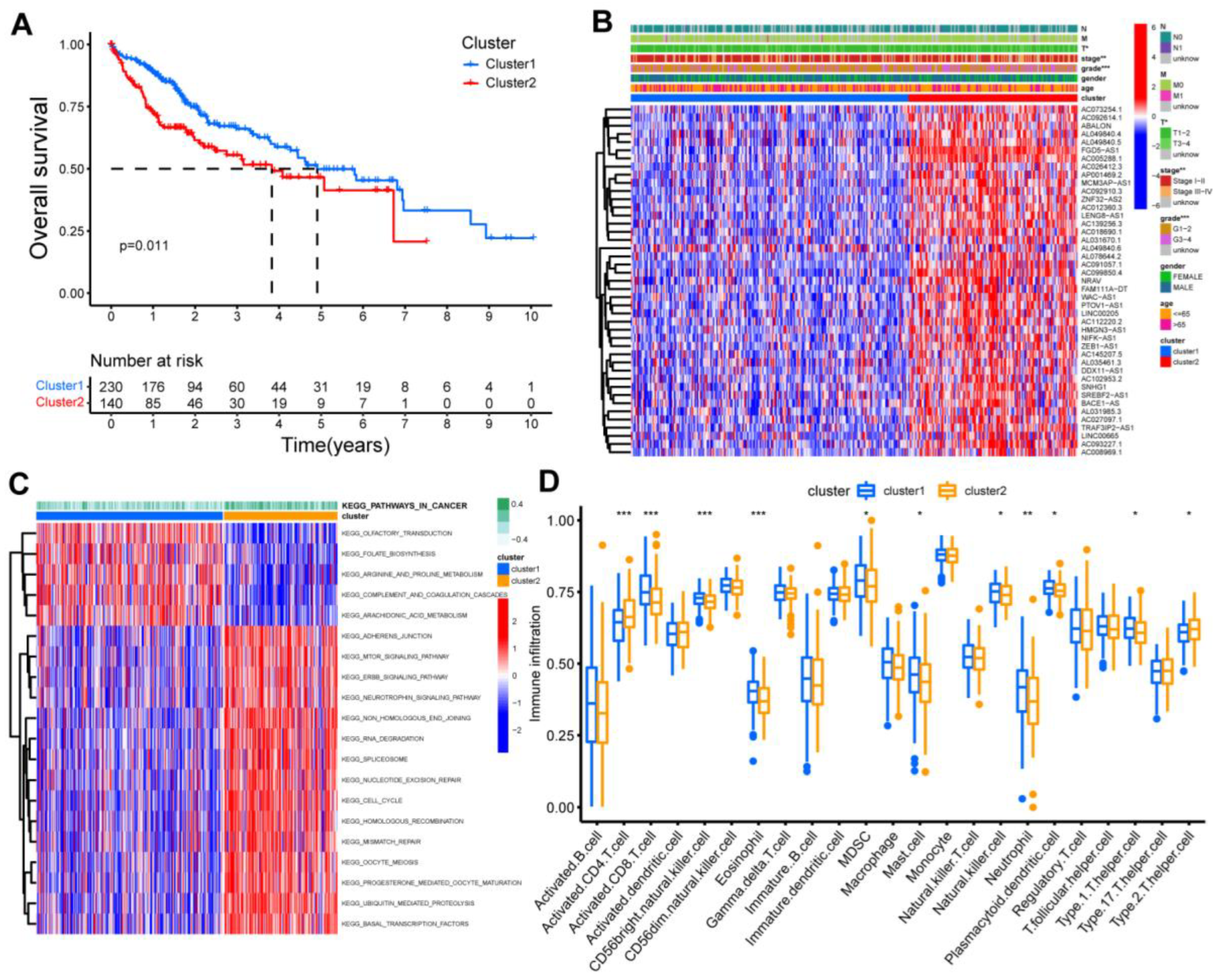
3.1. Identification of Two Clusters Based on the Co-Expressed lncRNAs
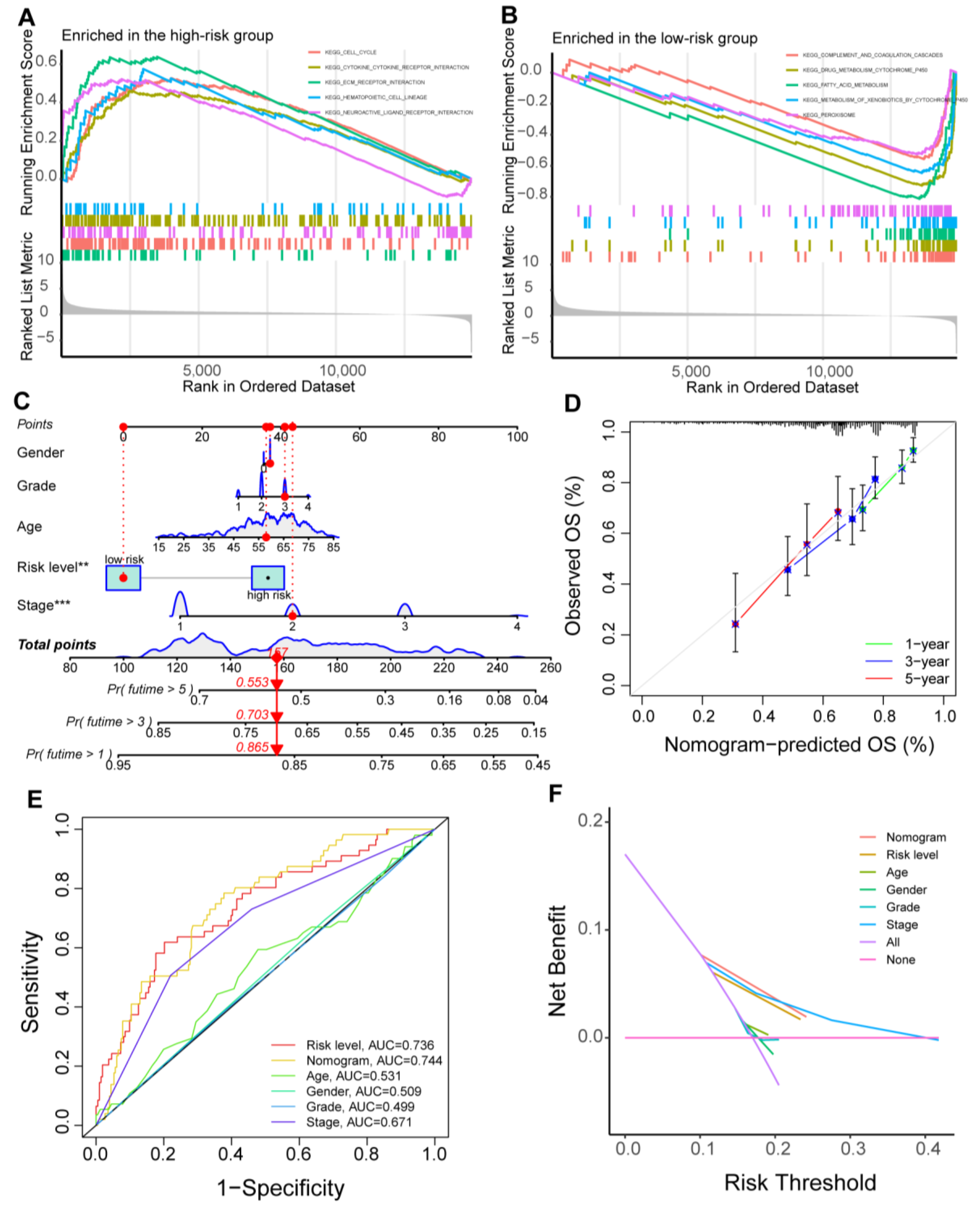
3.2. Generating an m1AScore for Prognostic Prediction
3.3. Function Analysis and Nomogram Construction
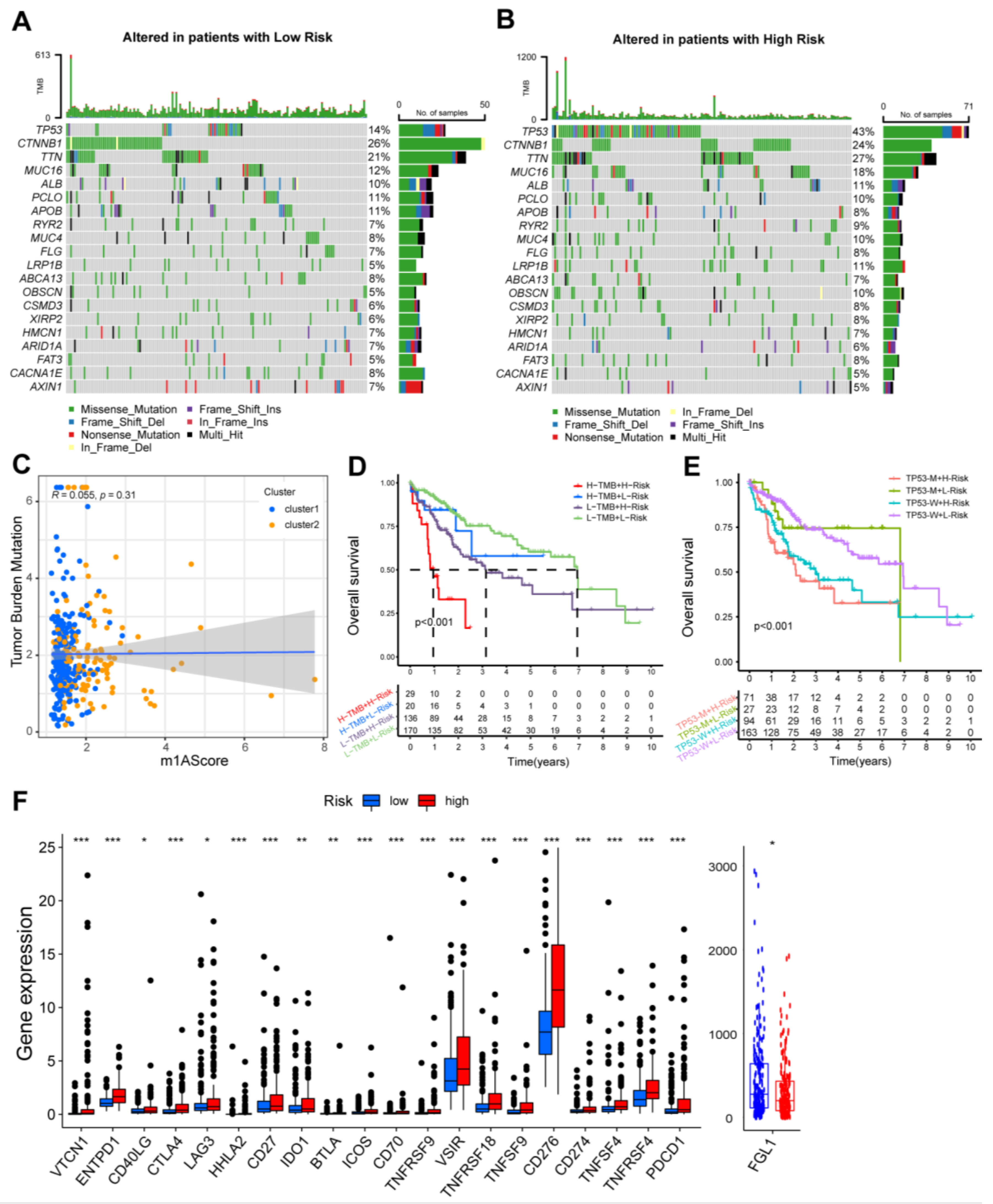
3.4. Survival Stratification Based on the Prognostic Signature and Single-Nucleotide Variant
3.5. The Immune Landscape of m1AScore
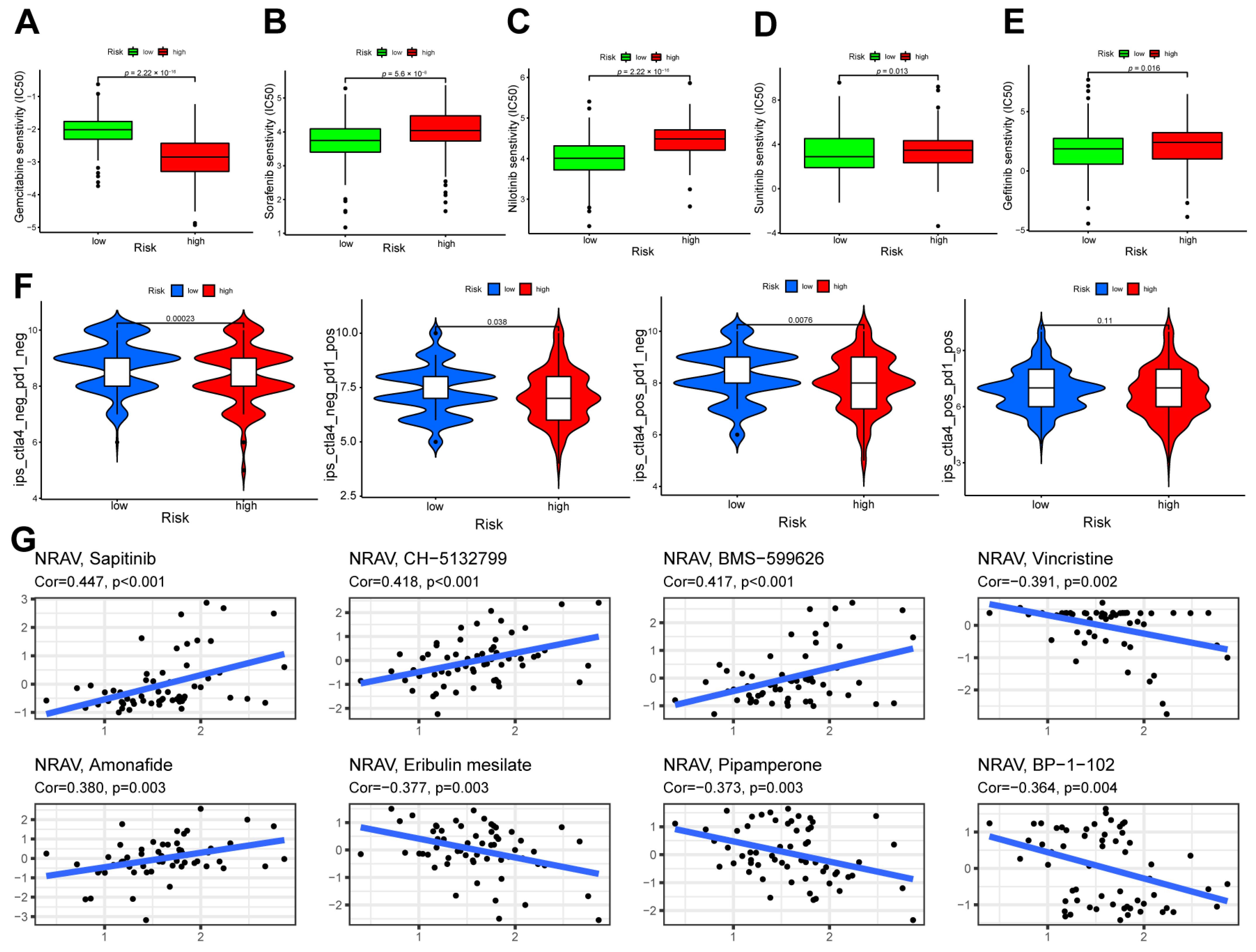
3.6. Therapeutic Response Assessment and Drug Sensitivity
3.7. Validation of the Expression Patterns of Five Screened lncRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Liang, D.; Li, D.; Shan, B.; Zheng, R.; Zhang, S.; Wei, W.; He, J. Incidence and mortality of laryngeal cancer in China, 2015. Chin. J. Cancer Res. 2020, 32, 10–17. [Google Scholar] [CrossRef]
- Levrero, M.; Zucman-Rossi, J. Mechanisms of HBV-induced hepatocellular carcinoma. J. Hepatol. 2016, 64, S84–S101. [Google Scholar] [CrossRef] [PubMed]
- Marengo, A.; Rosso, C.; Bugianesi, E. Liver Cancer: Connections with Obesity, Fatty Liver, and Cirrhosis. Annu. Rev. Med. 2016, 67, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Ganne-Carrié, N.; Nahon, P. Hepatocellular carcinoma in the setting of alcohol-related liver disease. J. Hepatol. 2019, 70, 284–293. [Google Scholar] [CrossRef]
- Zoller, H.; Tilg, H. Nonalcoholic fatty liver disease and hepatocellular carcinoma. Metabolism 2016, 65, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Flattau, A.; Cristallo, J.; Duggan, M.; Gbur, M.; Fabienne Daguilh, M.L.; Selwyn, P. Clinical Redeployment of an Academic Family Medicine Department in an Early, Severe COVID-19 Pandemic in the Bronx, NY. J. Am. Board Fam. Med. 2021, 34, 466–473. [Google Scholar] [CrossRef]
- Pinato, D.J.; Fessas, P.; Sapisochin, G.; Marron, T.U. Perspectives on the Neoadjuvant Use of Immunotherapy in Hepatocellular Carcinoma. Hepatology 2021, 74, 483–490. [Google Scholar] [CrossRef]
- Anwanwan, D.; Singh, S.K.; Singh, S.; Saikam, V.; Singh, R. Challenges in liver cancer and possible treatment approaches. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188314. [Google Scholar] [CrossRef]
- Dominissini, D.; Nachtergaele, S.; Moshitch-Moshkovitz, S.; Peer, E.; Kol, N.; Ben-Haim, M.S.; Dai, Q.; Di Segni, A.; Salmon-Divon, M.; Clark, W.C.; et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016, 530, 441–446. [Google Scholar] [CrossRef]
- Zhang, C.; Jia, G. Reversible RNA Modification N(1)-methyladenosine (m(1)A) in mRNA and tRNA. Genom. Proteom. Bioinform. 2018, 16, 155–161. [Google Scholar] [CrossRef]
- Safra, M.; Sas-Chen, A.; Nir, R.; Winkler, R.; Nachshon, A.; Bar-Yaacov, D.; Erlacher, M.; Rossmanith, W.; Stern-Ginossar, N.; Schwartz, S. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 2017, 551, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Xue, C.; Yuan, X.; He, Y.; Yu, Z. Gene signatures and prognostic values of m1A-related regulatory genes in hepatocellular carcinoma. Sci. Rep. 2020, 10, 15083. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Clark, W.; Luo, G.; Wang, X.; Fu, Y.; Wei, J.; Wang, X.; Hao, Z.; Dai, Q.; Zheng, G.; et al. ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell 2016, 167, 816–828.e816. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Wang, T.; Gonzalez, G.; Wang, Y. Identification of YTH Domain-Containing Proteins as the Readers for N1-Methyladenosine in RNA. Anal. Chem. 2018, 90, 6380–6384. [Google Scholar] [CrossRef]
- Engel, M.; Chen, A. The emerging role of mRNA methylation in normal and pathological behavior. Genes Brain Behav 2018, 17, e12428. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Li, X.; Xiong, X.; Wang, J.; Zhou, Z.; Zhu, X.; Gu, Y.; Dominissini, D.; He, L.; et al. N(1)-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat. Commun. 2021, 12, 6314. [Google Scholar] [CrossRef] [PubMed]
- Sanchez Calle, A.; Kawamura, Y.; Yamamoto, Y.; Takeshita, F.; Ochiya, T. Emerging roles of long non-coding RNA in cancer. Cancer Sci. 2018, 109, 2093–2100. [Google Scholar] [CrossRef]
- Wang, E.; Li, Y.; Ming, R.; Wei, J.; Du, P.; Zhou, P.; Zong, S.; Xiao, H. The Prognostic Value and Immune Landscapes of a m(6)A/m(5)C/m(1)A-Related LncRNAs Signature in Head and Neck Squamous Cell Carcinoma. Front. Cell Dev. Biol. 2021, 9, 718974. [Google Scholar] [CrossRef]
- Liu, J.; Chen, C.; Wang, Y.; Qian, C.; Wei, J.; Xing, Y.; Bai, J. Comprehensive of N1-Methyladenosine Modifications Patterns and Immunological Characteristics in Ovarian Cancer. Front. Immunol. 2021, 12, 746647. [Google Scholar] [CrossRef]
- Zheng, Q.; Yu, X.; Zhang, Q.; He, Y.; Guo, W. Genetic characteristics and prognostic implications of m1A regulators in pancreatic cancer. Biosci. Rep. 2021, 41, BSR20210337. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Shen, S.; Xue, C. A Novel m1A-Score Model Correlated With the Immune Microenvironment Predicts Prognosis in Hepatocellular Carcinoma. Front. Immunol. 2022, 13, 805967. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, Z.; Zhang, G.; Zhang, Z.; Luo, Y.; Wang, F.; Wang, S.; Che, Y.; Zeng, Q.; Sun, N.; et al. Clinical significance and inflammatory landscapes of a novel recurrence-associated immune signature in early-stage lung adenocarcinoma. Cancer Lett. 2020, 479, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Geeleher, P.; Cox, N.; Huang, R.S. pRRophetic: An R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE 2014, 9, e107468. [Google Scholar] [CrossRef]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef]
- Shankavaram, U.T.; Varma, S.; Kane, D.; Sunshine, M.; Chary, K.K.; Reinhold, W.C.; Pommier, Y.; Weinstein, J.N. CellMiner: A relational database and query tool for the NCI-60 cancer cell lines. BMC Genom. 2009, 10, 277. [Google Scholar] [CrossRef]
- Lee, T.L.; Xiao, A.; Rennert, O.M. Identification of novel long noncoding RNA transcripts in male germ cells. Methods Mol. Biol. 2012, 825, 105–114. [Google Scholar] [CrossRef]
- Huang, W.; Skanderup, A.J.; Lee, C.G. Advances in genomic hepatocellular carcinoma research. Gigascience 2018, 7, giy135. [Google Scholar] [CrossRef]
- Rizzo, A.; Ricci, A.D.; Brandi, G. PD-L1, TMB, MSI, and Other Predictors of Response to Immune Checkpoint Inhibitors in Biliary Tract Cancer. Cancers 2021, 13, 558. [Google Scholar] [CrossRef]
- Condelli, V.; Calice, G.; Cassano, A.; Basso, M.; Rodriquenz, M.G.; Zupa, A.; Maddalena, F.; Crispo, F.; Pietrafesa, M.; Aieta, M.; et al. Novel Epigenetic Eight-Gene Signature Predictive of Poor Prognosis and MSI-Like Phenotype in Human Metastatic Colorectal Carcinomas. Cancers 2021, 13, 158. [Google Scholar] [CrossRef]
- Shen, J.; Feng, X.P.; Hu, R.B.; Wang, H.; Wang, Y.L.; Qian, J.H.; Zhou, Y.X. N-methyladenosine reader YTHDF2-mediated long noncoding RNA FENDRR degradation promotes cell proliferation in endometrioid endometrial carcinoma. Lab. Investig. 2021, 101, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, Y.; Yao, B.; Sui, T.; Lai, L.; Li, Z. A novel N6-methyladenosine (m6A)-dependent fate decision for the lncRNA THOR. Cell Death Dis. 2020, 11, 613. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, J.H.; Wu, Q.N.; Jin, Y.; Wang, D.S.; Chen, Y.X.; Liu, J.; Luo, X.J.; Meng, Q.; Pu, H.Y.; et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol. Cancer 2019, 18, 174. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Wei, Y.; Zen, C.; Xiong, W.; Niu, Y.; Zhao, Y. Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol. Cancer 2020, 19, 171. [Google Scholar] [CrossRef]
- Wu, Z.H.; Li, Z.W.; Yang, D.L.; Liu, J. Development and Validation of a Pyroptosis-Related Long Non-coding RNA Signature for Hepatocellular Carcinoma. Front. Cell Dev. Biol. 2021, 9, 713925. [Google Scholar] [CrossRef]
- Xia, X.; Zhang, H.; Xia, P.; Zhu, Y.; Liu, J.; Xu, K.; Yuan, Y. Identification of Glycolysis-Related lncRNAs and the Novel lncRNA WAC-AS1 Promotes Glycolysis and Tumor Progression in Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 733595. [Google Scholar] [CrossRef]
- Zhou, P.; Lu, Y.; Zhang, Y.; Wang, L. Construction of an Immune-Related Six-lncRNA Signature to Predict the Outcomes, Immune Cell Infiltration, and Immunotherapy Response in Patients With Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 661758. [Google Scholar] [CrossRef]
- Chen, Z.A.; Tian, H.; Yao, D.M.; Zhang, Y.; Feng, Z.J.; Yang, C.J. Identification of a Ferroptosis-Related Signature Model Including mRNAs and lncRNAs for Predicting Prognosis and Immune Activity in Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 738477. [Google Scholar] [CrossRef]
- Müller, M.; Bird, T.G.; Nault, J.C. The landscape of gene mutations in cirrhosis and hepatocellular carcinoma. J. Hepatol. 2020, 72, 990–1002. [Google Scholar] [CrossRef]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021, 7, 6. [Google Scholar] [CrossRef]
- Zaanan, A.; Williet, N.; Hebbar, M.; Dabakuyo, T.S.; Fartoux, L.; Mansourbakht, T.; Dubreuil, O.; Rosmorduc, O.; Cattan, S.; Bonnetain, F.; et al. Gemcitabine plus oxaliplatin in advanced hepatocellular carcinoma: A large multicenter AGEO study. J. Hepatol. 2013, 58, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.C.; Lin, C.S.; Tai, W.T.; Liu, C.Y.; Shiau, C.W.; Chen, K.F. Nilotinib induces autophagy in hepatocellular carcinoma through AMPK activation. J. Biol. Chem. 2013, 288, 18249–18259. [Google Scholar] [CrossRef] [PubMed]
- Turpin, A.; de Baere, T.; Heurgué, A.; Le Malicot, K.; Ollivier-Hourmand, I.; Lecomte, T.; Perrier, H.; Vergniol, J.; Sefrioui, D.; Rinaldi, Y.; et al. Liver transarterial chemoembolization and sunitinib for unresectable hepatocellular carcinoma: Results of the PRODIGE 16 study. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101464. [Google Scholar] [CrossRef] [PubMed]
- Schiffer, E.; Housset, C.; Cacheux, W.; Wendum, D.; Desbois-Mouthon, C.; Rey, C.; Clergue, F.; Poupon, R.; Barbu, V.; Rosmorduc, O. Gefitinib, an EGFR inhibitor, prevents hepatocellular carcinoma development in the rat liver with cirrhosis. Hepatology 2005, 41, 307–314. [Google Scholar] [CrossRef]
- Scheipl, S.; Barnard, M.; Cottone, L.; Jorgensen, M.; Drewry, D.H.; Zuercher, W.J.; Turlais, F.; Ye, H.; Leite, A.P.; Smith, J.A.; et al. EGFR inhibitors identified as a potential treatment for chordoma in a focused compound screen. J. Pathol. 2016, 239, 320–334. [Google Scholar] [CrossRef] [PubMed]


| Gene | AL031985.3 | NRAV | WAC-AS1 | AC026412.3 | AC099850.4 |
| Coef | 0.2752020 | 0.0816187 | 0.0071720 | 0.5638878 | 0.0049139 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, D.; Wang, X.; Wang, Y.; Liang, W.; Luo, J.; Zheng, J.; Zhu, K. Integrated Analysis of N1-Methyladenosine Methylation Regulators-Related lncRNAs in Hepatocellular Carcinoma. Cancers 2023, 15, 1800. https://doi.org/10.3390/cancers15061800
Song D, Wang X, Wang Y, Liang W, Luo J, Zheng J, Zhu K. Integrated Analysis of N1-Methyladenosine Methylation Regulators-Related lncRNAs in Hepatocellular Carcinoma. Cancers. 2023; 15(6):1800. https://doi.org/10.3390/cancers15061800
Chicago/Turabian StyleSong, Danjun, Xi Wang, Yining Wang, Weiren Liang, Jun Luo, Jiaping Zheng, and Kai Zhu. 2023. "Integrated Analysis of N1-Methyladenosine Methylation Regulators-Related lncRNAs in Hepatocellular Carcinoma" Cancers 15, no. 6: 1800. https://doi.org/10.3390/cancers15061800
APA StyleSong, D., Wang, X., Wang, Y., Liang, W., Luo, J., Zheng, J., & Zhu, K. (2023). Integrated Analysis of N1-Methyladenosine Methylation Regulators-Related lncRNAs in Hepatocellular Carcinoma. Cancers, 15(6), 1800. https://doi.org/10.3390/cancers15061800




