Differential Spatial Gene and Protein Expression Associated with Recurrence Following Chemoradiation for Localized Anal Squamous Cell Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Patient Samples
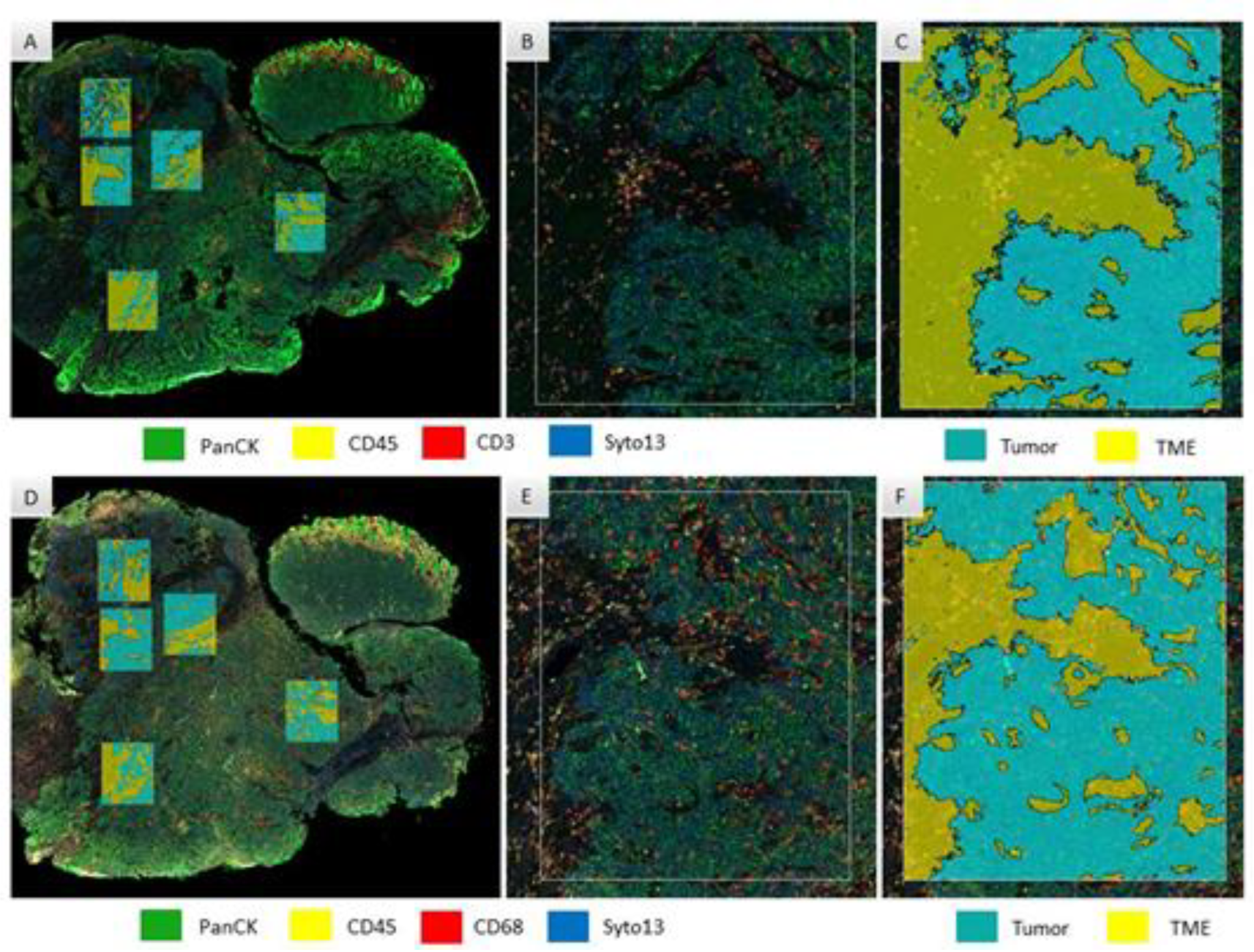
2.2. Digital Spatial Profiling
2.3. Selection of Regions of Interest (ROI)
2.4. Data Analysis
3. Results
3.1. Demographics
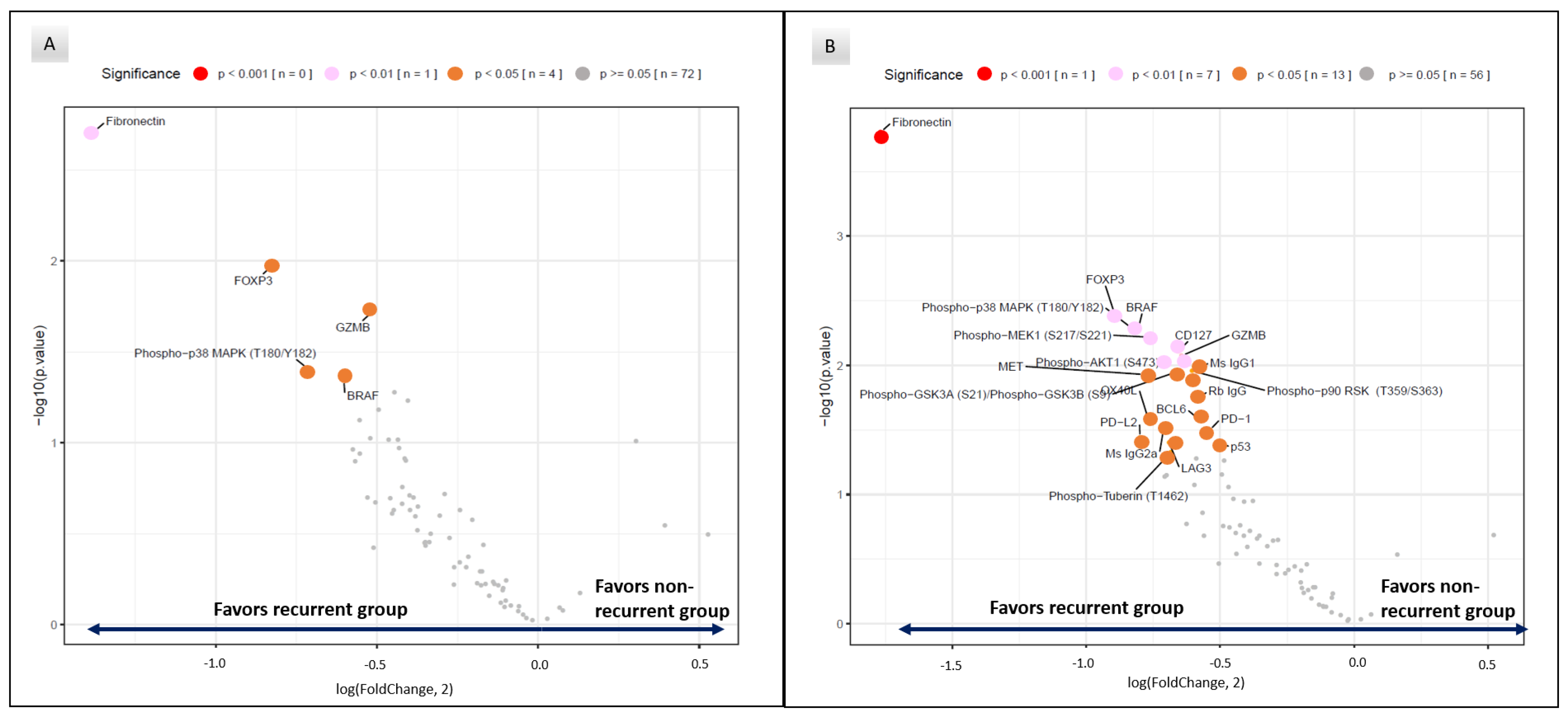
3.2. Differential Protein Expression between Recurrent and Non-Recurrent Patients
3.3. RNA Expression in Recurrent and Non-Recurrent Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Vuyst, H.; Clifford, G.M.; Nascimento, M.C.; Madeleine, M.M.; Franceschi, S. Prevalence and type distribution of human papillomavirus in carcinoma and intraepithelial neoplasia of the vulva, vagina and anus: A meta-analysis. Int. J. Cancer 2009, 124, 1626–1636. [Google Scholar] [CrossRef]
- Group, F.I.S. Quadrivalent vaccine against human papillomavirus to prevent high-grade cervical lesions. N. Engl. J. Med. 2007, 356, 1915–1927. [Google Scholar] [CrossRef]
- Garland, S.M.; Hernandez-Avila, M.; Wheeler, C.M.; Perez, G.; Harper, D.M.; Leodolter, S.; Tang, G.W.; Ferris, D.G.; Steben, M.; Bryan, J.; et al. Quadrivalent vaccine against human papillomavirus to prevent anogenital diseases. N. Engl. J. Med. 2007, 356, 1928–1943. [Google Scholar] [CrossRef]
- Giuliano, A.R.; Palefsky, J.M.; Goldstone, S.; Moreira, E.D., Jr.; Penny, M.E.; Aranda, C.; Vardas, E.; Moi, H.; Jessen, H.; Hillman, R.; et al. Efficacy of quadrivalent HPV vaccine against HPV Infection and disease in males. N. Engl. J. Med. 2011, 364, 401–411. [Google Scholar] [CrossRef]
- Deshmukh, A.A.; Suk, R.; Shiels, M.S.; Sonawane, K.; Nyitray, A.G.; Liu, Y.; Gaisa, M.M.; Palefsky, J.M.; Sigel, K. Recent Trends in Squamous Cell Carcinoma of the Anus Incidence and Mortality in the United States, 2001-2015. J. Natl. Cancer Inst. 2020, 112, 829–838. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Gunderson, L.L.; Winter, K.A.; Ajani, J.A.; Pedersen, J.E.; Moughan, J.; Benson, A.B., 3rd; Thomas, C.R., Jr.; Mayer, R.J.; Haddock, M.G.; Rich, T.A.; et al. Long-term update of US GI intergroup RTOG 98-11 phase III trial for anal carcinoma: Survival, relapse, and colostomy failure with concurrent chemoradiation involving fluorouracil/mitomycin versus fluorouracil/cisplatin. J. Clin. Oncol. 2012, 30, 4344–4351. [Google Scholar] [CrossRef]
- James, R.D.; Glynne-Jones, R.; Meadows, H.M.; Cunningham, D.; Myint, A.S.; Saunders, M.P.; Maughan, T.; McDonald, A.; Essapen, S.; Leslie, M.; et al. Mitomycin or cisplatin chemoradiation with or without maintenance chemotherapy for treatment of squamous-cell carcinoma of the anus (ACT II): A randomised, phase 3, open-label, 2 x 2 factorial trial. Lancet Oncol. 2013, 14, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Schiller, D.E.; Cummings, B.J.; Rai, S.; Le, L.W.; Last, L.; Davey, P.; Easson, A.; Smith, A.J.; Swallow, C.J. Outcomes of salvage surgery for squamous cell carcinoma of the anal canal. Ann. Surg. Oncol. 2007, 14, 2780–2789. [Google Scholar] [CrossRef] [PubMed]
- Mullen, J.T.; Rodriguez-Bigas, M.A.; Chang, G.J.; Barcenas, C.H.; Crane, C.H.; Skibber, J.M.; Feig, B.W. Results of surgical salvage after failed chemoradiation therapy for epidermoid carcinoma of the anal canal. Ann. Surg. Oncol. 2007, 14, 478–483. [Google Scholar] [CrossRef]
- Ajani, J.A.; Winter, K.A.; Gunderson, L.L.; Pedersen, J.; Benson, A.B., 3rd; Thomas, C.R., Jr.; Mayer, R.J.; Haddock, M.G.; Rich, T.A.; Willett, C. Fluorouracil, mitomycin, and radiotherapy vs fluorouracil, cisplatin, and radiotherapy for carcinoma of the anal canal: A randomized controlled trial. JAMA 2008, 299, 1914–1921. [Google Scholar] [CrossRef]
- Bartelink, H.; Roelofsen, F.; Eschwege, F.; Rougier, P.; Bosset, J.F.; Gonzalez, D.G.; Peiffert, D.; van Glabbeke, M.; Pierart, M. Concomitant radiotherapy and chemotherapy is superior to radiotherapy alone in the treatment of locally advanced anal cancer: Results of a phase III randomized trial of the European Organization for Research and Treatment of Cancer Radiotherapy and Gastrointestinal Cooperative Groups. J. Clin. Oncol. 1997, 15, 2040–2049. [Google Scholar] [CrossRef]
- Glynne-Jones, R.; Sebag-Montefiore, D.; Adams, R.; Gollins, S.; Harrison, M.; Meadows, H.M.; Jitlal, M.; United Kingdom Coordinating Committee on Cancer Research Anal Cancer Trial Working, P. Prognostic factors for recurrence and survival in anal cancer: Generating hypotheses from the mature outcomes of the first United Kingdom Coordinating Committee on Cancer Research Anal Cancer Trial (ACT I). Cancer 2013, 119, 748–755. [Google Scholar] [CrossRef]
- Das, P.; Bhatia, S.; Eng, C.; Ajani, J.A.; Skibber, J.M.; Rodriguez-Bigas, M.A.; Chang, G.J.; Bhosale, P.; Delclos, M.E.; Krishnan, S.; et al. Predictors and patterns of recurrence after definitive chemoradiation for anal cancer. Int. J. Radiat. Oncol. Biol. Phys. 2007, 68, 794–800. [Google Scholar] [CrossRef]
- Chung, J.H.; Sanford, E.; Johnson, A.; Klempner, S.J.; Schrock, A.B.; Palma, N.A.; Erlich, R.L.; Frampton, G.M.; Chalmers, Z.R.; Vergilio, J.; et al. Comprehensive genomic profiling of anal squamous cell carcinoma reveals distinct genomically defined classes. Ann. Oncol. 2016, 27, 1336–1341. [Google Scholar] [CrossRef] [PubMed]
- Morris, V.; Rao, X.; Pickering, C.; Foo, W.C.; Rashid, A.; Eterovic, K.; Kim, T.; Chen, K.; Wang, J.; Shaw, K.; et al. Comprehensive Genomic Profiling of Metastatic Squamous Cell Carcinoma of the Anal Canal. Mol. Cancer Res. 2017, 15, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Marabelle, A.; Fakih, M.; Lopez, J.; Shah, M.; Shapira-Frommer, R.; Nakagawa, K.; Chung, H.C.; Kindler, H.L.; Lopez-Martin, J.A.; Miller, W.H., Jr.; et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: Prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020, 21, 1353–1365. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Marabelle, A.; Cassier, P.A.; Fakih, M.; Guren, T.K.; Italiano, A.; Kao, S.C.-H.; Nielsen, D.; Ascierto, P.A.; Bariani, G.M.; Santoro, A.; et al. Pembrolizumab for advanced anal squamous cell carcinoma (ASCC): Results from the multicohort, phase II KEYNOTE-158 study. J. Clin. Oncol. 2020, 38 (Suppl. 4), 1. [Google Scholar] [CrossRef]
- Rao, S.; Anandappa, G.; Capdevila, J.; Dahan, L.; Evesque, L.; Kim, S.; Saunders, M.P.; Gilbert, D.C.; Jensen, L.H.; Samalin, E.; et al. A phase II study of retifanlimab (INCMGA00012) in patients with squamous carcinoma of the anal canal who have progressed following platinum-based chemotherapy (POD1UM-202). ESMO Open 2022, 7, 100529. [Google Scholar] [CrossRef]
- Morris, V.K.; Salem, M.E.; Nimeiri, H.; Iqbal, S.; Singh, P.; Ciombor, K.; Polite, B.; Deming, D.; Chan, E.; Wade, J.L.; et al. Nivolumab for previously treated unresectable metastatic anal cancer (NCI9673): A multicentre, single-arm, phase 2 study. Lancet Oncol. 2017, 18, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Merritt, C.R.; Ong, G.T.; Church, S.E.; Barker, K.; Danaher, P.; Geiss, G.; Hoang, M.; Jung, J.; Liang, Y.; McKay-Fleisch, J.; et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 2020, 38, 586–599. [Google Scholar] [CrossRef] [PubMed]
- NanoString, I. GeoMx® Protein Assays for Immuno-Oncology. Available online: https://nanostring.com/wp-content/uploads/PB_MK3350_DSP_IO_Protein_PB_R28.pdf (accessed on 5 September 2022).
- NanoString, I. GeoMx® DSP Automated Slide Preparation User Manual. Available online: https://university.nanostring.com/geomx-dsp-automated-slide-preparation-user-manual/1209595 (accessed on 27 June 2022).
- Zhou, Y.; Shu, C.; Huang, Y. Fibronectin promotes cervical cancer tumorigenesis through activating FAK signaling pathway. J. Cell. Biochem. 2019, 120, 10988–10997. [Google Scholar] [CrossRef] [PubMed]
- Gopal, S.; Veracini, L.; Grall, D.; Butori, C.; Schaub, S.; Audebert, S.; Camoin, L.; Baudelet, E.; Radwanska, A.; Beghelli-de la Forest Divonne, S.; et al. Fibronectin-guided migration of carcinoma collectives. Nat. Commun. 2017, 8, 14105. [Google Scholar] [CrossRef]
- Ramos Gde, O.; Bernardi, L.; Lauxen, I.; Sant’Ana Filho, M.; Horwitz, A.R.; Lamers, M.L. Fibronectin Modulates Cell Adhesion and Signaling to Promote Single Cell Migration of Highly Invasive Oral Squamous Cell Carcinoma. PLoS ONE 2016, 11, e0151338. [Google Scholar] [CrossRef]
- Zhou, W.H.; Du, W.D.; Li, Y.F.; Al-Aroomi, M.A.; Yan, C.; Wang, Y.; Zhang, Z.Y.; Liu, F.Y.; Sun, C.F. The Overexpression of Fibronectin 1 Promotes Cancer Progression and Associated with M2 Macrophages Polarization in Head and Neck Squamous Cell Carcinoma Patients. Int. J. Gen. Med. 2022, 15, 5027–5042. [Google Scholar] [CrossRef]
- Hynes, R.O.; Naba, A. Overview of the matrisome--an inventory of extracellular matrix constituents and functions. Cold Spring Harb. Perspect. Biol. 2012, 4, a004903. [Google Scholar] [CrossRef]
- Hori, S.; Sakaguchi, S. Foxp3: A critical regulator of the development and function of regulatory T cells. Microbes Infect. 2004, 6, 745–751. [Google Scholar] [CrossRef]
- Takahashi, T.; Kuniyasu, Y.; Toda, M.; Sakaguchi, N.; Itoh, M.; Iwata, M.; Shimizu, J.; Sakaguchi, S. Immunologic self-tolerance maintained by CD25+CD4+ naturally anergic and suppressive T cells: Induction of autoimmune disease by breaking their anergic/suppressive state. Int. Immunol 1998, 10, 1969–1980. [Google Scholar] [CrossRef]
- Thornton, A.M.; Shevach, E.M. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J. Exp. Med. 1998, 188, 287–296. [Google Scholar] [CrossRef]
- Morris, V.; Liu, S.; Johnson, B.; Prasad, S.; Mahvash, A.; Bhosale, P.; Rubin, M.L.; Rothschild, N.; Futreal, A.; Wistuba, I.I.; et al. 403MO Atezolizumab in combination with bevacizumab for patients with unresectable/metastatic anal cancer. Ann. Oncol. 2020, 31, S412. [Google Scholar] [CrossRef]
- Lonardi, S.; Prete, A.A.; Morano, F.; Messina, M.; Formica, V.; Corsi, D.C.; Orciuolo, C.; Frassineti, G.L.; Zampino, M.G.; Casagrande, M.; et al. Randomized phase II trial of avelumab alone or in combination with cetuximab for patients with previously treated, locally advanced, or metastatic squamous cell anal carcinoma: The CARACAS study. J. Immunother. Cancer 2021, 9, e002996. [Google Scholar] [CrossRef]
- Gupta, A.K.; McKenna, W.G.; Weber, C.N.; Feldman, M.D.; Goldsmith, J.D.; Mick, R.; Machtay, M.; Rosenthal, D.I.; Bakanauskas, V.J.; Cerniglia, G.J.; et al. Local recurrence in head and neck cancer: Relationship to radiation resistance and signal transduction. Clin. Cancer Res. 2002, 8, 885–892. [Google Scholar]
- Brognard, J.; Clark, A.S.; Ni, Y.; Dennis, P.A. Akt/protein kinase B is constitutively active in non-small cell lung cancer cells and promotes cellular survival and resistance to chemotherapy and radiation. Cancer Res. 2001, 61, 3986–3997. [Google Scholar]
- Tanno, S.; Yanagawa, N.; Habiro, A.; Koizumi, K.; Nakano, Y.; Osanai, M.; Mizukami, Y.; Okumura, T.; Testa, J.R.; Kohgo, Y. Serine/threonine kinase AKT is frequently activated in human bile duct cancer and is associated with increased radioresistance. Cancer Res. 2004, 64, 3486–3490. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, J.; Yuan, K.; Wu, Z.; Hu, L.; Lu, Y.; Li, K.; Guo, J.; Chen, J.; Ma, C.; et al. The oncoprotein BCL6 enables solid tumor cells to evade genotoxic stress. Elife 2022, 11, e69255. [Google Scholar] [CrossRef]
- Fabre, M.S.; Stanton, N.M.; Slatter, T.L.; Lee, S.; Senanayake, D.; Gordon, R.M.A.; Castro, M.L.; Rowe, M.R.; Taha, A.; Royds, J.A.; et al. The oncogene BCL6 is up-regulated in glioblastoma in response to DNA damage, and drives survival after therapy. PLoS ONE 2020, 15, e0231470. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Peng, J.; Zhang, H.; Mondesire, W.H.; Jian, W.; Mills, G.B.; Hung, M.C.; Meric-Bernstam, F. Role of glycogen synthase kinase 3beta in rapamycin-mediated cell cycle regulation and chemosensitivity. Cancer Res. 2005, 65, 1961–1972. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Wang, J.; Gao, Y.; Gao, T.W.; Chen, G.; Bower, K.A.; Odetallah, M.; Ding, M.; Ke, Z.; Luo, J. The role of glycogen synthase kinase 3beta in the transformation of epidermal cells. Cancer Res. 2007, 67, 7756–7764. [Google Scholar] [CrossRef] [PubMed]
- Garg, M.K.; Zhao, F.; Sparano, J.A.; Palefsky, J.; Whittington, R.; Mitchell, E.P.; Mulcahy, M.F.; Armstrong, K.I.; Nabbout, N.H.; Kalnicki, S.; et al. Cetuximab Plus Chemoradiotherapy in Immunocompetent Patients With Anal Carcinoma: A Phase II Eastern Cooperative Oncology Group-American College of Radiology Imaging Network Cancer Research Group Trial (E3205). J. Clin. Oncol. 2017, 35, 718–726. [Google Scholar] [CrossRef] [PubMed]
- Sparano, J.A.; Lee, J.Y.; Palefsky, J.; Henry, D.H.; Wachsman, W.; Rajdev, L.; Aboulafia, D.; Ratner, L.; Fitzgerald, T.J.; Kachnic, L.; et al. Cetuximab Plus Chemoradiotherapy for HIV-Associated Anal Carcinoma: A Phase II AIDS Malignancy Consortium Trial. J. Clin. Oncol. 2017, 35, 727–733. [Google Scholar] [CrossRef]
- Olivatto, L.O.; Vieira, F.M.; Pereira, B.V.; Victorino, A.P.; Bezerra, M.; Araujo, C.M.; Erlich, F.; Faroni, L.; Castro, L.; Lusis, E.C.; et al. Phase 1 study of cetuximab in combination with 5-fluorouracil, cisplatin, and radiotherapy in patients with locally advanced anal canal carcinoma. Cancer 2013, 119, 2973–2980. [Google Scholar] [CrossRef]
- Deutsch, E.; Lemanski, C.; Pignon, J.P.; Levy, A.; Delarochefordiere, A.; Martel-Lafay, I.; Rio, E.; Malka, D.; Conroy, T.; Miglianico, L.; et al. Unexpected toxicity of cetuximab combined with conventional chemoradiotherapy in patients with locally advanced anal cancer: Results of the UNICANCER ACCORD 16 phase II trial. Ann. Oncol. 2013, 24, 2834–2838. [Google Scholar] [CrossRef]
- Sklar, M.D. The ras oncogenes increase the intrinsic resistance of NIH 3T3 cells to ionizing radiation. Science 1988, 239, 645–647. [Google Scholar] [CrossRef]
- Bernhard, E.J.; Stanbridge, E.J.; Gupta, S.; Gupta, A.K.; Soto, D.; Bakanauskas, V.J.; Cerniglia, G.J.; Muschel, R.J.; McKenna, W.G. Direct evidence for the contribution of activated N-ras and K-ras oncogenes to increased intrinsic radiation resistance in human tumor cell lines. Cancer Res. 2000, 60, 6597–6600. [Google Scholar]
- McKenna, W.G.; Bernhard, E.J.; Markiewicz, D.A.; Rudoltz, M.S.; Maity, A.; Muschel, R.J. Regulation of radiation-induced apoptosis in oncogene-transfected fibroblasts: Influence of H-ras on the G2 delay. Oncogene 1996, 12, 237–245. [Google Scholar]
- Dror, S.; Sander, L.; Schwartz, H.; Sheinboim, D.; Barzilai, A.; Dishon, Y.; Apcher, S.; Golan, T.; Greenberger, S.; Barshack, I.; et al. Melanoma miRNA trafficking controls tumour primary niche formation. Nat. Cell Biol. 2016, 18, 1006–1017. [Google Scholar] [CrossRef] [PubMed]
- Wentzensen, N.; Sherman, M.E.; Schiffman, M.; Wang, S.S. Utility of methylation markers in cervical cancer early detection: Appraisal of the state-of-the-science. Gynecol. Oncol. 2009, 112, 293–299. [Google Scholar] [CrossRef]
| Non-Recurrent (N = 25) | Recurrent (N = 23) | p-Value * | |
|---|---|---|---|
| Age, years (SD) ** | 59.5 (10.0) | 56.6 (9.0) | 0.30 |
| Gender (%) | 0.10 | ||
| Female | 18 (72) | 21 (91) | |
| Male | 7 (28) | 2 (9) | |
| Ethnicity (%) *** | 0.22 | ||
| African-American | 0 (0) | 2 (9) | |
| Caucasian | 22 (88) | 20 (87) | |
| Hispanic | 3 (12) | 1 (4) | |
| Stage at Diagnosis (%) | 0.36 | ||
| I | 1 (4) | 0 (0) | |
| II | 9 (36) | 5 (22) | |
| III | 15 (60) | 17 (74) | |
| IV | 0 (0) | 1 (4) | |
| HPV status (%) | 10.0 | ||
| Positive | 18 (72) | 18 (78) | |
| Negative | 1 (4) | 1 (4) | |
| Not available | 6 (24) | 4 (17) | |
| HIV Status (%) | 0.46 | ||
| Negative | 25 (100) | 22 (96) | |
| Positive | 0 (0) | 1 (4) | |
| Differentiation (%) | 0.56 | ||
| Well | 1 (4) | 0 (0) | |
| Moderately | 10 (40) | 10 (43) | |
| Poorly | 11 (44) | 13 (57) | |
| Not available | 3 (12) | 0 (0) | |
| Coexisting autoimmune disease (%) | 0.36 | ||
| Absent | 22 (88) | 22 (94) | |
| Present | 3 (12) | 1 (4) | |
| Chemotherapy with radiation | 0.01 | ||
| Fluoropyrimidine + | |||
| mitomycin C | 5 (20) | 13 (57) | |
| Fluoropyrimidine + | |||
| cisplatin | 20 (80) | 10 (4) |
| Biomarker | Expression Fold Change (Recurrent: Non-Recurrent) | p-Value * |
|---|---|---|
| Fibronectin | 2.59 | 0.002 |
| FoxP3 | 1.77 | 0.005 |
| GZMB | 1.43 | 0.02 |
| Phospho−p38 MAPK (T180/Y182) | 1.64 | 0.04 |
| BRAF | 1.52 | 0.045 |
| Biomarker | Fold Change (Recurrent: Non-Recurrent) | p-Value * |
|---|---|---|
| Fibronectin | 3.41 | 0.0002 |
| FOXP3 | 1.86 | 0.004 |
| GZMB | 1.56 | 0.008 |
| Phospho−p38 MAPK (T180/Y182) | 1.76 | 0.006 |
| BRAF | 1.75 | 0.005 |
| CD127 | 1.58 | 0.008 |
| PD−L2 | 1.74 | 0.04 |
| OX40L | 1.70 | 0.03 |
| PD-1 | 1.45 | 0.03 |
| LAG3 | 1.61 | 0.04 |
| BCL6 | 1.50 | 0.03 |
| p53 | 1.42 | 0.04 |
| MET | 1.72 | 0.01 |
| Phospho−GSK3A (S21)/Phospho−GSK3B (S9) | 1.58 | 0.01 |
| Phospho−MEK1 (S217/S221) | 1.69 | 0.006 |
| Phospho−p90 RSK (T359/S363) | 1.52 | 0.01 |
| Phospho−AKT1 (S473) | 1.63 | 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernandez, S.; Das, P.; Holliday, E.B.; Shen, L.; Lu, W.; Johnson, B.; Messick, C.A.; Taniguchi, C.M.; Skibber, J.; Ludmir, E.B.; et al. Differential Spatial Gene and Protein Expression Associated with Recurrence Following Chemoradiation for Localized Anal Squamous Cell Cancer. Cancers 2023, 15, 1701. https://doi.org/10.3390/cancers15061701
Hernandez S, Das P, Holliday EB, Shen L, Lu W, Johnson B, Messick CA, Taniguchi CM, Skibber J, Ludmir EB, et al. Differential Spatial Gene and Protein Expression Associated with Recurrence Following Chemoradiation for Localized Anal Squamous Cell Cancer. Cancers. 2023; 15(6):1701. https://doi.org/10.3390/cancers15061701
Chicago/Turabian StyleHernandez, Sharia, Prajnan Das, Emma B. Holliday, Li Shen, Wei Lu, Benny Johnson, Craig A. Messick, Cullen M. Taniguchi, John Skibber, Ethan B. Ludmir, and et al. 2023. "Differential Spatial Gene and Protein Expression Associated with Recurrence Following Chemoradiation for Localized Anal Squamous Cell Cancer" Cancers 15, no. 6: 1701. https://doi.org/10.3390/cancers15061701
APA StyleHernandez, S., Das, P., Holliday, E. B., Shen, L., Lu, W., Johnson, B., Messick, C. A., Taniguchi, C. M., Skibber, J., Ludmir, E. B., You, Y. N., Smith, G. L., Bednarski, B., Kostousov, L., Koay, E. J., Minsky, B. D., Tillman, M., Portier, S., Eng, C., ... Morris, V. K. (2023). Differential Spatial Gene and Protein Expression Associated with Recurrence Following Chemoradiation for Localized Anal Squamous Cell Cancer. Cancers, 15(6), 1701. https://doi.org/10.3390/cancers15061701







