Interactions of SNPs in Folate Metabolism Related Genes on Prostate Cancer Aggressiveness in European Americans and African Americans
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Genotyping
2.3. Statistical Analyses
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.M.; Wender, R.C.; Etzioni, R.B.; Thompson, I.M.; D’Amico, A.V.; Volk, R.J.; Brooks, D.D.; Dash, C.; Guessous, I.; Andrews, K.; et al. American Cancer Society guideline for the early detection of prostate cancer: Update 2010. CA Cancer J. Clin. 2010, 60, 70–98. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Cancer statistics for African Americans, 2019. CA Cancer J. Clin. 2019, 69, 211–233. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Cancer Facts & Figures; American Cancer Society: Atlanta, GA, USA, 2021. [Google Scholar]
- Kim, Y.I. Folate: A magic bullet or a double edged sword for colorectal cancer prevention? Gut 2006, 55, 1387–1389. [Google Scholar] [CrossRef] [PubMed]
- Wien, T.N.; Pike, E.; Wisloff, T.; Staff, A.; Smeland, S.; Klemp, M. Cancer risk with folic acid supplements: A systematic review and meta-analysis. BMJ Open 2012, 2, e000653. [Google Scholar] [CrossRef]
- Figueiredo, J.C.; Grau, M.V.; Haile, R.W.; Sandler, R.S.; Summers, R.W.; Bresalier, R.S.; Burke, C.A.; McKeown-Eyssen, G.E.; Baron, J.A. Folic acid and risk of prostate cancer: Results from a randomized clinical trial. J. Natl. Cancer Inst. 2009, 101, 432–435. [Google Scholar] [CrossRef]
- Pelucchi, C.; Galeone, C.; Talamini, R.; Negri, E.; Parpinel, M.; Franceschi, S.; Montella, M.; La Vecchia, C. Dietary folate and risk of prostate cancer in Italy. Cancer Epidemiol. Biomark. Prev. 2005, 14, 944–948. [Google Scholar] [CrossRef]
- Weinstein, S.J.; Stolzenberg-Solomon, R.; Pietinen, P.; Taylor, P.R.; Virtamo, J.; Albanes, D. Dietary factors of one-carbon metabolism and prostate cancer risk. Am. J. Clin. Nutr. 2006, 84, 929–935. [Google Scholar] [CrossRef]
- Vlajinac, H.D.; Marinkovic, J.M.; Ilic, M.D.; Kocev, N.I. Diet and prostate cancer: A case-control study. Eur. J. Cancer 1997, 33, 101–107. [Google Scholar] [CrossRef]
- Stevens, V.L.; Rodriguez, C.; Pavluck, A.L.; McCullough, M.L.; Thun, M.J.; Calle, E.E. Folate nutrition and prostate cancer incidence in a large cohort of US men. Am. J. Epidemiol. 2006, 163, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Lawson, K.A.; Wright, M.E.; Subar, A.; Mouw, T.; Hollenbeck, A.; Schatzkin, A.; Leitzmann, M.F. Multivitamin use and risk of prostate cancer in the National Institutes of Health-AARP Diet and Health Study. J. Natl. Cancer Inst. 2007, 99, 754–764. [Google Scholar] [CrossRef] [PubMed]
- McCann, S.E.; Ambrosone, C.B.; Moysich, K.B.; Brasure, J.; Marshall, J.R.; Freudenheim, J.L.; Wilkinson, G.S.; Graham, S. Intakes of selected nutrients, foods, and phytochemicals and prostate cancer risk in western New York. Nutr. Cancer 2005, 53, 33–41. [Google Scholar] [CrossRef]
- Shannon, J.; Phoutrides, E.; Palma, A.; Farris, P.; Peters, L.; Forester, A.; Tillotson, C.J.; Garzotto, M. Folate intake and prostate cancer risk: A case-control study. Nutr. Cancer 2009, 61, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Aune, D.; Deneo-Pellegrini, H.; Ronco, A.L.; Boffetta, P.; Acosta, G.; Mendilaharsu, M.; De Stefani, E. Dietary folate intake and the risk of 11 types of cancer: A case-control study in Uruguay. Ann. Oncol. 2011, 22, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Hung, J.; Beilby, J.P.; Knuiman, M.W.; Divitini, M.L.; Bartholomew, H. Folate levels and cancer morbidity and mortality: Prospective cohort study from Busselton, Western Australia. Ann. Epidemiol. 2006, 16, 206–212. [Google Scholar] [CrossRef] [PubMed]
- American Institute for Cancer Research, World Cancer Research Fund. Continuous Update Project Report. Analysing Research on Cancer Prevention and Survival. Diet, Nutrition, Physical Activity and Prostate Cancer; American Institute for Cancer Research: Washington, DC, USA, 2014. [Google Scholar]
- Jackson, M.D.; Tulloch-Reid, M.K.; McFarlane-Anderson, N.; Watson, A.; Seers, V.; Bennett, F.I.; Egleston, B.; Ragin, C. Complex interaction between serum folate levels and genetic polymorphisms in folate pathway genes: Biomarkers of prostate cancer aggressiveness. Genes Nutr. 2013, 8, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Tio, M.; Andrici, J.; Cox, M.R.; Eslick, G.D. Folate intake and the risk of prostate cancer: A systematic review and meta-analysis. Prostate Cancer Prostatic Dis. 2014, 17, 213–219. [Google Scholar] [CrossRef]
- Vollset, S.E.; Clarke, R.; Lewington, S.; Ebbing, M.; Halsey, J.; Lonn, E.; Armitage, J.; Manson, J.E.; Hankey, G.J.; Spence, J.D.; et al. Effects of folic acid supplementation on overall and site-specific cancer incidence during the randomised trials: Meta-analyses of data on 50,000 individuals. Lancet 2013, 381, 1029–1036. [Google Scholar] [CrossRef]
- Qin, X.; Cui, Y.; Shen, L.; Sun, N.; Zhang, Y.; Li, J.; Xu, X.; Wang, B.; Xu, X.; Huo, Y.; et al. Folic acid supplementation and cancer risk: A meta-analysis of randomized controlled trials. Int. J. Cancer 2013, 133, 1033–1041. [Google Scholar] [CrossRef]
- Bailey, R.L.; Mills, J.L.; Yetley, E.A.; Gahche, J.J.; Pfeiffer, C.M.; Dwyer, J.T.; Dodd, K.W.; Sempos, C.T.; Betz, J.M.; Picciano, M.F. Serum unmetabolized folic acid in a nationally representative sample of adults >/=60 years in the United States, 2001–2002. Food Nutr. Res. 2012, 56, 5616. [Google Scholar] [CrossRef]
- Choi, S.W. (Ed.) Interaction between Folate and Methylene-Tetrahydrofolate Reductase Gene in Cancer; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Liu, A.Y.; Scherer, D.; Poole, E.; Potter, J.D.; Curtin, K.; Makar, K.; Slattery, M.L.; Caan, B.J.; Ulrich, C.M. Gene-diet-interactions in folate-mediated one-carbon metabolism modify colon cancer risk. Mol. Nutr. Food Res. 2013, 57, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, J.C.; Levine, A.J.; Crott, J.W.; Baurley, J.; Haile, R.W. Folate-genetics and colorectal neoplasia: What we know and need to know next. Mol. Nutr. Food Res. 2013, 57, 607–627. [Google Scholar] [CrossRef] [PubMed]
- Orjuela, M.A.; Cabrera-Munoz, L.; Paul, L.; Ramirez-Ortiz, M.A.; Liu, X.; Chen, J.; Mejia-Rodriguez, F.; Medina-Sanson, A.; Diaz-Carreno, S.; Suen, I.H.; et al. Risk of retinoblastoma is associated with a maternal polymorphism in dihydrofolatereductase (DHFR) and prenatal folic acid intake. Cancer 2012, 118, 5912–5919. [Google Scholar] [CrossRef] [PubMed]
- Kalmbach, R.D.; Choumenkovitch, S.F.; Troen, A.P.; Jacques, P.F.; D’Agostino, R.; Selhub, J. A 19-base pair deletion polymorphism in dihydrofolate reductase is associated with increased unmetabolized folic acid in plasma and decreased red blood cell folate. J. Nutr. 2008, 138, 2323–2327. [Google Scholar] [CrossRef] [PubMed]
- Kimura, F.; Franke, K.H.; Steinhoff, C.; Golka, K.; Roemer, H.C.; Anastasiadis, A.G.; Schulz, W.A. Methyl group metabolism gene polymorphisms and susceptibility to prostatic carcinoma. Prostate 2000, 45, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Cicek, M.S.; Nock, N.L.; Li, L.; Conti, D.V.; Casey, G.; Witte, J.S. Relationship between methylenetetrahydrofolate reductase C677T and A1298C genotypes and haplotypes and prostate cancer risk and aggressiveness. Cancer Epidemiol. Biomark. Prev. 2004, 13, 1331–1336. [Google Scholar] [CrossRef]
- Collin, S.M.; Metcalfe, C.; Zuccolo, L.; Lewis, S.J.; Chen, L.; Cox, A.; Davis, M.; Lane, J.A.; Donovan, J.; Smith, G.D.; et al. Association of folate-pathway gene polymorphisms with the risk of prostate cancer: A population-based nested case-control study, systematic review, and meta-analysis. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2528–2539. [Google Scholar] [CrossRef]
- Stevens, V.L.; Rodriguez, C.; Sun, J.; Talbot, J.T.; Thun, M.J.; Calle, E.E. No association of single nucleotide polymorphisms in one-carbon metabolism genes with prostate cancer risk. Cancer Epidemiol. Biomark. Prev. 2008, 17, 3612–3614. [Google Scholar] [CrossRef]
- Singal, R.; Ferdinand, L.; Das, P.M.; Reis, I.M.; Schlesselman, J.J. Polymorphisms in the methylenetetrahydrofolate reductase gene and prostate cancer risk. Int. J. Oncol. 2004, 25, 1465–1471. [Google Scholar] [CrossRef]
- Van Guelpen, B.R.; Wiren, S.M.; Bergh, A.R.; Hallmans, G.; Stattin, P.E.; Hultdin, J. Polymorphisms of methylenetetrahydrofolate reductase and the risk of prostate cancer: A nested case-control study. Eur. J. Cancer Prev. 2006, 15, 46–50. [Google Scholar] [CrossRef]
- Safarinejad, M.R.; Shafiei, N.; Safarinejad, S. Relationship between three polymorphisms of methylenetetrahydrofolate reductase (MTHFR C677T, A1298C, and G1793A) gene and risk of prostate cancer: A case-control study. Prostate 2010, 70, 1645–1657. [Google Scholar] [CrossRef] [PubMed]
- Heijmans, B.T.; Boer, J.M.; Suchiman, H.E.; Cornelisse, C.J.; Westendorp, R.G.; Kromhout, D.; Feskens, E.J.; Slagboom, P.E. A common variant of the methylenetetrahydrofolate reductase gene (1p36) is associated with an increased risk of cancer. Cancer Res. 2003, 63, 1249–1253. [Google Scholar] [PubMed]
- Hultdin, J.; Van Guelpen, B.; Bergh, A.; Hallmans, G.; Stattin, P. Plasma folate, vitamin B12, and homocysteine and prostate cancer risk: A prospective study. Int. J. Cancer 2005, 113, 819–824. [Google Scholar] [CrossRef] [PubMed]
- Fard-Esfahani, P.; Mohammadi Torbati, P.; Hashemi, Z.; Fayaz, S.; Golkar, M. Analysis of relation between C677T genotype in MTHFR gene and prostatic cancer in Iranian males. Acta Med. Iran 2012, 50, 657–663. [Google Scholar]
- Ducker, G.S.; Rabinowitz, J.D. One-Carbon Metabolism in Health and Disease. Cell Metab. 2017, 25, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Zhu, Z.; Zhou, J.; Li, W.; Dong, Y.; Qian, Y.; Wei, P.; Wu, M. Associations of one-carbon metabolism-related gene polymorphisms with breast cancer risk are modulated by diet, being higher when adherence to the Mediterranean dietary pattern is low. Breast Cancer Res. Treat. 2021, 187, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Lin, J.; Jiang, J.; Chen, Y.; Tang, W.; Liu, L. Association between methylenetetrahydrofolate reductase tagging polymorphisms and susceptibility of hepatocellular carcinoma: A case-control study. Biosci. Rep. 2019, 39, BSR20192517. [Google Scholar] [CrossRef] [PubMed]
- Weiner, A.S.; Oskina, N.A.; Lacarev, A.F.; Petrova, V.D.; Ganov, D.I.; Boyarskih, U.A.; Tonacheva, O.G.; Voronina, E.N.; Filipenko, M.L. Role of polymorphic variants of MTR gene A2756G and SHMT1 gene C1420T in the development of prostatic cancer in residents of the Western Siberian Region of Russia. Bull. Exp. Biol. Med. 2012, 152, 466–469. [Google Scholar] [CrossRef]
- Divyya, S.; Naushad, S.M.; Addlagatta, A.; Murthy, P.V.; Reddy Ch, R.; Digumarti, R.R.; Gottumukkala, S.R.; Subbarao, S.A.; Kutala, V.K. Association of glutamate carboxypeptidase II (GCPII) haplotypes with breast and prostate cancer risk. Gene 2013, 516, 76–81. [Google Scholar] [CrossRef]
- Yang, Q.H.; Botto, L.D.; Gallagher, M.; Friedman, J.M.; Sanders, C.L.; Koontz, D.; Nikolova, S.; Erickson, J.D.; Steinberg, K. Prevalence and effects of gene-gene and gene-nutrient interactions on serum folate and serum total homocysteine concentrations in the United States: Findings from the third National Health and Nutrition Examination Survey DNA Bank. Am. J. Clin. Nutr. 2008, 88, 232–246. [Google Scholar] [CrossRef]
- Hayashi, H.; Horino, M.; Morishita, M.; Tazoe, Y.; Tsuboi, S.; Matsuyama, T.; Kosuge, K.; Yamada, H.; Tsuji, D.; Inoue, K.; et al. Dihydrofolate reductase gene intronic 19-bp deletion polymorphisms in a Japanese population. Drug. Metab. Pharmacokinet. 2010, 25, 516–518. [Google Scholar] [CrossRef] [PubMed]
- Metayer, C.; Scelo, G.; Chokkalingam, A.P.; Barcellos, L.F.; Aldrich, M.C.; Chang, J.S.; Guha, N.; Urayama, K.Y.; Hansen, H.M.; Block, G.; et al. Genetic variants in the folate pathway and risk of childhood acute lymphoblastic leukemia. Cancer Causes Control 2011, 22, 1243–1258. [Google Scholar] [CrossRef] [PubMed]
- Parle-McDermott, A.; Pangilinan, F.; Mills, J.L.; Kirke, P.N.; Gibney, E.R.; Troendle, J.; O'Leary, V.B.; Molloy, A.M.; Conley, M.; Scott, J.M.; et al. The 19-bp deletion polymorphism in intron-1 of dihydrofolate reductase (DHFR) may decrease rather than increase risk for spina bifida in the Irish population. Am. J. Med. Genet. A 2007, 143A, 1174–1180. [Google Scholar] [CrossRef] [PubMed]
- Philip, D.; Buch, A.; Moorthy, D.; Scott, T.M.; Parnell, L.D.; Lai, C.Q.; Ordovas, J.M.; Selhub, J.; Rosenberg, I.H.; Tucker, K.L.; et al. Dihydrofolate reductase 19-bp deletion polymorphism modifies the association of folate status with memory in a cross-sectional multi-ethnic study of adults. Am. J. Clin. Nutr. 2015, 102, 1279–1288. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, L.; Wang, J.; Qiu, L.; Mi, Y.; Ma, X.; Xiao, Z. Polymorphisms in folate-related genes: Impact on risk of adult acute lymphoblastic leukemia rather than pediatric in Han Chinese. Leuk. Lymphoma 2011, 52, 1770–1776. [Google Scholar] [CrossRef] [PubMed]
- Keith, S.W.; Kwabi-Addo, B.; Zeigler-Johnson, C. Interactions Between Obesity and One-Carbon Metabolism Genes in Predicting Prostate Cancer Outcomes Among White and Black Patients. J. Racial. Ethn. Health Disparities 2021, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.S.; Li, X.T.; Yu, L.G.; Wang, L.; Shi, Z.Y.; Guo, X.L. Roles of galectin-3 in metabolic disorders and tumor cell metabolism. Int. J. Biol. Macromol. 2020, 142, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Carroll, C.P.; Bolland, H.; Vancauwenberghe, E.; Collier, P.; Ritchie, A.A.; Clarke, P.A.; Grabowska, A.M.; Harris, A.L.; McIntyre, A. Targeting hypoxia regulated sodium driven bicarbonate transporters reduces triple negative breast cancer metastasis. Neoplasia 2022, 25, 41–52. [Google Scholar] [CrossRef]
- Bery, F.; Figiel, S.; Kouba, S.; Fontaine, D.; Gueguinou, M.; Potier-Cartereau, M.; Vandier, C.; Guibon, R.; Bruyere, F.; Fromont, G.; et al. Hypoxia Promotes Prostate Cancer Aggressiveness by Upregulating EMT-Activator Zeb1 and SK3 Channel Expression. Int. J. Mol. Sci. 2020, 21, 4786. [Google Scholar] [CrossRef]
- Schroeder, J.C.; Bensen, J.T.; Su, L.J.; Mishel, M.; Ivanova, A.; Smith, G.J.; Godley, P.A.; Fontham, E.T.; Mohler, J.L. The North Carolina-Louisiana Prostate Cancer Project (PCaP): Methods and design of a multidisciplinary population-based cohort study of racial differences in prostate cancer outcomes. Prostate 2006, 66, 1162–1176. [Google Scholar] [CrossRef]
- Sucheston, L.E.; Bensen, J.T.; Xu, Z.; Singh, P.K.; Preus, L.; Mohler, J.L.; Su, L.J.; Fontham, E.T.; Ruiz, B.; Smith, G.J.; et al. Genetic ancestry, self-reported race and ethnicity in African Americans and European Americans in the PCaP cohort. PLoS ONE 2012, 7, e30950. [Google Scholar] [CrossRef] [PubMed]
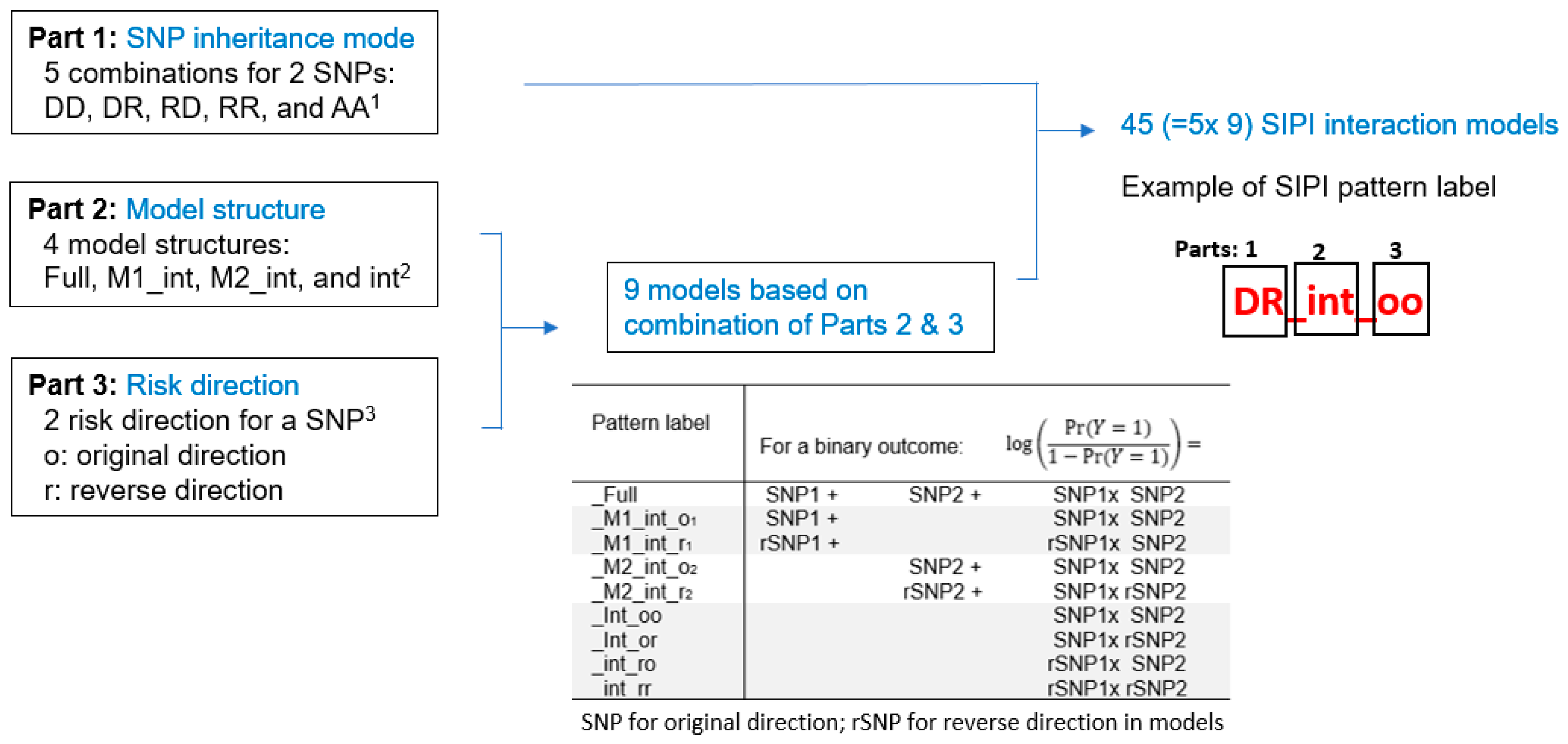
- Lin, H.Y.; Chen, D.T.; Huang, P.Y.; Liu, Y.H.; Ochoa, A.; Zabaleta, J.; Mercante, D.E.; Fang, Z.; Sellers, T.A.; Pow-Sang, J.M.; et al. SNP interaction pattern identifier (SIPI): An intensive search for SNP-SNP interaction patterns. Bioinformatics 2017, 33, 822–833. [Google Scholar] [CrossRef] [PubMed]
- Laurin, C.; Boomsma, D.; Lubke, G. The use of vector bootstrapping to improve variable selection precision in Lasso models. Stat. Appl. Genet. Mol. Biol. 2016, 15, 305–320. [Google Scholar] [CrossRef] [PubMed]
- Marchal, C.; Redondo, M.; Reyes-Engel, A.; Perea-Milla, E.; Gaitan, M.J.; Machuca, J.; Diaz, F.; Caballero, J.; Carnero, J. Association between polymorphisms of folate-metabolizing enzymes and risk of prostate cancer. Eur. J. Surg. Oncol. 2008, 34, 805–810. [Google Scholar] [CrossRef]
- Lin, V.C.; Lu, T.L.; Yin, H.L.; Yang, S.F.; Lee, Y.C.; Liu, C.C.; Huang, C.Y.; Yu, C.C.; Chang, T.Y.; Huang, S.P.; et al. Prognostic Relevance of Methylenetetrahydrofolate Reductase Polymorphisms for Prostate Cancer. Int. J. Mol. Sci. 2016, 17, 1996. [Google Scholar] [CrossRef]
- Dutta, H.K.; Borbora, D.; Baruah, M.; Narain, K. Evidence of gene-gene interactions between MTHFD1 and MTHFR in relation to anterior encephalocele susceptibility in Northeast India. Birth Defects Res. 2017, 109, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Machado, R.A.; de Oliveira Silva, C.; Martelli-Junior, H.; das Neves, L.T.; Coletta, R.D. Machine learning in prediction of genetic risk of nonsyndromic oral clefts in the Brazilian population. Clin. Oral. Investig. 2021, 25, 1273–1280. [Google Scholar] [CrossRef]
- Wang, Y.; Nangia-Makker, P.; Tait, L.; Balan, V.; Hogan, V.; Pienta, K.J.; Raz, A. Regulation of prostate cancer progression by galectin-3. Am. J. Pathol. 2009, 174, 1515–1523. [Google Scholar] [CrossRef]
- Saraswati, S.; Block, A.S.; Davidson, M.K.; Rank, R.G.; Mahadevan, M.; Diekman, A.B. Galectin-3 is a substrate for prostate specific antigen (PSA) in human seminal plasma. Prostate 2011, 71, 197–208. [Google Scholar] [CrossRef]
- Nangia-Makker, P.; Balan, V.; Raz, A. LGALS3 (Lectin, Galactoside-Binding, Soluble, 3). Available online: https://atlasgeneticsoncology.org/gene/44396/lgals3-(lectin-galactoside-binding-soluble-3) (accessed on 1 February 2023).
- Morgan, A.A.; Rubenstein, E. Proline: The distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS ONE 2013, 8, e53785. [Google Scholar] [CrossRef]
- Caputo, S.; Grioni, M.; Brambillasca, C.S.; Monno, A.; Brevi, A.; Freschi, M.; Piras, I.S.; Elia, A.R.; Pieri, V.; Baccega, T.; et al. Galectin-3 in Prostate Cancer Stem-Like Cells Is Immunosuppressive and Drives Early Metastasis. Front. Immunol. 2020, 11, 1820. [Google Scholar] [CrossRef] [PubMed]
- Oczkowski, M.; Dziendzikowska, K.; Pasternak-Winiarska, A.; Wlodarek, D.; Gromadzka-Ostrowska, J. Dietary Factors and Prostate Cancer Development, Progression, and Reduction. Nutrients 2021, 13, 496. [Google Scholar] [CrossRef]
- Sargsyan, A.; Dubasi, H.B. Milk Consumption and Prostate Cancer: A Systematic Review. World J. Mens Health 2021, 39, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Vinnai, J.R.; Cumming, R.C.; Thompson, G.J.; Timoshenko, A.V. The association between oxidative stress-induced galectins and differentiation of human promyelocytic HL-60 cells. Exp. Cell Res. 2017, 355, 113–123. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, J.T.; Ribeiro, C.; Barros, R.; Gomes, C.; de Matos, A.J.; Reis, C.A.; Rutteman, G.R.; Gartner, F. Hypoxia Up-Regulates Galectin-3 in Mammary Tumor Progression and Metastasis. PLoS ONE 2015, 10, e0134458. [Google Scholar] [CrossRef] [PubMed]
- Pao, P.C.; Patnaik, D.; Watson, L.A.; Gao, F.; Pan, L.; Wang, J.; Adaikkan, C.; Penney, J.; Cam, H.P.; Huang, W.C.; et al. HDAC1 modulates OGG1-initiated oxidative DNA damage repair in the aging brain and Alzheimer's disease. Nat. Commun. 2020, 11, 2484. [Google Scholar] [CrossRef]
- Pravenec, M.; Kozich, V.; Krijt, J.; Sokolova, J.; Zidek, V.; Landa, V.; Simakova, M.; Mlejnek, P.; Silhavy, J.; Oliyarnyk, O.; et al. Folate deficiency is associated with oxidative stress, increased blood pressure, and insulin resistance in spontaneously hypertensive rats. Am. J. Hypertens. 2013, 26, 135–140. [Google Scholar] [CrossRef]
- Padmanabhan, S.; Waly, M.I.; Taranikanti, V.; Guizani, N.; Ali, A.; Rahman, M.S.; Al-Attabi, Z.; Al-Malky, R.N.; Al-Maskari, S.N.M.; Al-Ruqaishi, B.R.S.; et al. Folate/Vitamin B12 Supplementation Combats Oxidative Stress-Associated Carcinogenesis in a Rat Model of Colon Cancer. Nutr. Cancer 2019, 71, 100–110. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.; Sirotkin, K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999, 9, 677–679. [Google Scholar] [CrossRef]
| Factor | European Americans (N = 690) N (%) | African Americans (n = 604) N (%) | p-Value 1 |
|---|---|---|---|
| Age (year) | |||
| Mean ± standard deviation | 64.0 ± 7.7 | 61.8 ± 7.8 | <0.001 |
| Study site | |||
| Louisiana | 371 (53.8) | 335 (55.5) | 0.541 |
| North Carolina | 319 (46.2) | 269 (44.5) | |
| Genetic ancestry % | |||
| Mean ± standard deviation | 96.7% ± 7.3% | 90.6% ± 15.5% | - |
| Prostate cancer aggressiveness | |||
| No | 542 (78.6) | 419 (69.4) | <0.001 |
| Yes | 148 (21.4) | 185 (30.6) |
| Gene Symbol 1 | Gene Full Name (Location) | Functional Annotation 2 |
|---|---|---|
| DHFR | dihydrofolate reductase (5q14.1) | metabolic process, cellular process, multicellular organismal process, developmental process, single-organism process, response to stimulus, biological regulation, cellular component organization or biogenesis |
| MTR | 5-methyltetrahydrofolate-homocysteine methyltransferase (1q43) | metabolic process, cellular process, multicellular organismal process, developmental process, single-organism process, response to stimulus, cellular component organization or biogenesis |
| MTRR | 5-methyltetrahydrofolate-homocysteine methyltransferase reductase (5p15.31) | metabolic process, cellular process, single-organism process, biological regulation |
| MTHFR | methylenetetrahydrofolate reductase (1p36.22) | metabolic process, cellular process, multicellular organismal process, developmental process, single-organism process, response to stimulus, biological regulation, cellular component organization or biogenesis |
| MTHFD1 | methylenetetrahydrofolate dehydrogenase, cyclohydrolase, and formyltetrahydrofolate synthetase 1 (14q23.3) | immune system process, metabolic process, cellular process, multicellular organismal process, developmental process, single-organism process, biological regulation |
| MTHFS | methenyltetrahydrofolate synthetase (15q25.1) | metabolic process, cellular process, single-organism process |
| SLC4A5 | solute carrier family 4 member 5 (2p13.1) | metabolic process, cellular process, multicellular organismal process, developmental process, single-organism process, localization, biological regulation, cellular component organization or biogenesis |
| LGALS3 | galectin 3 (14q22.3) | cell killing, immune system process, metabolic process, cellular process, biological adhesion, signaling, developmental process, locomotion, single-organism process, response to stimulus, localization, multi-organism process, biological regulation, cellular component organization or biogenesis |
| European Americans (n = 690) | African Americans (n = 604) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP /Polymorphism 4 | Chr | Position (GRCh38) | Gene | Min < Maj (MAF) 1 | Mode | OR (95% CI) 2 | p | Min < Maj (MAF) 1 | Mode | OR (95% CI) 2 | p |
| rs2274976 | 1 | 11790870 | MTHFR | A < G (0.05) | Dom | 0.88 (0.46–1.69) | 0.702 | A < G (0.03) | Dom | 0.79 (0.37, 1.65) | 0.525 |
| rs1801131 | 1 | 11794419 | MTHFR | C < A (0.33) | Dom | 0.89 (0.61, 1.29) | 0.532 | C < A (0.17) | Rec | 0.83 (0.26, 2.64) | 0.746 |
| rs1801133 | 1 | 11796321 | MTHFR | A < G (0.33) | Dom | 0.79 (0.55, 1.15) | 0.223 | A < G (0.13) | Add | 0.72 (0.49, 1.07) | 0.104 |
| rs1805087 | 1 | 236885200 | MTR | G < A (0.2) | Rec | 1.33 (0.57, 3.14) | 0.510 | G < A (0.29) | Add | 0.91 (0.69, 1.19) | 0.495 |
| rs7587117 | 2 | 74221528 | SLC4A5 | C < T (0.32) | Add | 1.24 (0.95, 1.62) | 0.112 | C < T (0.16) | Rec | 1.51 (0.57, 3.98) | 0.408 |
| rs10380 | 5 | 7897078 | MTRR | T < C (0.1) | Dom | 1.12 (0.7, 1.78) | 0.645 | T < C (0.34) | Rec | 1.49 (0.88, 2.53) | 0.139 |
| rs4644 | 14 | 55138217 | LGALS3 | A < C (0.39) | Add | 1.06 (0.81, 1.38) | 0.683 | A < C (0.26) | Rec | 0.53 (0.24, 1.18) | 0.120 |
| rs4652 5 | 14 | 55138318 | LGALS3 | C < A (0.42) | Dom | 1.09 (0.74, 1.62) | 0.661 | A < C (0.16) | Rec | 1.78 (0.71, 4.42) | 0.217 |
| rs2236225 | 14 | 64442127 | MTHFD1 | T < C (0.43) | Rec | 1.46 (0.94, 2.28) | 0.096 | T < C (0.22) | Rec | 1.84 (0.85, 3.99) | 0.123 |
| rs622506 | 15 | 79846853 | MTHFS | C < A (0.35) | Rec | 0.81 (0.43, 1.54) | 0.529 | C < A (0.18) | Rec | 1.63 (0.75, 3.55) | 0.219 |
| DHFR-19bp 3 | 19 | DHFR | Del < Ins (0.43) | Dom | 0.88 (0.59, 1.31) | 0.531 | Ins < Del (0.45) | Dom | 1.30 (0.88, 1.91) | 0.189 | |
| SNP Pair | Gene1 | Gene2 | Pattern | OR (95% CI) 1 | p-Value 1 | Significance% 2 |
|---|---|---|---|---|---|---|
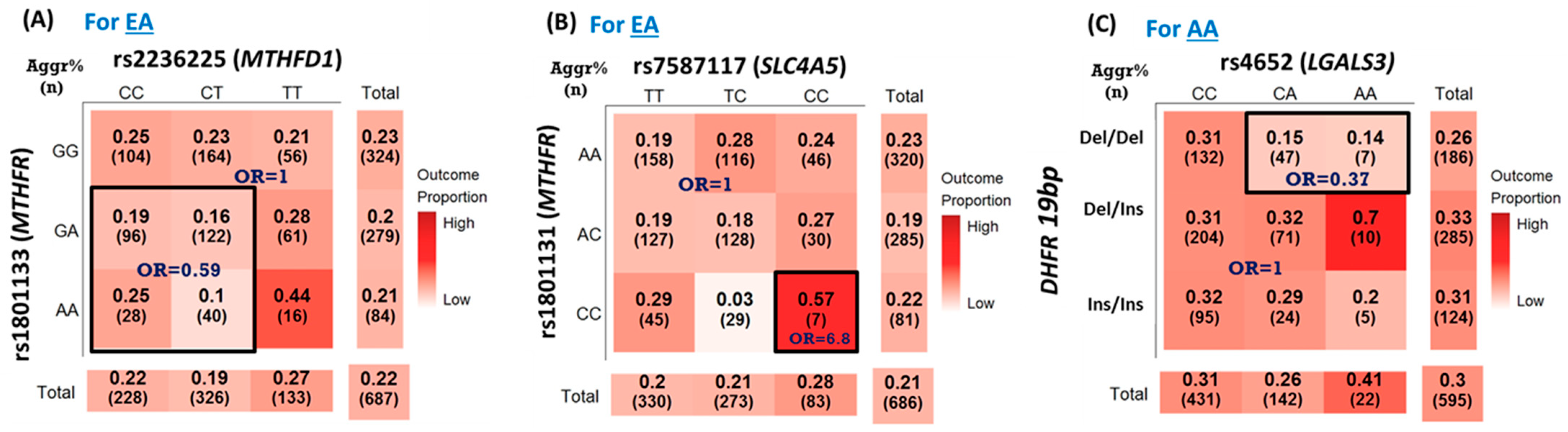
| rs1801133_rs2236225 | MTHFR | MTHFD1 | DR_int_or | 0.59 (0.40, 0.88) | 0.009 | 68.8 |
| rs1801133_rs4644 | MTHFR | LGALS3 | RD_int_or | 0.22 (0.06, 0.73) | 0.013 | 55.2 |
| rs2236225_rs7587117 | MTHFD1 | SLC4A5 | RR_int_rr | 0.61 (0.41, 0.91) | 0.014 | 58.2 |
| rs1801133_ rs4652 | MTHFR | LGALS3 | RD_int_or | 0.23 (0.07, 0.77) | 0.018 | 52.2 |
| DHFR-19bp_rs1805087 | DHFR | MTR | RR_int_oo | 6.26 (1.36, 28.76) | 0.018 | 40.8 |
| rs1801131_ rs7587117 | MTHFR | SLC4A5 | RR_int_oo | 6.80 (1.39, 33.28) | 0.018 | 69.0 |
| rs1801133_rs1805087 | MTHFR | MTR | DD_int_or | 0.64 (0.43, 0.97) | 0.034 | 50.8 |
| rs1805087_rs2236225 | MTR | MTHFD1 | DR_int_oo | 1.92 (1.04, 3.56) | 0.038 | 44.8 |
| SNP Pair | Gene1 | Gene2 | Pattern | OR (95% CI) 1 | p-Value 1 | Significance% 2 |
|---|---|---|---|---|---|---|
| DHFR-19bp_rs4644 | DHFR | LGALS3 | DD_int_rr | 0.49 (0.29, 0.85) | 0.011 | 52.2 |
| rs1805087_rs2236225 | MTR | MTHFD1 | DR_int_ro | 4.02 (1.38, 11.68) | 0.011 | 57.4 |
| DHFR-19bp_rs4652 | DHFR | LGALS3 | DD_int_ro | 0.37 (0.17, 0.81) | 0.012 | 65.4 |
| rs1801131_rs4652 | MTHFR | LGALS3 | DR_int_ro | 2.86 (1.07, 7.67) | 0.037 | 36.4 |
| rs10380_rs1805087 | MTRR | MTR | RD_int_oo | 1.98 (1.02, 3.83) | 0.043 | 42.6 |
| DHFR-19bp_rs10380 | DHFR | MTRR | RR_int_ro | 1.79 (1.02, 3.14) | 0.043 | 40.6 |
| rs10380_rs2236225 | MTRR | MTHFD1 | RD_int_oo | 2.43 (1.02, 5.79) | 0.045 | 43.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, H.-Y.; Steck, S.E.; Sarkar, I.; Fontham, E.T.H.; Diekman, A.; Rogers, L.J.; Ratliff, C.T.; Bensen, J.T.; Mohler, J.L.; Su, L.J. Interactions of SNPs in Folate Metabolism Related Genes on Prostate Cancer Aggressiveness in European Americans and African Americans. Cancers 2023, 15, 1699. https://doi.org/10.3390/cancers15061699
Lin H-Y, Steck SE, Sarkar I, Fontham ETH, Diekman A, Rogers LJ, Ratliff CT, Bensen JT, Mohler JL, Su LJ. Interactions of SNPs in Folate Metabolism Related Genes on Prostate Cancer Aggressiveness in European Americans and African Americans. Cancers. 2023; 15(6):1699. https://doi.org/10.3390/cancers15061699
Chicago/Turabian StyleLin, Hui-Yi, Susan E. Steck, Indrani Sarkar, Elizabeth T. H. Fontham, Alan Diekman, Lora J. Rogers, Calvin T. Ratliff, Jeannette T. Bensen, James L. Mohler, and L. Joseph Su. 2023. "Interactions of SNPs in Folate Metabolism Related Genes on Prostate Cancer Aggressiveness in European Americans and African Americans" Cancers 15, no. 6: 1699. https://doi.org/10.3390/cancers15061699
APA StyleLin, H.-Y., Steck, S. E., Sarkar, I., Fontham, E. T. H., Diekman, A., Rogers, L. J., Ratliff, C. T., Bensen, J. T., Mohler, J. L., & Su, L. J. (2023). Interactions of SNPs in Folate Metabolism Related Genes on Prostate Cancer Aggressiveness in European Americans and African Americans. Cancers, 15(6), 1699. https://doi.org/10.3390/cancers15061699





