Precision Medicine Approach Based on Molecular Alterations for Patients with Relapsed or Refractory Multiple Myeloma: Results from the MM-EP1 Study
Abstract
Simple Summary
Abstract
1. Introduction
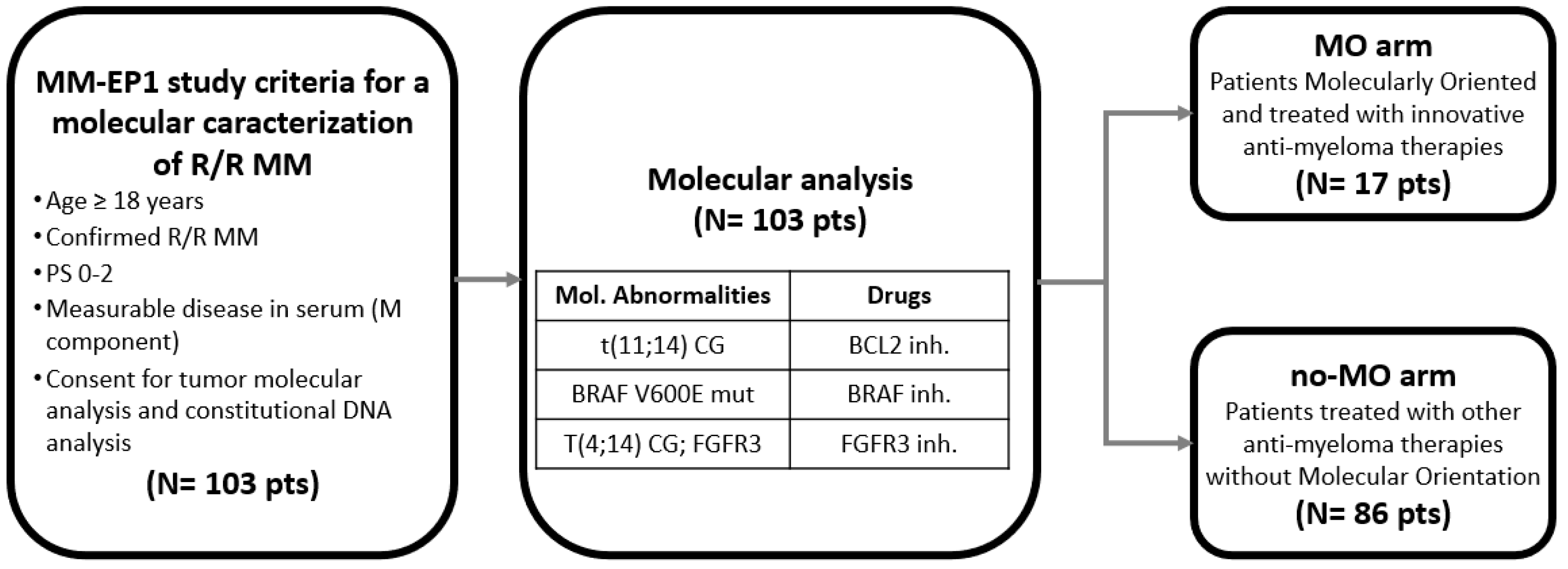
2. Materials and Methods
3. Results
3.1. Patients’ Characteristics
3.2. Molecular Characteristics of r/r MM
3.3. Potentially Actionable Molecular Target Identification
3.4. Molecularly Oriented and Treated Patients
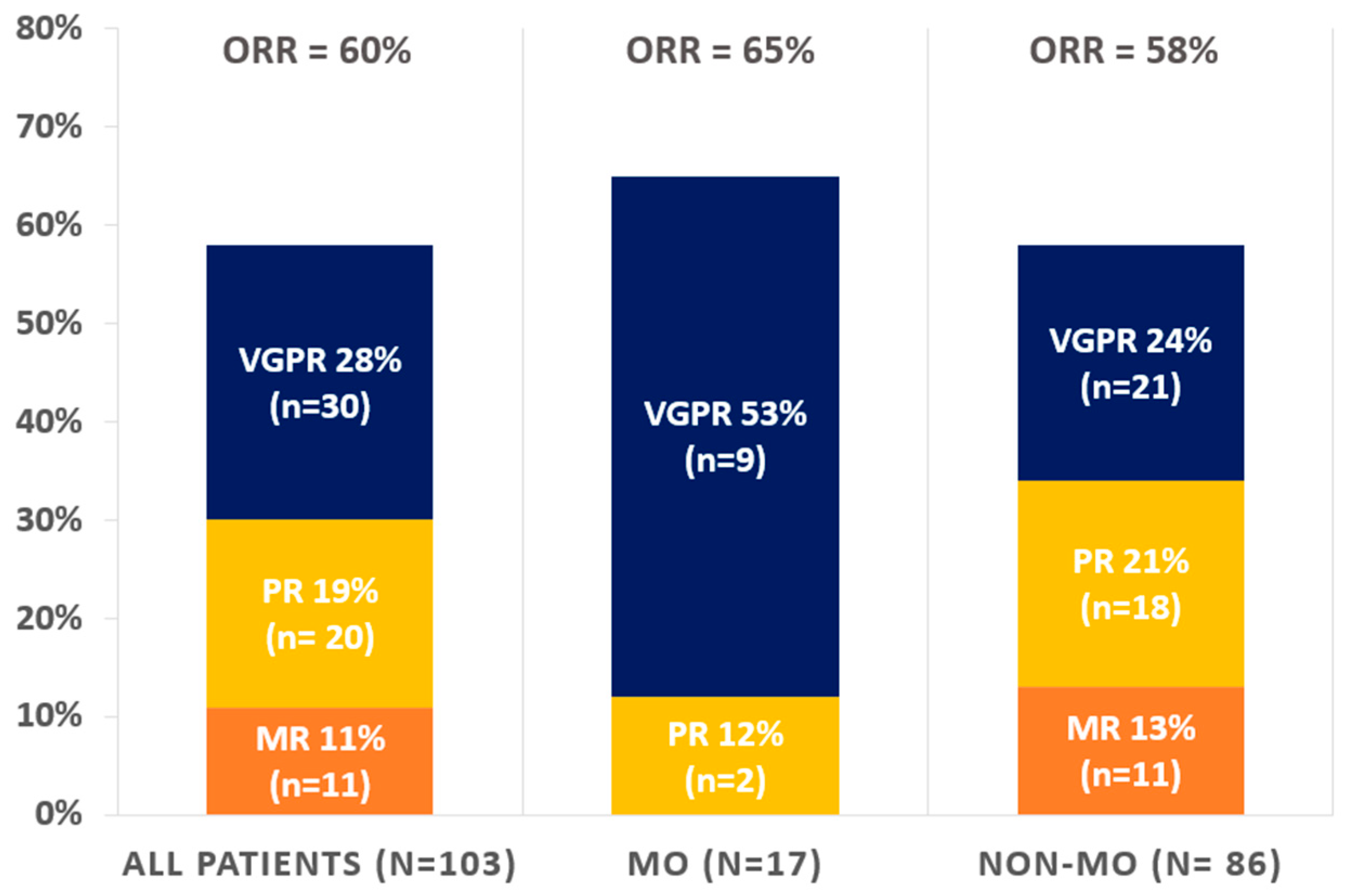
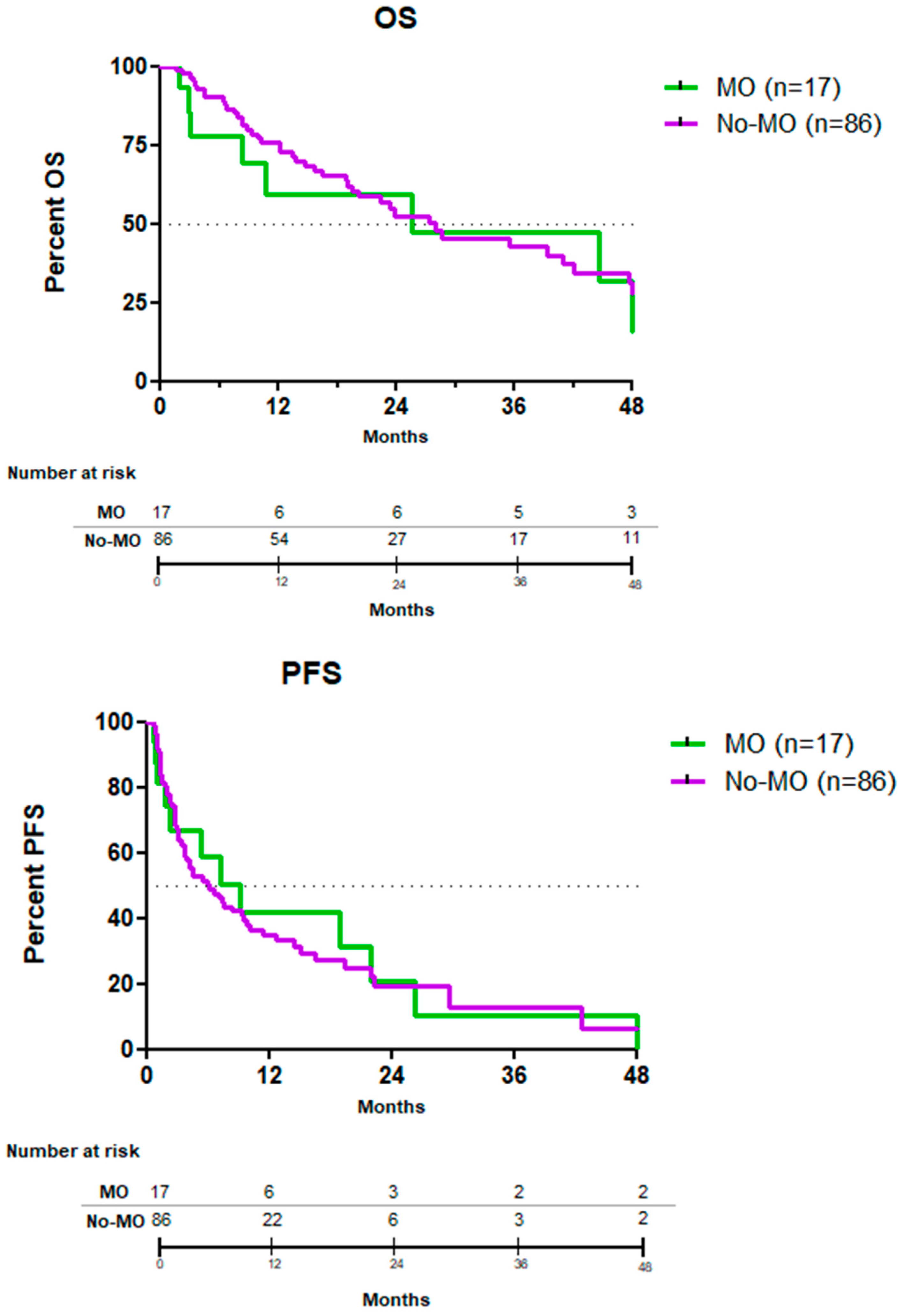
3.5. Efficacy Endpoints
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Slamon, D.J.; Clark, G.M.; Wong, S.G.; Levin, W.J.; Ullrich, A.; McGuire, W.L. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987, 235, 177–182. [Google Scholar] [CrossRef]
- Beau-Faller, M.; Prim, N.; Ruppert, A.-M.; Nanni-Metéllus, I.; Lacave, R.; Lacroix, L.; Escande, F.; Lizard, S.; Pretet, J.-L.; Rouquette, I.; et al. Rare EGFR exon 18 and exon 20 mutations in non-small-cell lung cancer on 10 117 patients: A multicentre observational study by the French ERMETIC-IFCT network. Ann. Oncol. 2014, 25, 126–131. [Google Scholar] [CrossRef]
- Dawood, S.; Broglio, K.; Buzdar, A.U.; Hortobagyi, G.N.; Giordano, S.H. Prognosis of Women with Metastatic Breast Cancer by HER2 Status and Trastuzumab Treatment: An Institutional-Based Review. J. Clin. Oncol. 2010, 28, 92–98. [Google Scholar] [CrossRef]
- Soria, J.-C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef]
- Mateo, J.; Chakravarty, D.; Dienstmann, R.; Jezdic, S.; Gonzalez-Perez, A.; Lopez-Bigas, N.; Ng, C.; Bedard, P.; Tortora, G.; Douillard, J.-Y.; et al. A framework to rank genomic alterations as targets for cancer precision medicine: The ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann. Oncol. 2018, 29, 1895–1902. [Google Scholar] [CrossRef]
- Landgren, O.; Morgan, G.J. Biologic Frontiers in Multiple Myeloma: From Biomarker Identification to Clinical Practice. Clin. Cancer Res. 2014, 20, 804–813. [Google Scholar] [CrossRef]
- Bolli, N.; Genuardi, E.; Ziccheddu, B.; Martello, M.; Oliva, S.; Terragna, C. Next-Generation Sequencing for Clinical Management of Multiple Myeloma: Ready for Prime Time? Front. Oncol. 2020, 10, 189. [Google Scholar] [CrossRef]
- Manier, S.; Salem, K.Z.; Park, J.; Landau, D.A.; Getz, G.; Ghobrial, I.M. Genomic complexity of multiple myeloma and its clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, M.; Barbieri, M.; Todoerti, K.; Agnelli, L.; Marzorati, S.; Fabris, S.; Ciceri, G.; Galletti, S.; Milesi, G.; Manzoni, M.; et al. Molecular spectrum of BRAF, NRAS and KRAS gene mutations in plasma cell dyscrasias: Implication for MEK-ERK pathway activation. Oncotarget 2015, 6, 24205–24217. [Google Scholar] [CrossRef] [PubMed]
- Andrulis, M.; Lehners, N.; Capper, D.; Penzel, R.; Heining, C.; Huellein, J.; Zenz, T.; von Deimling, A.; Schirmacher, P.; Ho, A.D.; et al. Targeting the BRAF V600E Mutation in Multiple Myeloma. Cancer Discov. 2013, 3, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Salama, A.K.S.; Li, S.; Macrae, E.R.; Park, J.-I.; Mitchell, E.P.; Zwiebel, J.A.; Chen, H.X.; Gray, R.J.; McShane, L.M.; Rubinstein, L.V.; et al. Dabrafenib and Trametinib in Patients With Tumors With BRAFV600E Mutations: Results of the NCI-MATCH Trial Subprotocol H. J. Clin. Oncol. 2020, 38, 3895–3904. [Google Scholar] [CrossRef] [PubMed]
- Gulla’, A.; Anderson, K.C. Multiple myeloma: The (r)evolution of current therapy and a glance into future. Haematologica 2020, 105, 2358–2367. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kaufman, J.L.; Gasparetto, C.; Mikhael, J.; Vij, R.; Pegourie, B.; Benboubker, L.; Facon, T.; Amiot, M.; Moreau, P.; et al. Efficacy of venetoclax as targeted therapy for relapsed/refractory t(11;14) multiple myeloma. Blood 2017, 130, 2401–2409. [Google Scholar] [CrossRef]
- Kumar, S.K.; Harrison, S.J.; Cavo, M.; de la Rubia, J.; Popat, R.; Gasparetto, C.; Hungria, V.; Salwender, H.; Suzuki, K.; Kim, I.; et al. Venetoclax or placebo in combination with bortezomib and dexamethasone in patients with relapsed or refractory multiple myeloma (BELLINI): A randomised, double-blind, multicentre, phase 3 trial. Lancet Oncol. 2020, 21, 1630–1642. [Google Scholar] [CrossRef]
- Kommalapati, A.; Tella, S.; Borad, M.; Javle, M.; Mahipal, A. FGFR Inhibitors in Oncology: Insight on the Management of Toxicities in Clinical Practice. Cancers 2021, 13, 2968. [Google Scholar] [CrossRef] [PubMed]
- Ronchetti, D.; Greco, A.; Compasso, S.; Colombo, G.; Dell’Era, P.; Otsuki, T.; Lombardi, L.; Neri, A. Deregulated FGFR3 mutants in multiple myeloma cell lines with t(4;14): Comparative analysis of Y373C, K650E and the novel G384D mutations. Oncogene 2001, 20, 3553–3562. [Google Scholar] [CrossRef]
- Szalat, R.; Munshi, N.C. Next-Generation Sequencing Informing Therapeutic Decisions and Personalized Approaches. Am. Soc. Clin. Oncol. Educ. Book 2016, 35, e442–e448. [Google Scholar] [CrossRef]
- Pawlyn, C.; Davies, F.E. Toward personalized treatment in multiple myeloma based on molecular characteristics. Blood 2019, 133, 660–675. [Google Scholar] [CrossRef]
- Auclair, D.; Anderson, K.C.; Avigan, D.; Bianchi, G.; Biran, N.; Chaudhry, M.; Cho, H.J.; Furlong, M.; Hofmeister, C.C.; Kansagra, A.J.; et al. The myeloma-developing regimens using genomics (MyDRUG) master protocol. J. Clin. Oncol. 2019, 37, TPS8057. [Google Scholar] [CrossRef]
- Pasca, S.; Tomuleasa, C.; Teodorescu, P.; Ghiaur, G.; Dima, D.; Moisoiu, V.; Berce, C.; Stefan, C.; Ciechanover, A.; Einsele, H. KRAS/NRAS/BRAF Mutations as Potential Targets in Multiple Myeloma. Front. Oncol. 2019, 9, 01137. [Google Scholar] [CrossRef]
- Anwer, F.; Gee, K.M.; Iftikhar, A.; Baig, M.; Russ, A.D.; Saeed, S.; Abu Zar, M.; Razzaq, F.; Carew, J.; Nawrocki, S.; et al. Future of Personalized Therapy Targeting Aberrant Signaling Pathways in Multiple Myeloma. Clin. Lymphoma Myeloma Leuk. 2019, 19, 397–405. [Google Scholar] [CrossRef]
- Kumar, S.; Paiva, B.; Anderson, K.C.; Durie, B.; Landgren, O.; Moreau, P.; Munshi, N.; Lonial, S.; Bladé, J.; Mateos, M.-V.; et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016, 17, e328–e346. [Google Scholar] [CrossRef]
- Lohr, J.G.; Stojanov, P.; Carter, S.L.; Cruz-Gordillo, P.; Lawrence, M.S.; Auclair, D.; Sougnez, C.; Knoechel, B.; Gould, J.; Saksena, G.; et al. Widespread Genetic Heterogeneity in Multiple Myeloma: Implications for Targeted Therapy. Cancer Cell 2014, 25, 91–101. [Google Scholar] [CrossRef]
- Walker, B.; Boyle, E.M.; Wardell, C.; Murison, A.; Begum, D.B.; Dahir, N.M.; Proszek, P.Z.; Johnson, D.C.; Kaiser, M.F.; Melchor, L.; et al. Mutational Spectrum, Copy Number Changes, and Outcome: Results of a Sequencing Study of Patients with Newly Diagnosed Myeloma. J. Clin. Oncol. 2015, 33, 3911–3920. [Google Scholar] [CrossRef] [PubMed]
- Le Tourneau, C.; Belin, L.; Paoletti, X.; Bièche, I.; Kamal, M. Precision medicine: Lessons learned from the SHIVA trial—Authors’ reply. Lancet Oncol. 2015, 16, e581–e582. [Google Scholar] [CrossRef] [PubMed]
- Raje, N.; Chau, I.; Hyman, D.M.; Ribrag, V.; Blay, J.-Y.; Tabernero, J.; Elez, E.; Wolf, J.; Yee, A.J.; Kaiser, M.; et al. Vemurafenib in Patients with Relapsed Refractory Multiple Myeloma Harboring BRAFV600 Mutations: A Cohort of the Histology-Independent VE-BASKET Study. JCO Precis. Oncol. 2018, 2, 1–9. [Google Scholar] [CrossRef]
- Pan, D.; Richter, J. Where We Stand with Precision Therapeutics in Myeloma: Prosperity, Promises, and Pipedreams. Front. Oncol. 2022, 11, 819127. [Google Scholar] [CrossRef]
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Wu, J.; Chen, Y.; Nie, J.; Chen, C. Activation of PI3K/AKT/mTOR Pathway Causes Drug Resistance in Breast Cancer. Front. Pharmacol. 2021, 12, 628690. [Google Scholar] [CrossRef]
- Bhinder, B.; Gilvary, C.; Madhukar, N.S.; Elemento, O. Artificial Intelligence in Cancer Research and Precision Medicine. Cancer Discov. 2021, 11, 900–915. [Google Scholar] [CrossRef]
- Ernst, T.; Aebi, S.; Zander, A.; Zander, T. Partial response to dabrafenib and trametinib in relapsed BRAF V600E-Mutated multiple myeloma and possible mechanisms of resistance. BMJ Case Rep. 2022, 15, e246264. [Google Scholar] [CrossRef] [PubMed]
- Waldschmidt, J.M.; Kloeber, J.A.; Anand, P.; Frede, J.; Kokkalis, A.; Dimitrova, V.; Potdar, S.; Nair, M.S.; Vijaykumar, T.; Im, N.G.; et al. Single-Cell Profiling Reveals Metabolic Reprogramming as a Resistance Mechanism inBRAF-Mutated Multiple Myeloma. Clin. Cancer Res. 2021, 27, 6432–6444. [Google Scholar] [CrossRef]
- Le Calvez, B.; Le Bris, Y.; Herbreteau, G.; Jamet, B.; Bossard, C.; Tessoulin, B.; Gastinne, T.; Mahé, B.; Dubruille, V.; Blin, N.; et al. RAS mutation leading to acquired resistance to dabrafenib and trametinib therapy in a multiple myeloma patient harboring BRAF mutation. Ejhaem 2020, 1, 318–322. [Google Scholar] [CrossRef]
- Martinez-Garcia, E.; Popovic, R.; Min, D.-J.; Sweet, S.; Thomas, P.; Zamdborg, L.; Heffner, A.; Will, C.; Lamy, L.; Staudt, L.M.; et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood 2011, 117, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.P.; Pfeiffer, R.M.; Ahn, I.E.; Mailankody, S.; Sonneveld, P.; van Duin, M.; Munshi, N.C.; Walker, B.A.; Morgan, G.; Landgren, O. Impact of Genes Highly Correlated with MMSET Myeloma on the Survival of Non-MMSET Myeloma Patients. Clin. Cancer Res. 2016, 22, 4039–4044. [Google Scholar] [CrossRef] [PubMed]
- Gazeau, N.; Beauvais, D.; Yakoub-Agha, I.; Mitra, S.; Campbell, T.B.; Facon, T.; Manier, S. Effective anti-BCMA retreatment in multiple myeloma. Blood Adv. 2021, 5, 3016–3020. [Google Scholar] [CrossRef]
- Bergaggio, E.; Riganti, C.; Garaffo, G.; Vitale, N.; Mereu, E.; Bandini, C.; Pellegrino, E.; Pullano, V.; Omedè, P.; Todoerti, K.; et al. IDH2 inhibition enhances proteasome inhibitor responsiveness in hematological malignancies. Blood 2019, 133, 156–167. [Google Scholar] [CrossRef]
- Leverson, J.D.; Zhang, H.; Chen, J.; Tahir, S.K.; Phillips, D.C.; Xue, J.; Nimmer, P.; Jin, S.; Smith, M.; Xiao, Y.; et al. Potent and selective small-molecule MCL-1 inhibitors demonstrate on-target cancer cell killing activity as single agents and in combination with ABT-263 (navitoclax). Cell Death Dis. 2015, 6, e1590. [Google Scholar] [CrossRef]



| All Patients (n = 103) | MO (n = 17) | No-MO (n = 86) | |
|---|---|---|---|
| Number, M/F | 102, 56/47 | 17, 7/10 | 86, 49/37 |
| Median age [range] | 67 yo [44; 85] | 59 yo [48; 81] | 68 yo [44; 85] |
| Involved chain | |||
| IgG | 60% (n = 62) | 41% (n = 7) | 63% (n = 54) |
| IgA | 17% (n = 18) | 24% (n = 4) | 16% (n = 14) |
| FLC | 23% (n = 24) | 35% (n = 6) | 21% (n = 18) |
| Kappa | 17% (n = 18) | 29% (n = 5) | 15% (n = 13) |
| Lambda | 6% (n = 6) | 6% (n = 1) | 6% (n = 5) |
| ISS | |||
| I | 28% (n = 29) | 29% (n = 5) | 28% (n = 24) |
| II | 22% (n = 23) | 41% (n = 7) | 17% (n = 15) |
| III | 23% (n = 24) | 12% (n = 2) | 26% (n = 22) |
| NA | 27% (n = 28) | 18% (n = 3) | 29% (n = 25) |
| Cytogenetic risk | |||
| t(4;14) | 20% (n = 21) | 24% (n = 4) | 20% (n = 17) |
| t(11;14) | 18% (n = 19) | 53% (n = 9) | 12% (n = 10) |
| chr1 abnorm | 19% (n = 20) | 24% (n = 4) | 20% (n = 17) |
| del13q14 | 3% (n = 3) | 6% (n = 1) | 2% (n = 2) |
| del17p | 12% (n = 12) | 18% (n = 3) | 11% (n = 9) |
| NA | 11% (n = 11) | 12% (n = 2) | 11% (n = 9) |
| Number prior lines median [range] | 4 [1–8] | 5 [1–11] | 3 [1–8] |
| Prior IMID | 100% (n = 103) | 100% (n = 17) | 100% (n = 86) |
| Prior alkylating agent | 96% (n = 99) | 100% (n = 17) | 95% (n = 82) |
| Prior PI | 96% (n = 99) | 100% (n = 17) | 95% (n = 82) |
| Prior dara (anti-CD38) | 23% (n = 24) | 58% (n = 6) | 21% (n = 18) |
| HSCT auto/allo | 72%/6% (n = 73/6) | 69%/19% (n = 11/3) | 72%/4% (n = 62/3) |
| Treatment considered (no. of patients) | BRAFi-based therapy: vemurafenib (3) dabrafenib (3) BCL2i-based therapy: venetoclax (9) FGFR3i-based therapy: erdafitinib (2) | Anti-CD38 (27) IMID-based (34) CPI (13) Belantamab (8) PI (20) MDM2 inhib (2) Cobimetinib (3) mTOR (1) Isatuximab (3) Other (1) | |
| All Patients (n = 103) | MO (n = 17) | No-MO (n = 86) | p-Value | |
|---|---|---|---|---|
| Time to best response | 7 ± 1 months | 9 ± 4 months | 6 ± 1 months | 0.47 |
| Response | ||||
| VGPR | 28% (n = 30) | 53% (n = 9) | 24% (n= 21) | 0.053 |
| PR | 19% (n = 20) | 12% (n = 2) | 21% (n = 18) | |
| MR | 11% (n = 11) | 0% (n = 0) | 13% (n= 11) | |
| SD | 21% (n = 22) | 12% (n = 2) | 23% (n = 20) | |
| PD | 20% (n = 21) | 24% (n = 4) | 20% (n= 17) | |
| Δ MC (median) | −34% | −91% | −30% | 0.33 |
| PFS (median) | 7 months | 9 months | 6 months | 0.88 |
| OS (median) | 28 months | 26 months | 28 months | 0.98 |
| Deaths | 52% (n = 54) | 47% (n = 8) | 53% (n = 46) | 0.23 |
| MM-related | 85% (prop: 45/54) | 88% (prop: 7/8) | 85% (prop: 39/46) |
| Pt No° | Involved Chain | Cytogenetic/Molecular Profile | Therapy | M-Composant Variation |
|---|---|---|---|---|
| 1 | κ | t (4;14); t (11;14); chr. 1 abn; Δ17p; TP53 mut | VEN-based | +882 |
| 2 | IgG, κ | BRAF mut V600E | BRAFi based (vemurafenib) | +296 |
| 3 | IgG, λ | t (4;14) | FGFRi based (erdafitinib) | +181 |
| 4 | λ | t (11;14); chr. 1 abn | VEN-based | +56 |
| 5 | IgG, κ | t (4;14) | FGFRi based (erdafitinib) | +2.3 |
| 6 | IgG, κ | t (11; 14); BRAF mut G464E; NRAS mut Q61R | Ven-based | −8 |
| 7 | IgA, κ | BRAF mut V600E | BRAFi based (dabrafenib) | −55 |
| 8 | IgA, κ | BRAF mut V600E | BRAFi based (vemurafenib) | −83 |
| 9 | IgG, λ | t (11;14); BRAF mut V600E | BRAFi based (dabrafenib) | −91 |
| 10 | IgA, κ | t (4;14); t (11;14) | VEN-based | −94 |
| 11 | IgG, λ | BRAF mut V600E | BRAFi based (vemurafenib) | −94 |
| 12 | κ | t (11;14) | VEN-based | −97 |
| 13 | IgA, λ | BRAF mut V600E | BRAFi based (Dabrafenib) | −97 |
| 14 | κ | t (11;14); chr. 1 abn; Δ17p | VEN-based | −98 |
| 15 | κ | t (11;14) | VEN-based | −99 |
| 16 | κ | t (11;14) | VEN-based | −100 |
| 17 | IgG, λ | t (11;14) | VEN-based | −100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andreozzi, F.; Dragani, M.; Quivoron, C.; Le Bras, F.; Assi, T.; Danu, A.; Belhadj, K.; Lazarovici, J.; Cotteret, S.; Bernard, O.A.; et al. Precision Medicine Approach Based on Molecular Alterations for Patients with Relapsed or Refractory Multiple Myeloma: Results from the MM-EP1 Study. Cancers 2023, 15, 1508. https://doi.org/10.3390/cancers15051508
Andreozzi F, Dragani M, Quivoron C, Le Bras F, Assi T, Danu A, Belhadj K, Lazarovici J, Cotteret S, Bernard OA, et al. Precision Medicine Approach Based on Molecular Alterations for Patients with Relapsed or Refractory Multiple Myeloma: Results from the MM-EP1 Study. Cancers. 2023; 15(5):1508. https://doi.org/10.3390/cancers15051508
Chicago/Turabian StyleAndreozzi, Fabio, Matteo Dragani, Cyril Quivoron, Fabien Le Bras, Tarek Assi, Alina Danu, Karim Belhadj, Julien Lazarovici, Sophie Cotteret, Olivier A. Bernard, and et al. 2023. "Precision Medicine Approach Based on Molecular Alterations for Patients with Relapsed or Refractory Multiple Myeloma: Results from the MM-EP1 Study" Cancers 15, no. 5: 1508. https://doi.org/10.3390/cancers15051508
APA StyleAndreozzi, F., Dragani, M., Quivoron, C., Le Bras, F., Assi, T., Danu, A., Belhadj, K., Lazarovici, J., Cotteret, S., Bernard, O. A., Ribrag, V., & Michot, J.-M. (2023). Precision Medicine Approach Based on Molecular Alterations for Patients with Relapsed or Refractory Multiple Myeloma: Results from the MM-EP1 Study. Cancers, 15(5), 1508. https://doi.org/10.3390/cancers15051508







