Immune Phenotypic Characterization of a TRAIL-Knockout Mouse
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Cell Isolation and Culture
2.3. Flow Cytometry
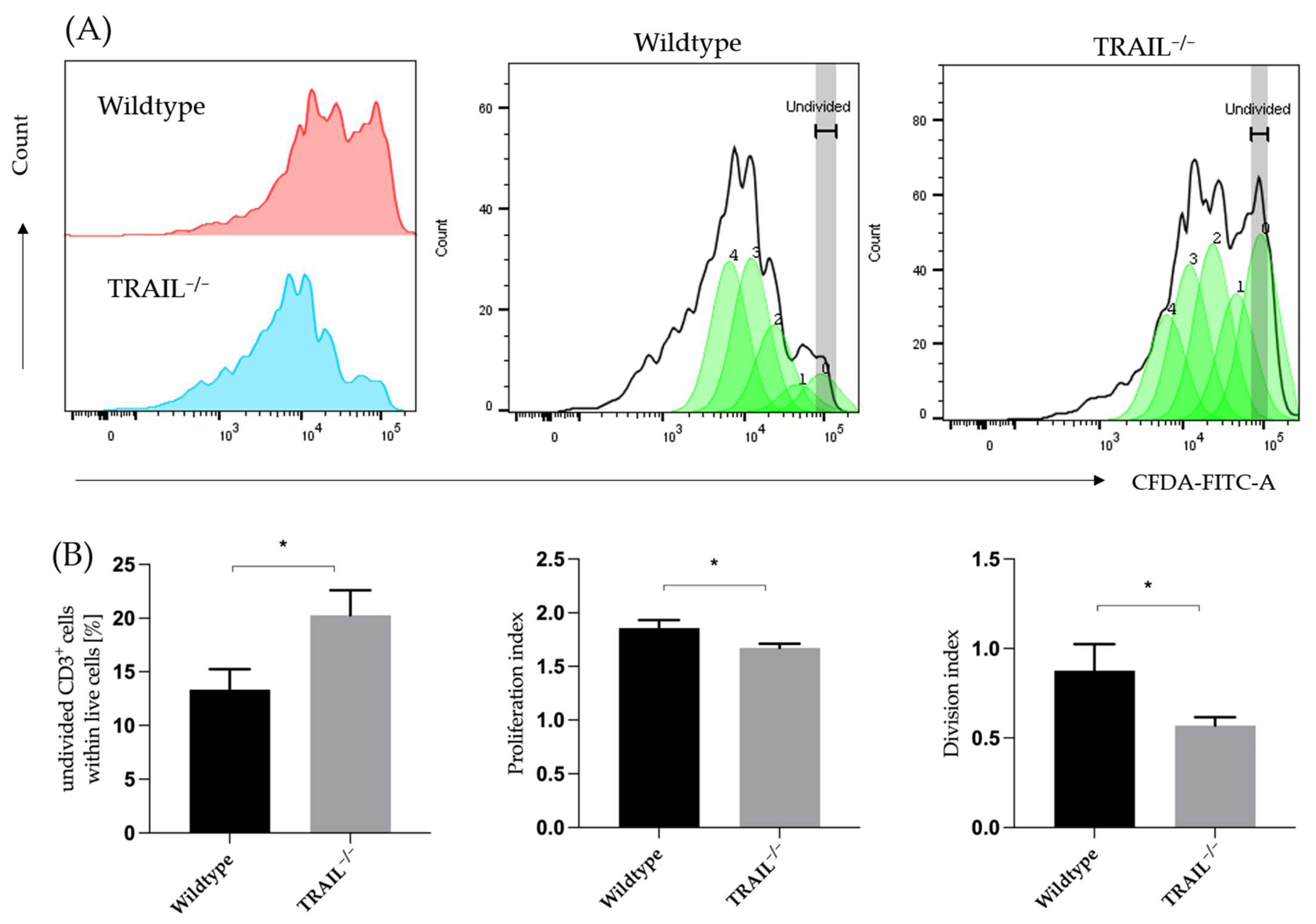
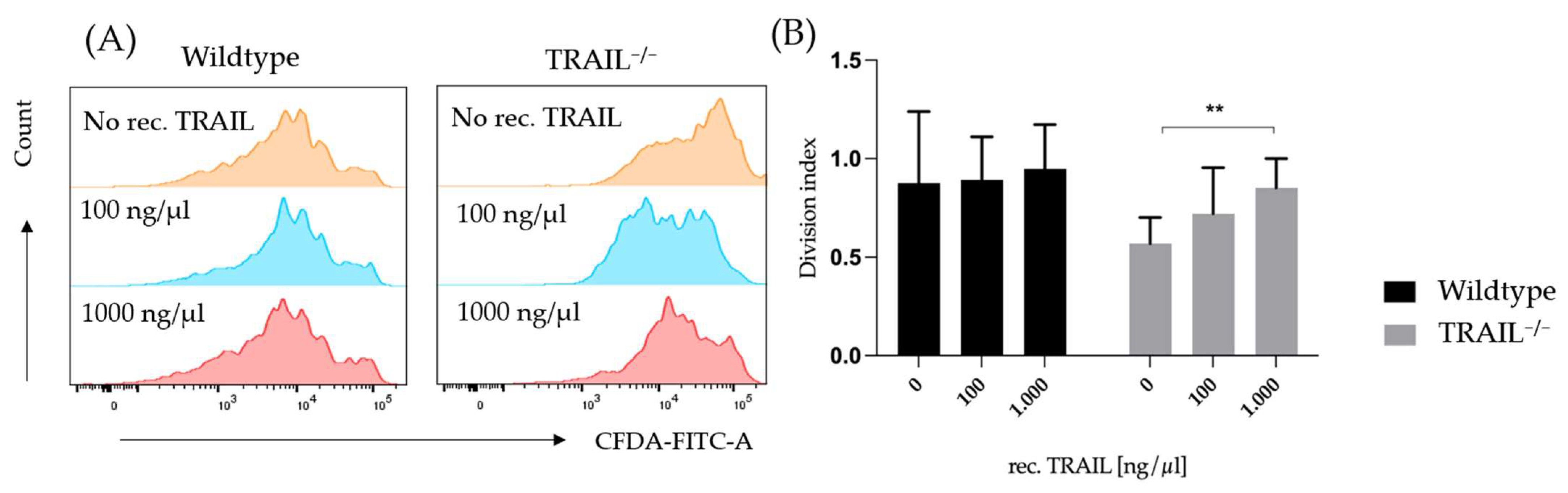
2.4. Proliferation Assay of CD3+ Cells
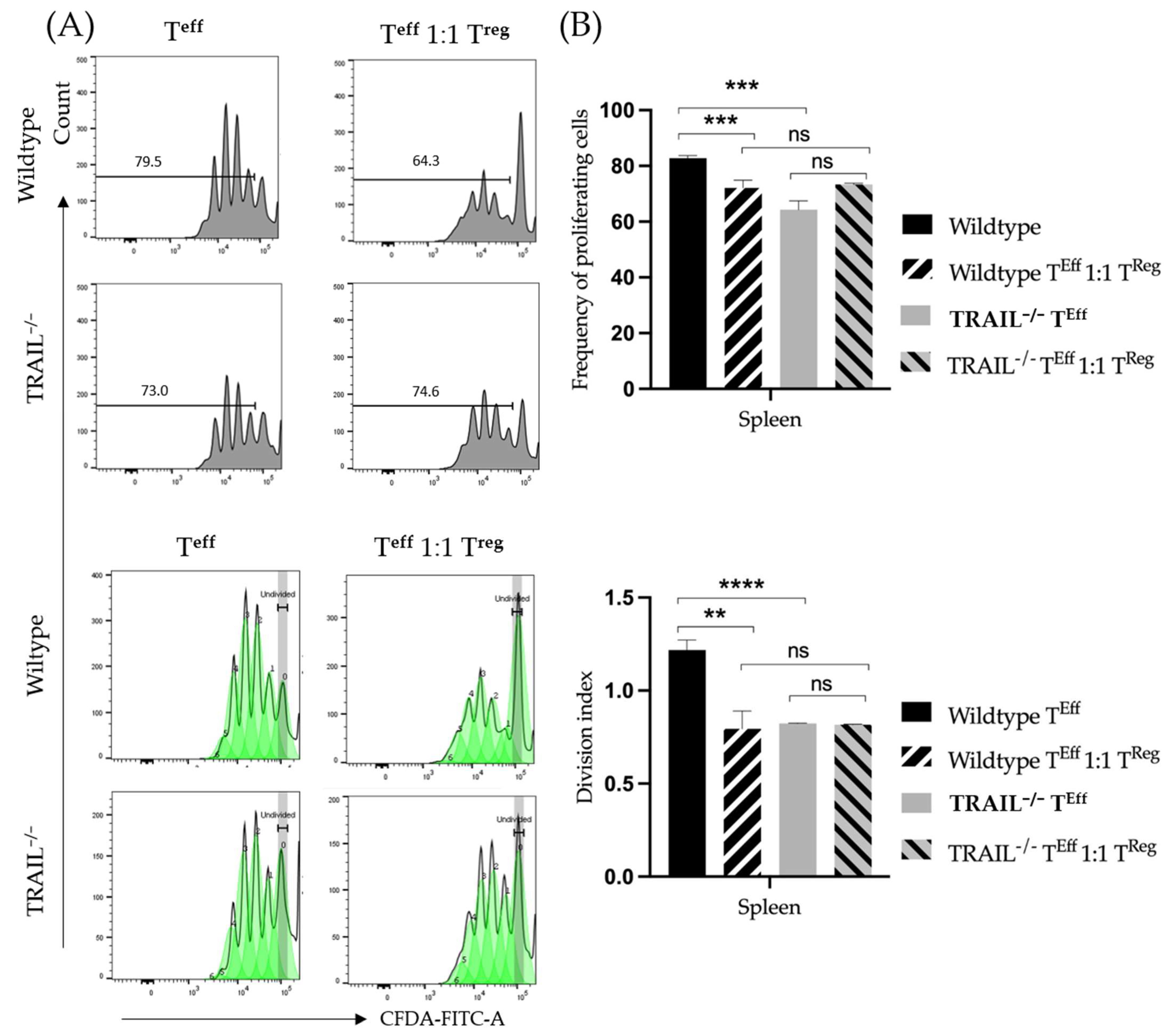
2.5. Suppression Assay
2.6. Statistical Methods
3. Results
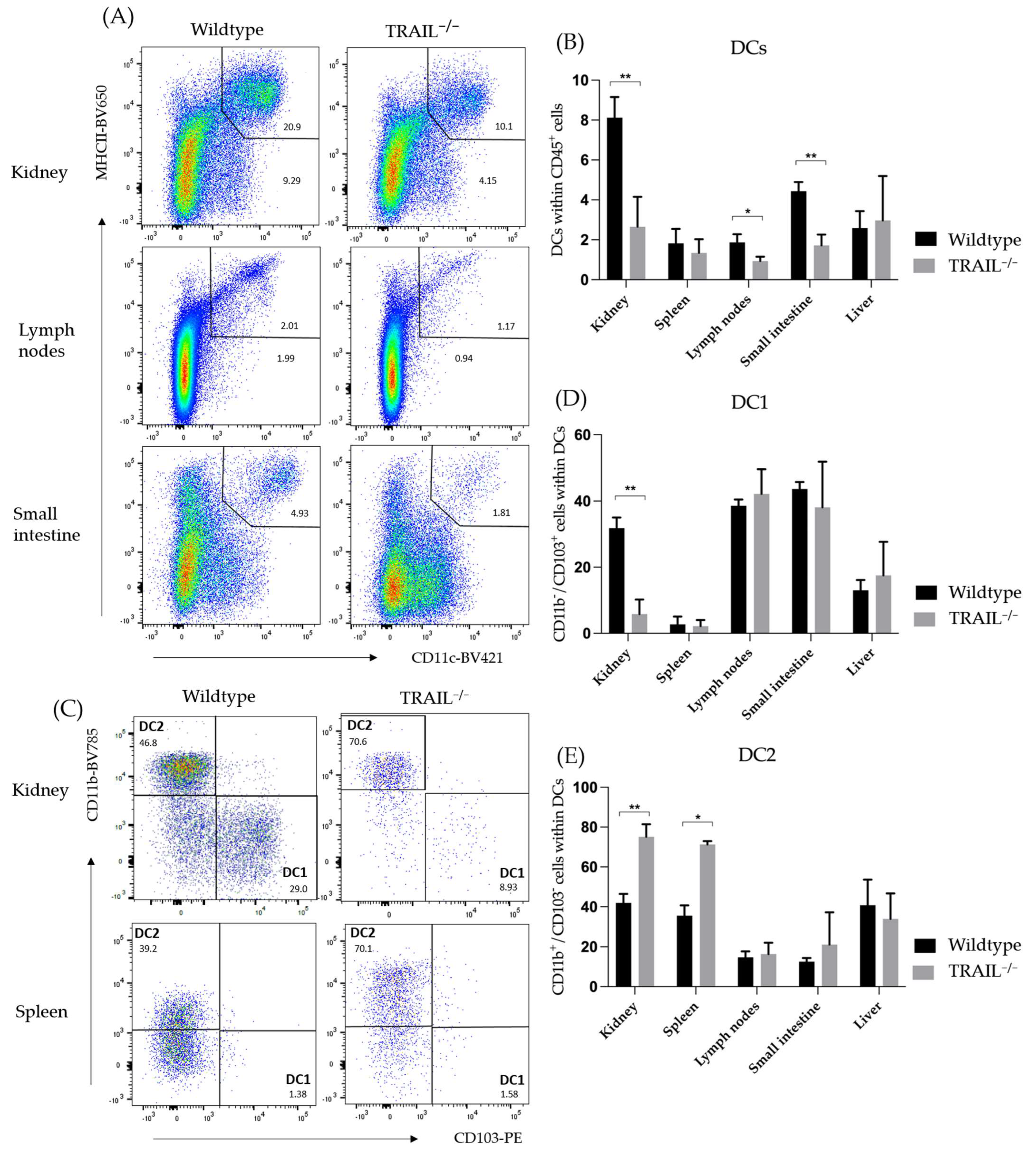
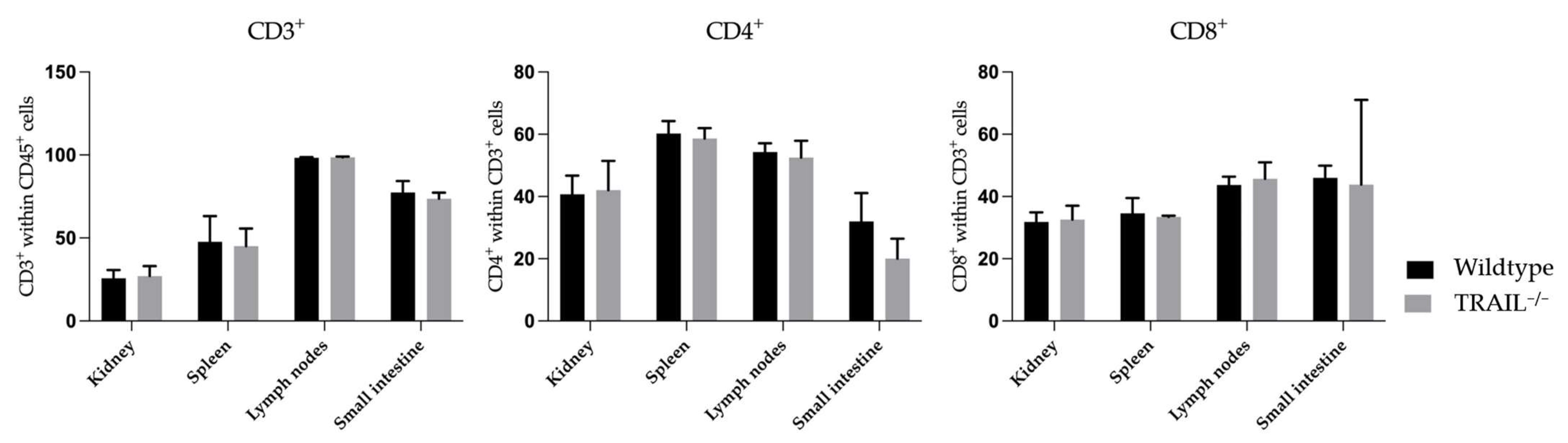
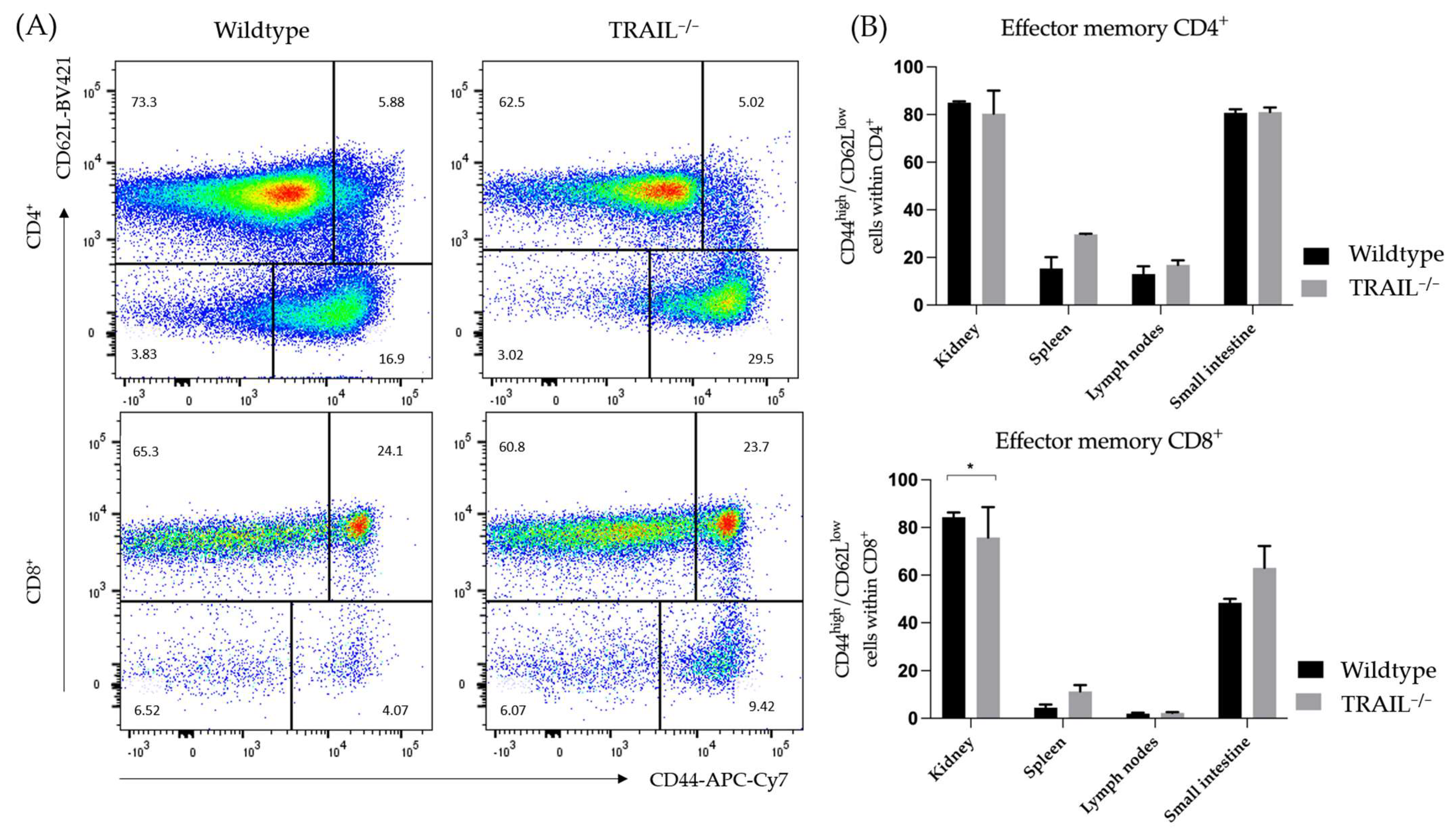
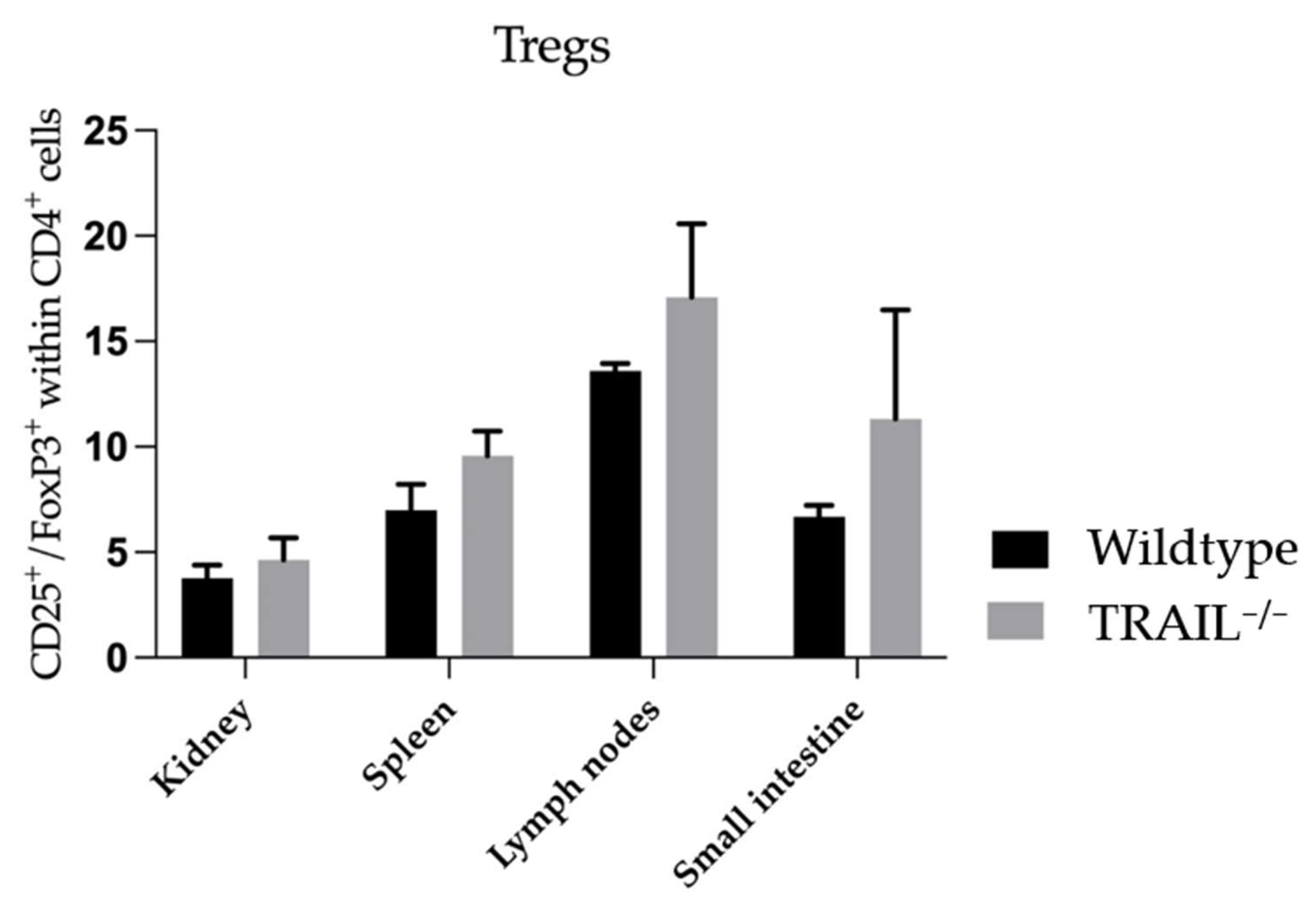
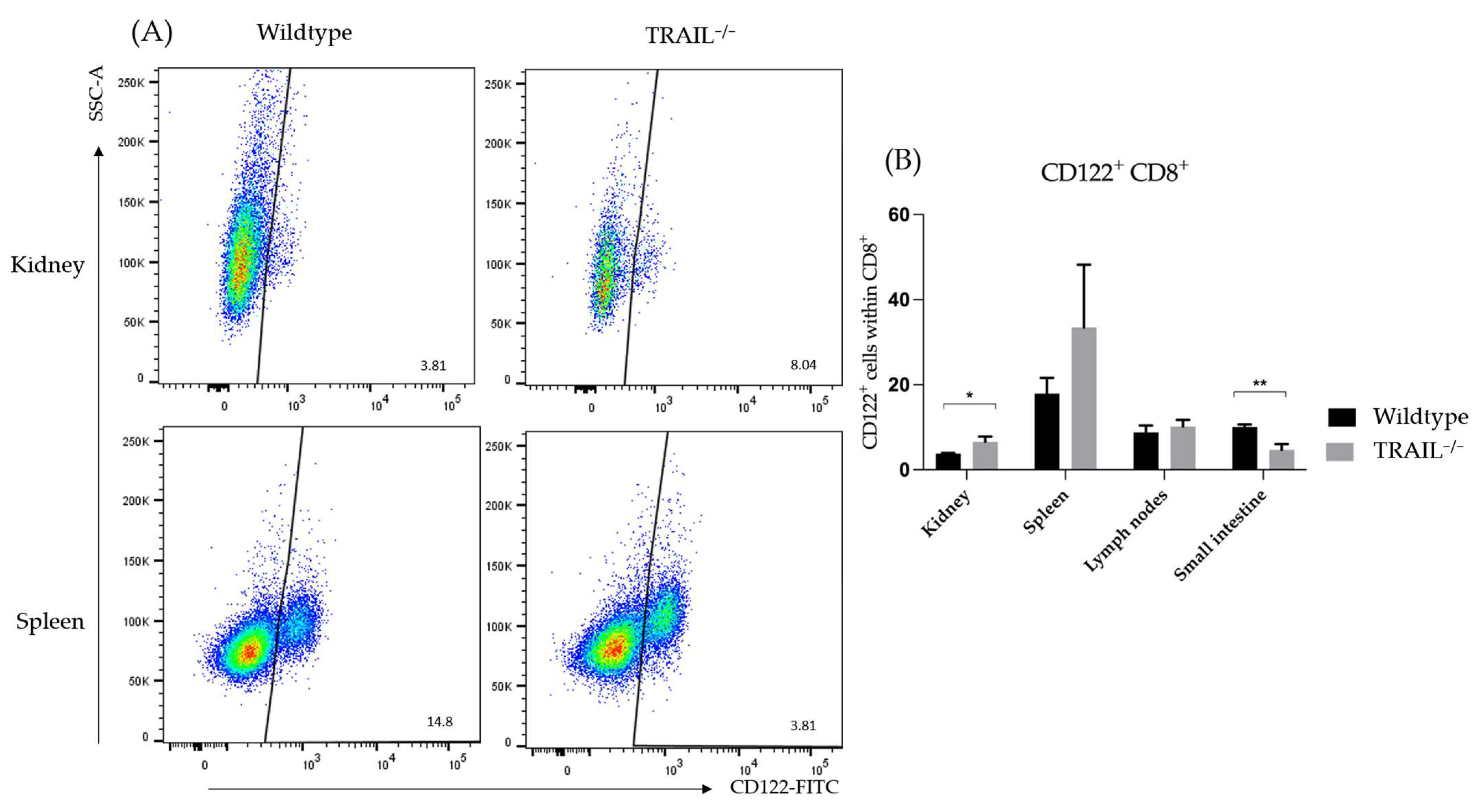
3.1. Immune Phenotyping of TRAIL−/− Mice
Frequency of Dendritic Cell (DC) Populations in a TRAIL−/− Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cretney, E.; Takeda, K.; Yagita, H.; Glaccum, M.; Peschon, J.J.; Smyth, M.J. Increased Susceptibility to Tumor Initiation and Metastasis in TNF-Related Apoptosis-Inducing Ligand-Deficient Mice. J. Immunol. 2002, 168, 1356–1361. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh Zeinebad, H.; Szegezdi, E. TRAIL in the Treatment of Cancer: From Soluble Cytokine to Nanosystems. Cancers 2022, 14, 5125. [Google Scholar] [CrossRef]
- Wu, G.S.; Burns, T.F.; Zhan, Y.; Alnemri, E.S.; El-Deiry, W.S. Molecular Cloning and Functional Analysis of the Mouse Homologue of the KILLER/DR5 Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand (TRAIL) Death Receptor. Cancer Res. 1999, 59, 2770–2775. [Google Scholar] [PubMed]
- Johnstone, R.W.; Frew, A.J.; Smyth, M.J. The TRAIL Apoptotic Pathway in Cancer Onset, Progression and Therapy. Nat. Rev. Cancer 2008, 8, 782–798. [Google Scholar] [CrossRef] [PubMed]
- Cullen, S.P.; Martin, S.J. Fas and TRAIL “death Receptors” as Initiators of Inflammation: Implications for Cancer. Semin. Cell Dev. Biol. 2015, 39, 26–34. [Google Scholar] [CrossRef]
- Zhou, D.-H.; Trauzold, A.; Röder, C.; Pan, G.; Zheng, C.; Kalthoff, H. The Potential Molecular Mechanism of Overexpression of UPA, IL-8, MMP-7 and MMP-9 Induced by TRAIL in Pancreatic Cancer Cell. Hepatobiliary Pancreat. Dis. Int. HBPD INT 2008, 7, 201–209. [Google Scholar]
- Chyuan, I.T.; Tsai, H.F.; Wu, C.S.; Hsu, P.N. TRAIL Suppresses Gut Inflammation and Inhibits Colitogeic T-Cell Activation in Experimental Colitis via an Apoptosis-Independent Pathway. Mucosal Immunol. 2019, 12, 980–989. [Google Scholar] [CrossRef]
- Chyuan, I.-T.; Tsai, H.-F.; Wu, C.-S.; Sung, C.-C.; Hsu, P.-N. TRAIL-Mediated Suppression of T Cell Receptor Signaling Inhibits T Cell Activation and Inflammation in Experimental Autoimmune Encephalomyelitis. Front. Immunol. 2018, 9, 15. [Google Scholar] [CrossRef]
- Chyuan, I.-T.; Tsai, H.-F.; Liao, H.-J.; Wu, C.-S.; Hsu, P.-N. An Apoptosis-Independent Role of TRAIL in Suppressing Joint Inflammation and Inhibiting T-Cell Activation in Inflammatory Arthritis. Cell. Mol. Immunol. 2018, 15, 846–857. [Google Scholar] [CrossRef]
- Chyuan, I.-T.; Hsu, P.-N. TRAIL Regulates T Cell Activation and Suppresses Inflammation in Autoimmune Diseases. Cell. Mol. Immunol. 2020, 17, 1281–1283. [Google Scholar] [CrossRef]
- Lehnert, C.; Weiswange, M.; Jeremias, I.; Bayer, C.; Grunert, M.; Debatin, K.-M.; Strauss, G. TRAIL–Receptor Costimulation Inhibits Proximal TCR Signaling and Suppresses Human T Cell Activation and Proliferation. J. Immunol. 2014, 193, 4021–4031. [Google Scholar] [CrossRef] [PubMed]
- Palucka, K.; Banchereau, J. Dendritic Cells: A Link between Innate and Adaptive Immunity. J. Clin. Immunol. 1999, 19, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Legge, K.L.; Gregg, R.K.; Maldonado-Lopez, R.; Li, L.; Caprio, J.C.; Moser, M.; Zaghouani, H. On the Role of Dendritic Cells in Peripheral T Cell Tolerance and Modulation of Autoimmunity. J. Exp. Med. 2002, 196, 217–227. [Google Scholar] [CrossRef]
- Del Rio, M.-L.; Bernhardt, G.; Rodriguez-Barbosa, J.-I.; Förster, R. Development and Functional Specialization of CD103+ Dendritic Cells. Immunol. Rev. 2010, 234, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Ganesan, A.; Okoye, I.; Arutyunova, E.; Elahi, S.; Lemieux, M.J.; Barakat, K. Targeting B7-1 in Immunotherapy. Med. Res. Rev. 2020, 40, 654–682. [Google Scholar] [CrossRef] [PubMed]
- Chambers, B.J.; Salcedo, M.; Ljunggren, H.G. Triggering of Natural Killer Cells by the Costimulatory Molecule CD80 (B7-1). Immunity 1996, 5, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Imasuen, I.; Bozeman, E.; He, S.; Patel, J.; Selvaraj, P. Increased B7-1 (CD80) Expression Reduces Overall Tumorigenicity and Metastatic Potential of the Murine Pancreatic Cancer Cell Model Pan02 (P2085). J. Immunol. 2013, 190, 53-43. [Google Scholar] [CrossRef]
- Vackova, J.; Polakova, I.; Johari, S.D.; Smahel, M. CD80 Expression on Tumor Cells Alters Tumor Microenvironment and Efficacy of Cancer Immunotherapy by CTLA-4 Blockade. Cancers 2021, 13, 1935. [Google Scholar] [CrossRef]
- Zhang, J.; Patel, J.M. Role of the CX3CL1-CX3CR1 Axis in Chronic Inflammatory Lung Diseases. Int. J. Clin. Exp. Med. 2010, 3, 233–244. [Google Scholar]
- Conroy, M.J.; Lysaght, J. CX3CL1 Signaling in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1231, 1–12. [Google Scholar] [CrossRef]
- Niess, J.H.; Brand, S.; Gu, X.; Landsman, L.; Jung, S.; McCormick, B.A.; Vyas, J.M.; Boes, M.; Ploegh, H.L.; Fox, J.G.; et al. CX3CR1-Mediated Dendritic Cell Access to the Intestinal Lumen and Bacterial Clearance. Science 2005, 307, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Bertani, F.R.; Mozetic, P.; Fioramonti, M.; Iuliani, M.; Ribelli, G.; Pantano, F.; Santini, D.; Tonini, G.; Trombetta, M.; Businaro, L.; et al. Classification of M1/M2-Polarized Human Macrophages by Label-Free Hyperspectral Reflectance Confocal Microscopy and Multivariate Analysis. Sci. Rep. 2017, 7, 8965. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Yang, M.; Shao, J.; Miao, Y.; Han, J.; Du, J. Chemokine Receptor CX3CR1 Contributes to Macrophage Survival in Tumor Metastasis. Mol. Cancer 2013, 12, 141. [Google Scholar] [CrossRef] [PubMed]
- Degli-Esposti, M.A.; Smolak, P.J.; Walczak, H.; Waugh, J.; Huang, C.-P.; DuBose, R.F.; Goodwin, R.G.; Smith, C.A. Cloning and Characterization of TRAIL-R3, a Novel Member of the Emerging TRAIL Receptor Family. J. Exp. Med. 1997, 186, 1165–1170. [Google Scholar] [CrossRef]
- Liu, J.; Chen, D.; Nie, G.D.; Dai, Z. CD8+CD122+ T-Cells: A Newly Emerging Regulator with Central Memory Cell Phenotypes. Front. Immunol. 2015, 6, 494. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Hirata, S.; Fukushima, S.; Matsunaga, Y.; Ito, T.; Uchino, M.; Nishimura, Y.; Senju, S. Dual Effects of TRAIL in Suppression of Autoimmunity: The Inhibition of Th1 Cells and the Promotion of Regulatory T Cells. J. Immunol. 2010, 185, 5259–5267. [Google Scholar] [CrossRef] [PubMed]
- Finnberg, N.; Klein-Szanto, A.J.P.; El-Deiry, W.S. TRAIL-R Deficiency in Mice Promotes Susceptibility to Chronic Inflammation and Tumorigenesis. J. Clin. Investig. 2008, 118, 111–123. [Google Scholar] [CrossRef]
- Seki, N.; Hayakawa, Y.; Brooks, A.D.; Wine, J.; Wiltrout, R.H.; Yagita, H.; Tanner, J.E.; Smyth, M.J.; Sayers, T.J. Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand-Mediated Apoptosis Is an Important Endogenous Mechanism for Resistance to Liver Metastases in Murine Renal Cancer. Cancer Res. 2003, 63, 207–213. [Google Scholar]
- Lamhamedi-Cherradi, S.-E.; Zheng, S.-J.; Maguschak, K.A.; Peschon, J.; Chen, Y.H. Defective Thymocyte Apoptosis and Accelerated Autoimmune Diseases in TRAIL-/- Mice. Nat. Immunol. 2003, 4, 255–260. [Google Scholar] [CrossRef]
- Smyth, M.J.; Cretney, E.; Takeda, K.; Wiltrout, R.H.; Sedger, L.M.; Kayagaki, N.; Yagita, H.; Okumura, K. Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand (TRAIL) Contributes to Interferon Gamma-Dependent Natural Killer Cell Protection from Tumor Metastasis. J. Exp. Med. 2001, 193, 661–670. [Google Scholar] [CrossRef]
- Beyer, K.; Normann, L.; Sendler, M.; Käding, A.; Heidecke, C.-D.; Partecke, L.I.; von Bernstorff, W. TRAIL Promotes Tumor Growth in a Syngeneic Murine Orthotopic Pancreatic Cancer Model and Affects the Host Immune Response. Pancreas 2016, 45, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Adams, A.B.; Williams, M.A.; Jones, T.R.; Shirasugi, N.; Durham, M.M.; Kaech, S.M.; Wherry, E.J.; Onami, T.; Lanier, J.G.; Kokko, K.E.; et al. Heterologous Immunity Provides a Potent Barrier to Transplantation Tolerance. J. Clin. Investig. 2003, 111, 1887–1895. [Google Scholar] [CrossRef] [PubMed]
- Lerret, N.M.; Houlihan, J.L.; Kheradmand, T.; Pothoven, K.L.; Zhang, Z.J.; Luo, X. Donor-Specific CD8+Foxp3+ T Cells Protect Skin Allografts and Facilitate Induction of Conventional CD4+Foxp3+ Regulatory T Cells. Am. J. Transplant. 2012, 12, 2335–2347. [Google Scholar] [CrossRef] [PubMed]
- Klebanoff, C.A.; Gattinoni, L.; Torabi-Parizi, P.; Kerstann, K.; Cardones, A.R.; Finkelstein, S.E.; Palmer, D.C.; Antony, P.A.; Hwang, S.T.; Rosenberg, S.A.; et al. Central Memory Self/Tumor-Reactive CD8+ T Cells Confer Superior Antitumor Immunity Compared with Effector Memory T Cells. Proc. Natl. Acad. Sci. USA 2005, 102, 9571–9576. [Google Scholar] [CrossRef] [PubMed]
- Plesca, I.; Benešová, I.; Beer, C.; Sommer, U.; Müller, L.; Wehner, R.; Heiduk, M.; Aust, D.; Baretton, G.; Bachmann, M.P.; et al. Clinical Significance of Tumor-Infiltrating Conventional and Plasmacytoid Dendritic Cells in Pancreatic Ductal Adenocarcinoma. Cancers 2022, 14, 1216. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, D.; Liu, D.; Liu, M.; Ge, Y.; Jiang, M.; Liu, Y.; Zheng, D. Tumor Necrosis Factor–Related Apoptosis-Inducing Ligand Induces the Expression of Proinflammatory Cytokines in Macrophages and Re-Educates Tumor-Associated Macrophages to an Antitumor Phenotype. Mol. Biol. Cell 2015, 26, 3178–3189. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.T.; Kim, J.H.; Kwon, S.J.; Stein, M.N.; Hong, J.H.; Nagaya, N.; Billakanti, S.; Kim, M.M.; Kim, W.-J.; Kim, I.Y. Dihydrotestosterone Increases Cytotoxic Activity of Macrophages on Prostate Cancer Cells via TRAIL. Endocrinology 2019, 160, 2049–2060. [Google Scholar] [CrossRef]
- Voigt, S.; Philipp, S.; Davarnia, P.; Winoto-Morbach, S.; Röder, C.; Arenz, C.; Trauzold, A.; Kabelitz, D.; Schütze, S.; Kalthoff, H.; et al. TRAIL-Induced Programmed Necrosis as a Novel Approach to Eliminate Tumor Cells. BMC Cancer 2014, 14, 74. [Google Scholar] [CrossRef]
- Eom, Y.W.; Akter, R.; Li, W.; Lee, S.; Hwang, S.; Kim, J.; Cho, M.-Y. M1 Macrophages Promote TRAIL Expression in Adipose Tissue-Derived Stem Cells, Which Suppresses Colitis-Associated Colon Cancer by Increasing Apoptosis of CD133+ Cancer Stem Cells and Decreasing M2 Macrophage Population. Int. J. Mol. Sci. 2020, 21, 3887. [Google Scholar] [CrossRef]
- Dominguez, G.A.; Condamine, T.; Mony, S.; Hashimoto, A.; Wang, F.; Liu, Q.; Forero, A.; Bendell, J.; Witt, R.; Hockstein, N.; et al. Selective Targeting of Myeloid-Derived Suppressor Cells in Cancer Patients Using DS-8273a, an Agonistic TRAIL-R2 Antibody. Clin. Cancer Res. 2017, 23, 2942–2950. [Google Scholar] [CrossRef]
- Sag, D.; Ayyildiz, Z.O.; Gunalp, S.; Wingender, G. The Role of TRAIL/DRs in the Modulation of Immune Cells and Responses. Cancers 2019, 11, 1469. [Google Scholar] [CrossRef] [PubMed]
- Liguori, M.; Buracchi, C.; Pasqualini, F.; Bergomas, F.; Pesce, S.; Sironi, M.; Grizzi, F.; Mantovani, A.; Belgiovine, C.; Allavena, P. Functional TRAIL Receptors in Monocytes and Tumor-Associated Macrophages: A Possible Targeting Pathway in the Tumor Microenvironment. Oncotarget 2016, 7, 41662–41676. [Google Scholar] [CrossRef] [PubMed]




| Marker | Fluorophore | Company |
|---|---|---|
| CD3 | PerCP | Miltenyi Biotec GmbH, Bergisch Gladbach, Germany |
| CD8 | BV605 | BioLegend, San Diego, CA, USA |
| CD44 | APC-eFluor780 | eBioScience, Waltham, MA, USA |
| CD62L | BV421 | BioLegend, San Diego, CA, USA |
| FoxP3 | Alexa-fluor-647 | BD Biosciences, San Jose, CA, USA |
| CD25 | PE | Miltenyi Biotec GmbH, Bergisch Gladbach, Germany |
| CD45 | BUV395 | BD Biosciences, San Jose, CA, USA |
| CD122 | FITC | BioLegend, San Diego, CA, USA |
| CD127 | BV785 | BioLegend, San Diego, CA, USA |
| DR5 | VioBrightB515 (FITC) | Miltenyi Biotec GmbH, Bergisch Gladbach, Germany |
| Dcr1 | APC | Miltenyi Biotec GmbH, Bergisch Gladbach, Germany |
| DcR2 | PE | BioLegend, San Diego, CA, USA |
| Ki67 | PE-eFluor 610 | ThermoFisher, Waltham, MA, USA |
| B220 * | BV510 | BioLegend, San Diego, CA, USA |
| Live/dead * | BV510 | BioLegend, San Diego, CA, USA |
| CD4 | PeCy7 | BD Biosciences, San Jose, CA, USA |
| BV711 | BioLegend, San Diego, CA, USA |
| Marker | Fluorophore | Company |
|---|---|---|
| CD3 * | BV510 | BioLegend, San Diego, CA, USA |
| NKp60 * | BV510 | BioLegend, San Diego, CA, USA |
| B220 * | BV510 | BioLegend, San Diego, CA, USA |
| CD45 | UV395 | BD Biosciences, San Jose, CA, USA |
| CD11c | BV421 | BioLegend, San Diego, CA, USA |
| CD11b | BV785 | BioLegend, San Diego, CA, USA |
| MHCII | BV650 | BioLegend, San Diego, CA, USA |
| F4/80 | APCCy7 | BioLegend, San Diego, CA, USA |
| CD64 | Pe-Cy7 | BioLegend, San Diego, CA, USA |
| Ly6C | PE-Dazzle | BioLegend, San Diego, CA, USA |
| CD80 | FITC | BioLegend, San Diego, CA, USA |
| CD8a | BV605 | BioLegend, San Diego, CA, USA |
| CD103 | PE | BioLegend, San Diego, CA, USA |
| CX3CR1 | PerCPCy5.5 | BioLegend, San Diego, CA, USA |
| CD38 | BV711 | BD Biosciences, San Jose, CA, USA |
| MHCI H2KD | Alexa700 | BioLegend, San Diego, CA, USA |
| Live/dead * | BV510 | BioLegend, San Diego, CA, USA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stoyanova, A.K.; Sattler, A.; Hahn, E.M.; Hering, N.A.; Arndt, M.; Lauscher, J.C.; Speichinger-Hillenberg, F.; Kotsch, K.; Berg, A.-K.; Beyer, K. Immune Phenotypic Characterization of a TRAIL-Knockout Mouse. Cancers 2023, 15, 1475. https://doi.org/10.3390/cancers15051475
Stoyanova AK, Sattler A, Hahn EM, Hering NA, Arndt M, Lauscher JC, Speichinger-Hillenberg F, Kotsch K, Berg A-K, Beyer K. Immune Phenotypic Characterization of a TRAIL-Knockout Mouse. Cancers. 2023; 15(5):1475. https://doi.org/10.3390/cancers15051475
Chicago/Turabian StyleStoyanova, Ani K., Arne Sattler, Elisabeth M. Hahn, Nina A. Hering, Marco Arndt, Johannes Christian Lauscher, Fiona Speichinger-Hillenberg, Katja Kotsch, Ann-Kathrin Berg, and Katharina Beyer. 2023. "Immune Phenotypic Characterization of a TRAIL-Knockout Mouse" Cancers 15, no. 5: 1475. https://doi.org/10.3390/cancers15051475
APA StyleStoyanova, A. K., Sattler, A., Hahn, E. M., Hering, N. A., Arndt, M., Lauscher, J. C., Speichinger-Hillenberg, F., Kotsch, K., Berg, A.-K., & Beyer, K. (2023). Immune Phenotypic Characterization of a TRAIL-Knockout Mouse. Cancers, 15(5), 1475. https://doi.org/10.3390/cancers15051475







