Interferon Gamma-Inducible NAMPT in Melanoma Cells Serves as a Mechanism of Resistance to Enhance Tumor Growth
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Cell Cultures and Reagents
2.2. In Vitro Activation, NAMPT Inhibition, and Agonist Treatment
2.3. RNA Isolation from Cultured Cells and RT-qPCR
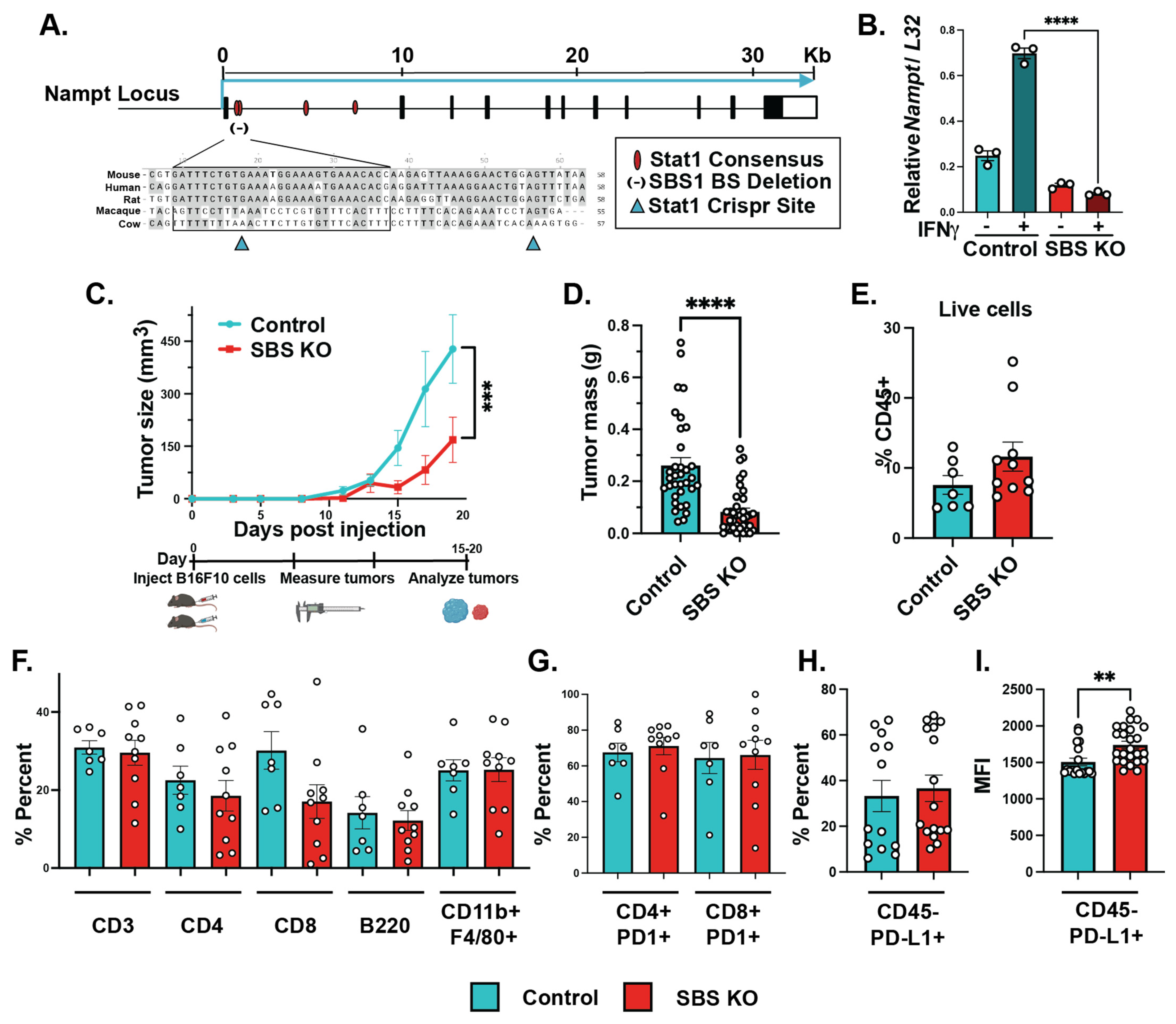
2.4. CRISPR/Cas9-Mediated Gene Deletion of the STAT1 Binding Site in Tumor Cell Lines
2.5. Western Blots
2.6. Mice
2.7. B16-F10 Melanoma Experiments
2.8. SBS Mutation Sequencing
2.9. Flow Cytometry
2.10. Incucyte Live Image Analysis
2.11. Statistical Analysis
3. Results
3.1. IFNs Induce NAMPT in Melanoma Cells
3.2. IFNγ-Inducible NAMPT Plays an Important Role in Melanoma Cell Growth
3.3. Mutations in NAMPT STAT1 Binding Sites Lead to Significantly Lower Levels of IFNγ-Dependent NAMPT Induction
3.4. IFNγ-Dependent NAMPT Induction Plays a Tumor Cell-Intrinsic Role in Promoting Melanoma Growth In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Cancer Society Cancer Facts & Figures 2017. Available online: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2017/cancer-facts-and-figures-2017.pdf (accessed on 15 August 2022).
- Domingues, B.; Lopes, J.; Soares, P.; Populo, H. Melanoma treatment in review. Immunotargets Ther. 2018, 7, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Kozar, I.; Margue, C.; Rothengatter, S.; Haan, C.; Kreis, S. Many ways to resistance: How melanoma cells evade targeted therapies. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Garbe, C.; Krasagakis, K. Effects of interferons and cytokines on melanoma cells. J. Investig. Dermatol. 1993, 100, 239S–244S. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Ashkar, A. The Dual Nature of Type I and Type II Interferons. Front. Immunol. 2018, 9, 2061. [Google Scholar] [CrossRef]
- Schiller, J.; Willson, J.; Bittner, G.; Wolberg, W.; Hawkins, M.; Borden, E.C. Antiproliferative effects of interferons on human melanoma cells in the human tumor colony-forming assay. J. Interferon. Res. 1986, 6, 615–625. [Google Scholar] [CrossRef]
- Castro, F.; Cardoso, A.; Goncalves, R.; Serre, K.; Oliveira, M.J. Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion. Front. Immunol. 2018, 9, 847. [Google Scholar] [CrossRef]
- Musella, M.; Manic, G.; De Maria, R.; Vitale, I.; Sistigu, A. Type-I-interferons in infection and cancer: Unanticipated dynamics with therapeutic implications. Oncoimmunology 2017, 6, e1314424. [Google Scholar] [CrossRef]
- Hou, W.; Medynski, D.; Wu, S.; Lin, X.; Li, L.Y. VEGI-192, a new isoform of TNFSF15, specifically eliminates tumor vascular endothelial cells and suppresses tumor growth. Clin. Cancer Res. 2005, 11, 5595–5602. [Google Scholar] [CrossRef]
- Schreiner, B.; Mitsdoerffer, M.; Kieseier, B.; Chen, L.; Hartung, H.; Weller, M.; Wiendl, H. Interferon-beta enhances monocyte and dendritic cell expression of B7-H1 (PD-L1), a strong inhibitor of autologous T-cell activation: Relevance for the immune modulatory effect in multiple sclerosis. J. Neuroimmunol. 2004, 155, 172–182. [Google Scholar] [CrossRef]
- Jewell, A.; Worman, C.; Lydyard, P.; Yong, K.; Giles, F.; Goldstone, A.H. Interferon-alpha up-regulates bcl-2 expression and protects B-CLL cells from apoptosis in vitro and in vivo. Br. J. Haematol. 1994, 88, 268–274. [Google Scholar] [CrossRef]
- Garten, A.; Schuster, S.; Penke, M.; Gorski, T.; de Giorgis, T.; Kiess, W. Physiological and pathophysiological roles of NAMPT and NAD metabolism. Nat. Rev. Endocrinol. 2015, 11, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Verdin, E. NAD(+) in aging, metabolism, and neurodegeneration. Science 2015, 350, 1208–1213. [Google Scholar] [CrossRef] [PubMed]
- Garrido, A.; Djouder, N. NAD(+) Deficits in Age-Related Diseases and Cancer. Trends Cancer 2017, 3, 593–610. [Google Scholar] [CrossRef] [PubMed]
- Maldi, E.; Travelli, C.; Caldarelli, A.; Agazzone, N.; Cintura, S.; Galli, U.; Scatolini, M.; Ostano, P.; Miglino, B.; Chiorino, G.; et al. Nicotinamide phosphoribosyltransferase (NAMPT) is over-expressed in melanoma lesions. Pigment. Cell Melanoma Res. 2013, 26, 144–146. [Google Scholar] [CrossRef]
- Ohanna, M.; Cerezo, M.; Nottet, N.; Bille, K.; Didier, R.; Beranger, G.; Mograbi, B.; Rocchi, S.; Yvan-Charvet, L.; Ballotti, R.; et al. Pivotal role of NAMPT in the switch of melanoma cells toward an invasive and drug-resistant phenotype. Genes Dev. 2018, 32, 448–461. [Google Scholar] [CrossRef]
- Espindola-Netto, J.; Chini, C.; Tarrago, M.; Wang, E.; Dutta, S.; Pal, K.; Mukhopadhyay, D.; Sola-Penna, M.; Chini, E.N. Preclinical efficacy of the novel competitive NAMPT inhibitor STF-118804 in pancreatic cancer. Oncotarget 2017, 8, 85054–85067. [Google Scholar] [CrossRef]
- Huffaker, T.; Ekiz, H.; Barba, C.; Lee, S.; Runtsch, M.; Nelson, M.; Bauer, K.; Tang, W.; Mosbruger, T.; Cox, J.; et al. A Stat1 bound enhancer promotes Nampt expression and function within tumor associated macrophages. Nat. Commun. 2021, 12, 2620. [Google Scholar] [CrossRef]
- Wallace, J.; Hu, R.; Mosbruger, T.; Dahlem, T.; Stephens, W.; Rao, D.; Round, J.; O’Connell, R.M. Genome-Wide CRISPR-Cas9 Screen Identifies MicroRNAs That Regulate Myeloid Leukemia Cell Growth. PLoS ONE 2016, 11, e0153689. [Google Scholar] [CrossRef]
- Wallace, J.; Kagele, D.; Eiring, A.; Kim, C.; Hu, R.; Runtsch, M.; Alexander, M.; Huffaker, T.; Lee, S.-H.; Patel, A.; et al. miR-155 promotes FLT3-ITD-induced myeloproliferative disease through inhibition of the interferon response. Blood 2017, 129, 3074–3086. [Google Scholar] [CrossRef]
- Meeth, K.; Wang, J.; Micevic, G.; Damsky, W.; Bosenberg, M.W. The YUMM lines: A series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment. Cell Melanoma Res. 2016, 29, 590–597. [Google Scholar] [CrossRef]
- Borden, E.C. Interferons alpha and beta in cancer: Therapeutic opportunities from new insights. Nat. Rev. Drug Discov. 2019, 18, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Puthenveetil, A.; Dubey, S. Metabolic reprograming of tumor-associated macrophages. Ann. Transl. Med. 2020, 8, 1030. [Google Scholar] [CrossRef] [PubMed]
- Meares, G.; Qin, H.; Liu, Y.; Holdbrooks, A.; Benveniste, E.N. AMP-activated protein kinase restricts IFN-gamma signaling. J. Immunol. 2013, 190, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Yu, Y.; Zhong, Y.; Giannopoulou, E.; Hu, X.; Liu, H.; Cross, J.; Ratsch, G.; Rice, C.; Ivashkiv, L.B. Interferon-gamma regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat. Immunol. 2015, 16, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Grolla, A.; Travelli, C.; Genazzani, A.; Sethi, J.K. Extracellular nicotinamide phosphoribosyltransferase, a new cancer metabokine. Br. J. Pharmacol. 2016, 173, 2182–2194. [Google Scholar] [CrossRef]
- Colombo, G.; Travelli, C.; Porta, C.; Genazzani, A.A. Extracellular nicotinamide phosphoribosyltransferase boosts IFNgamma-induced macrophage polarization independently of TLR4. iScience 2022, 25, 104147. [Google Scholar]
- Grasso, C.; Tsoi, J.; Onyshchenko, M.; Abril-Rodriguez, G.; Ross-Macdonald, P.; Wind-Rotolo, M.; Champhekar, A.; Medina, E.; Torrejon, D.; Shin, D.; et al. Conserved Interferon-gamma Signaling Drives Clinical Response to Immune Checkpoint Blockade Therapy in Melanoma. Cancer Cell 2021, 39, 122. [Google Scholar] [CrossRef]
- Benci, J.; Xu, B.; Qiu, Y.; Wu, T.; Dada, H.; Victor, C.T.-S.; Cucolo, L.; Lee, D.; Pauken, K.; Huang, A.; et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell 2016, 167, 1540.e1512–1554.e1512. [Google Scholar] [CrossRef]
- Topalian, S.; Hodi, F.; Brahmer, J.; Gettinger, S.; Smith, D.; McDermott, D.; Powderly, J.; Carvajal, R.; Sosman, J.; Atkins, M.; et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef]
- Lv, H.; Lv, G.; Chen, C.; Zong, Q.; Jiang, G.; Ye, D.; Cui, X.; He, Y.; Xiang, W.; Han, Q.; et al. NAD(+) Metabolism Maintains Inducible PD-L1 Expression to Drive Tumor Immune Evasion. Cell Metab. 2021, 33, 110.e115–127.e115. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barba, C.; Ekiz, H.A.; Tang, W.W.; Ghazaryan, A.; Hansen, M.; Lee, S.-H.; Voth, W.P.; O’Connell, R.M. Interferon Gamma-Inducible NAMPT in Melanoma Cells Serves as a Mechanism of Resistance to Enhance Tumor Growth. Cancers 2023, 15, 1411. https://doi.org/10.3390/cancers15051411
Barba C, Ekiz HA, Tang WW, Ghazaryan A, Hansen M, Lee S-H, Voth WP, O’Connell RM. Interferon Gamma-Inducible NAMPT in Melanoma Cells Serves as a Mechanism of Resistance to Enhance Tumor Growth. Cancers. 2023; 15(5):1411. https://doi.org/10.3390/cancers15051411
Chicago/Turabian StyleBarba, Cindy, H. Atakan Ekiz, William Weihao Tang, Arevik Ghazaryan, Mason Hansen, Soh-Hyun Lee, Warren Peter Voth, and Ryan Michael O’Connell. 2023. "Interferon Gamma-Inducible NAMPT in Melanoma Cells Serves as a Mechanism of Resistance to Enhance Tumor Growth" Cancers 15, no. 5: 1411. https://doi.org/10.3390/cancers15051411
APA StyleBarba, C., Ekiz, H. A., Tang, W. W., Ghazaryan, A., Hansen, M., Lee, S.-H., Voth, W. P., & O’Connell, R. M. (2023). Interferon Gamma-Inducible NAMPT in Melanoma Cells Serves as a Mechanism of Resistance to Enhance Tumor Growth. Cancers, 15(5), 1411. https://doi.org/10.3390/cancers15051411





