Serum Response Factor-Regulated IDO1/Kyn-Ahr Pathway Promotes Tumorigenesis of Oral Squamous Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tumor Samples
2.2. Cell Culture and Reagents
2.3. Immunohistochemical Staining
2.4. Creation of Cell Lines Stably Overexpressing SRF
2.5. Tumorigenicity Assays in Nude Mice
2.6. Western Blotting
2.7. Wound Healing Assays
2.8. Migration and Invasion Assays
2.9. Sequencing RNA and Hybridization
2.10. RNA Isolation and qRT-PCR
2.11. Kynurenine Measurements
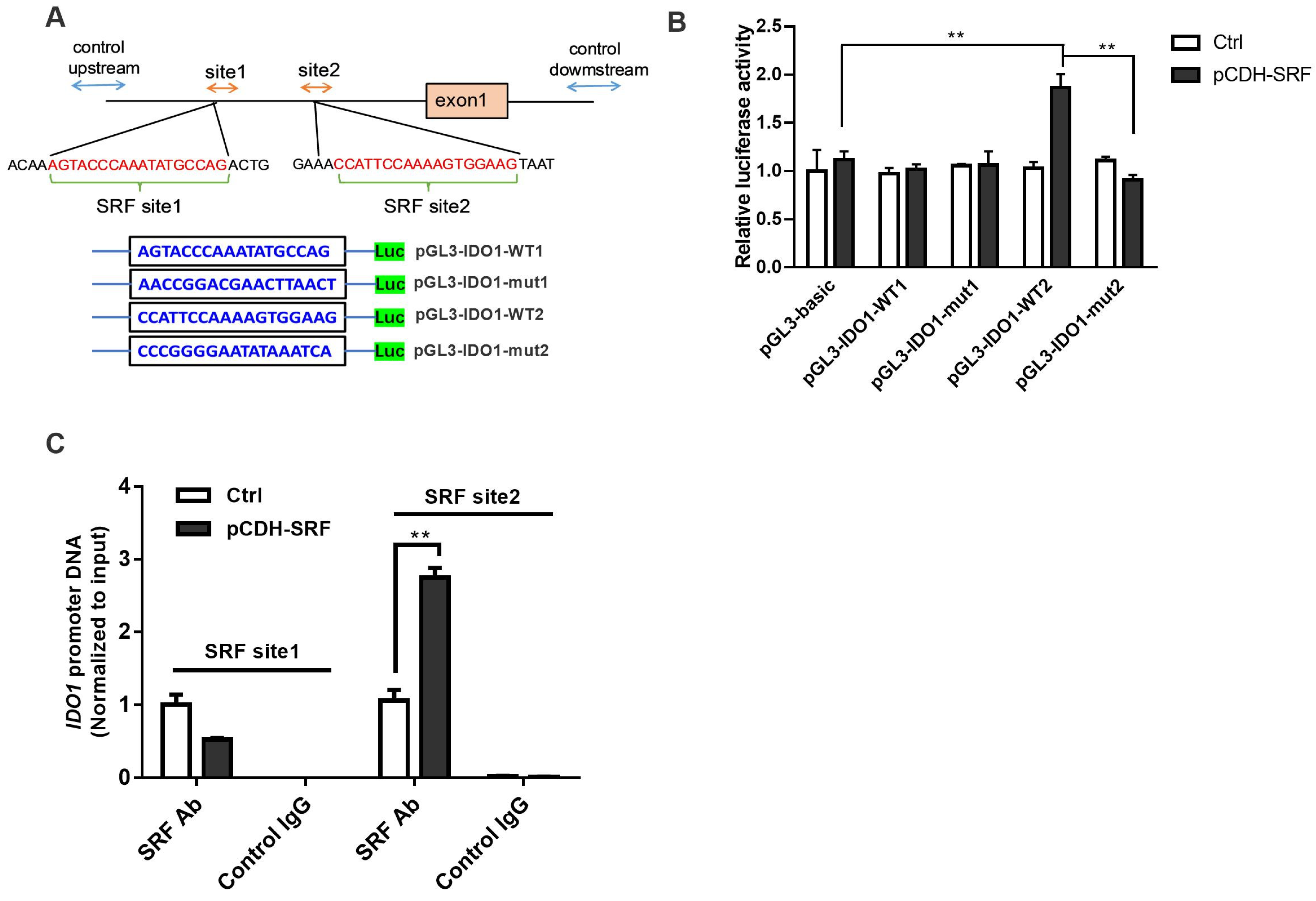
2.12. Dual Luciferase Reporter Assays
2.13. Chromatin Immunoprecipitation (ChIP) Assay
2.14. Signal Transduction Assays
2.15. Statistical Analysis
3. Results
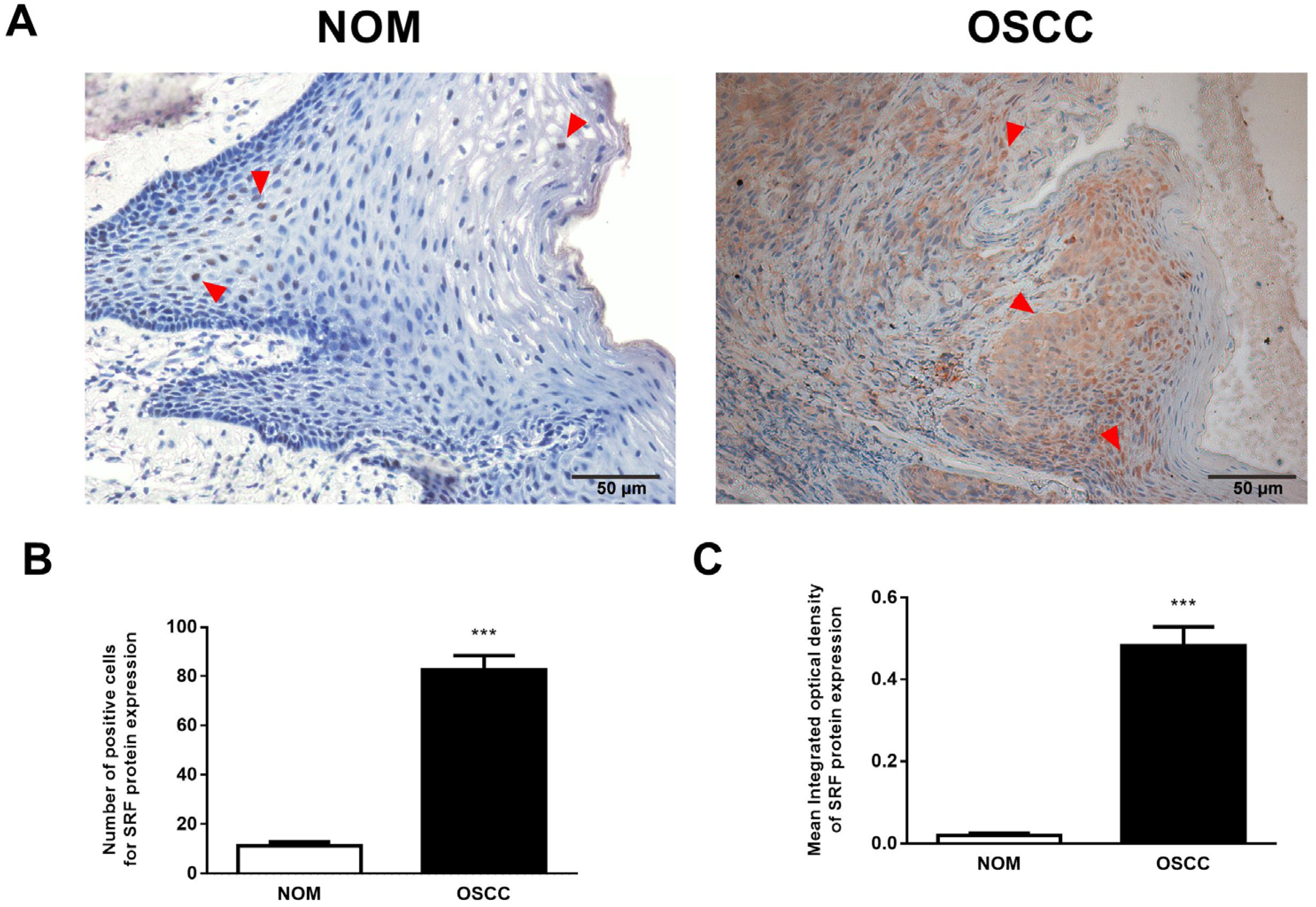
3.1. Upregulated SRF Expression Is Correlated with Tumor Invasion and Metastasis in Human OSCC Tissues
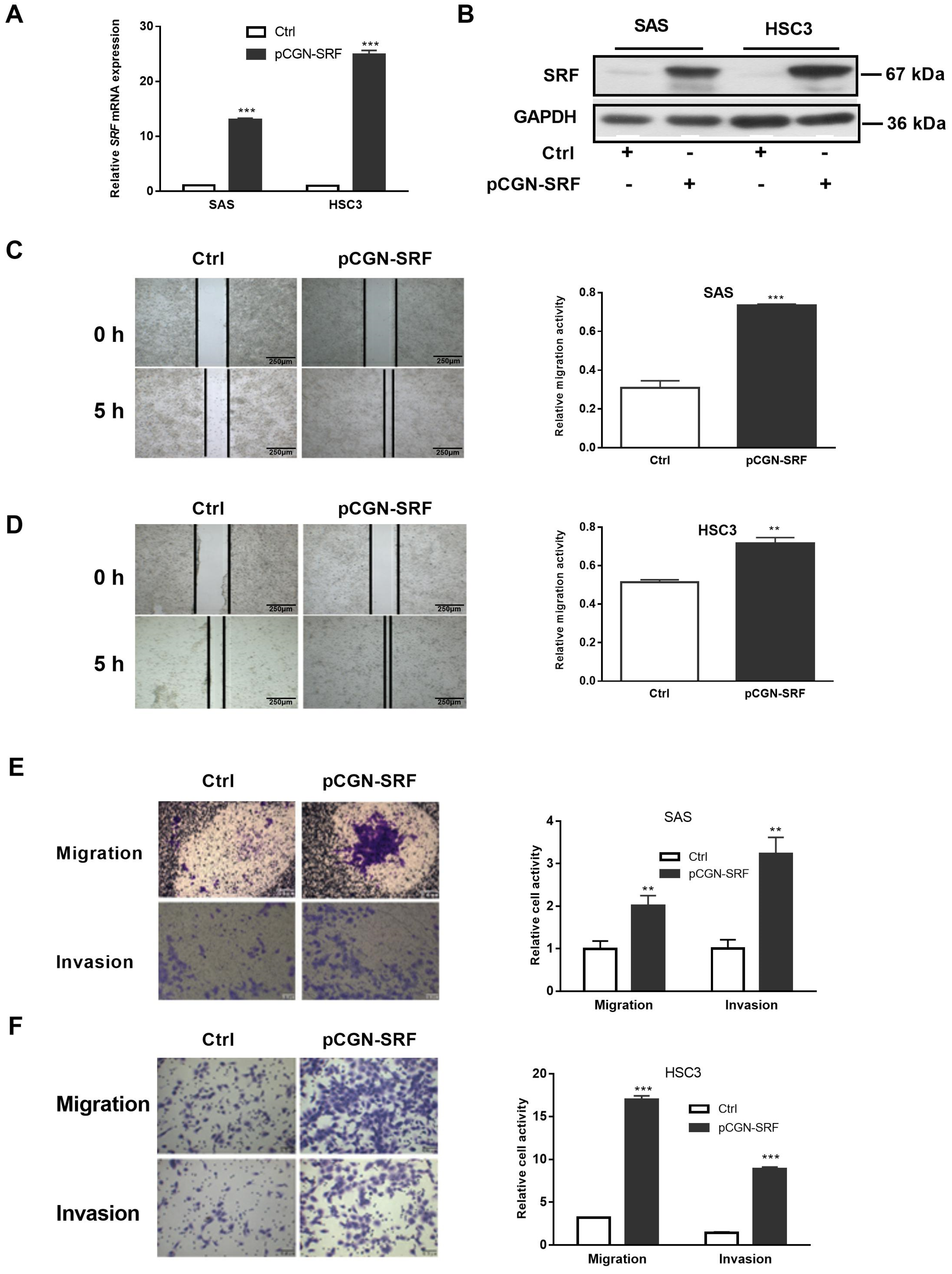
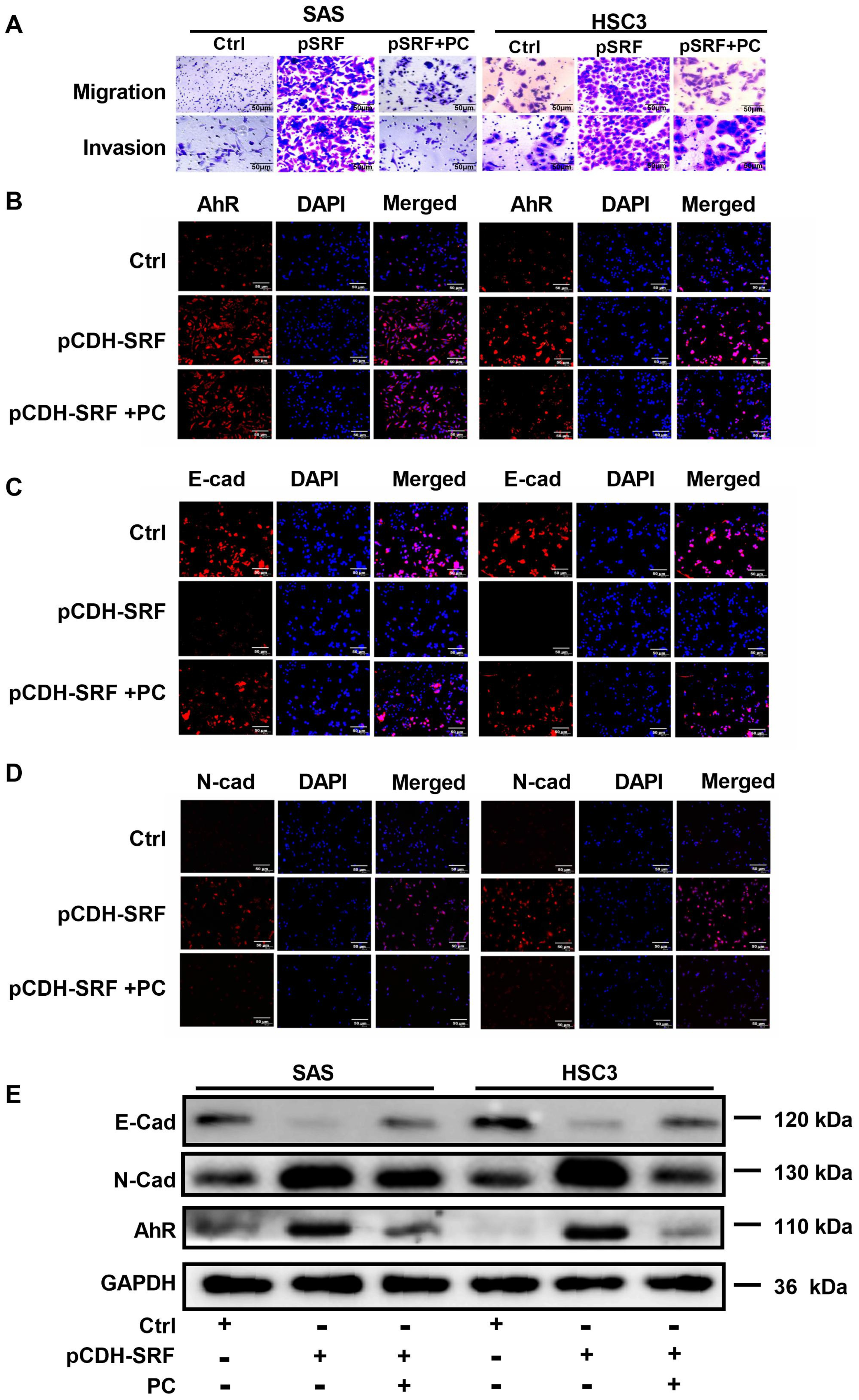
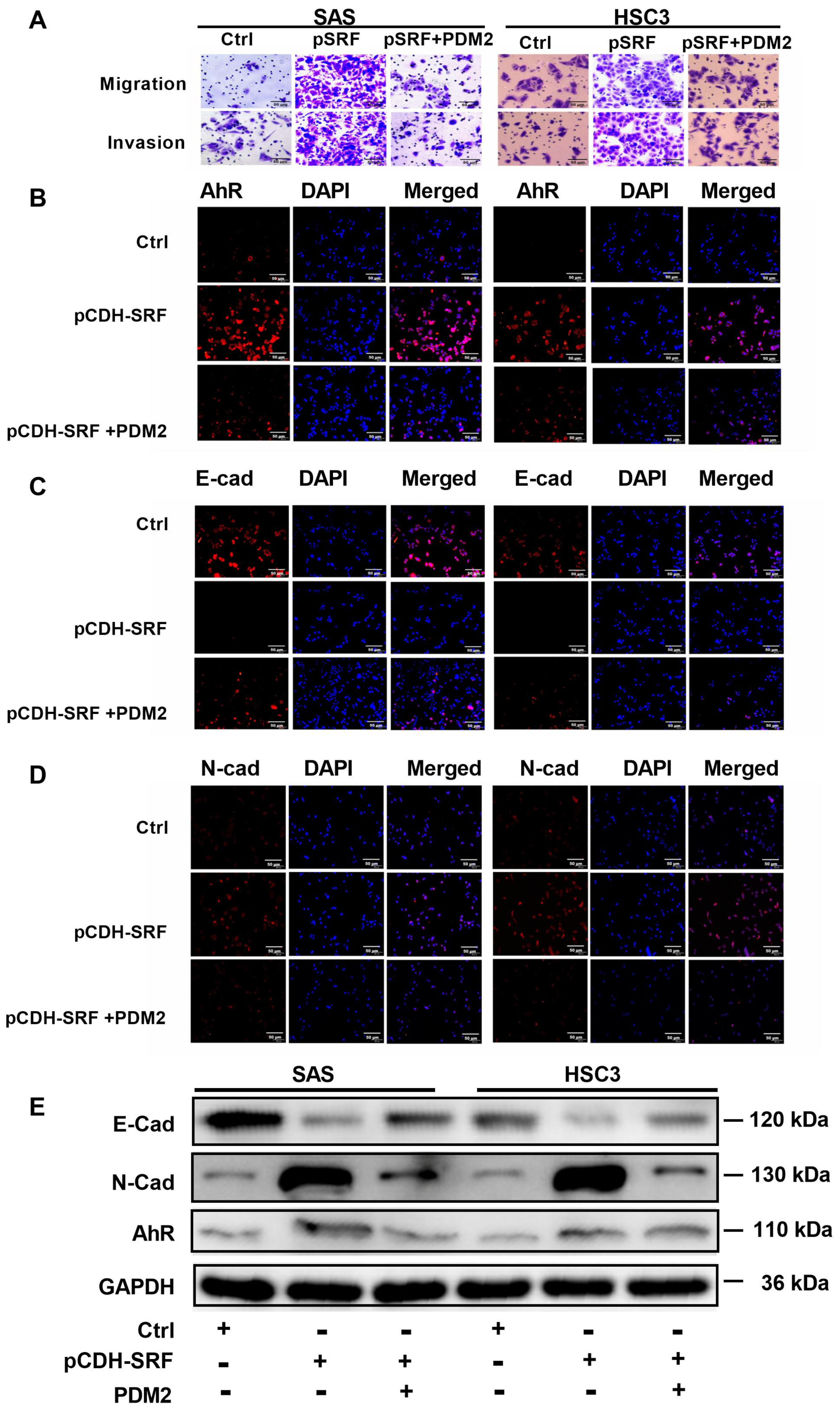
3.2. Overexpression of SRF Facilitates the Migration and Invasion of OSCC Cells
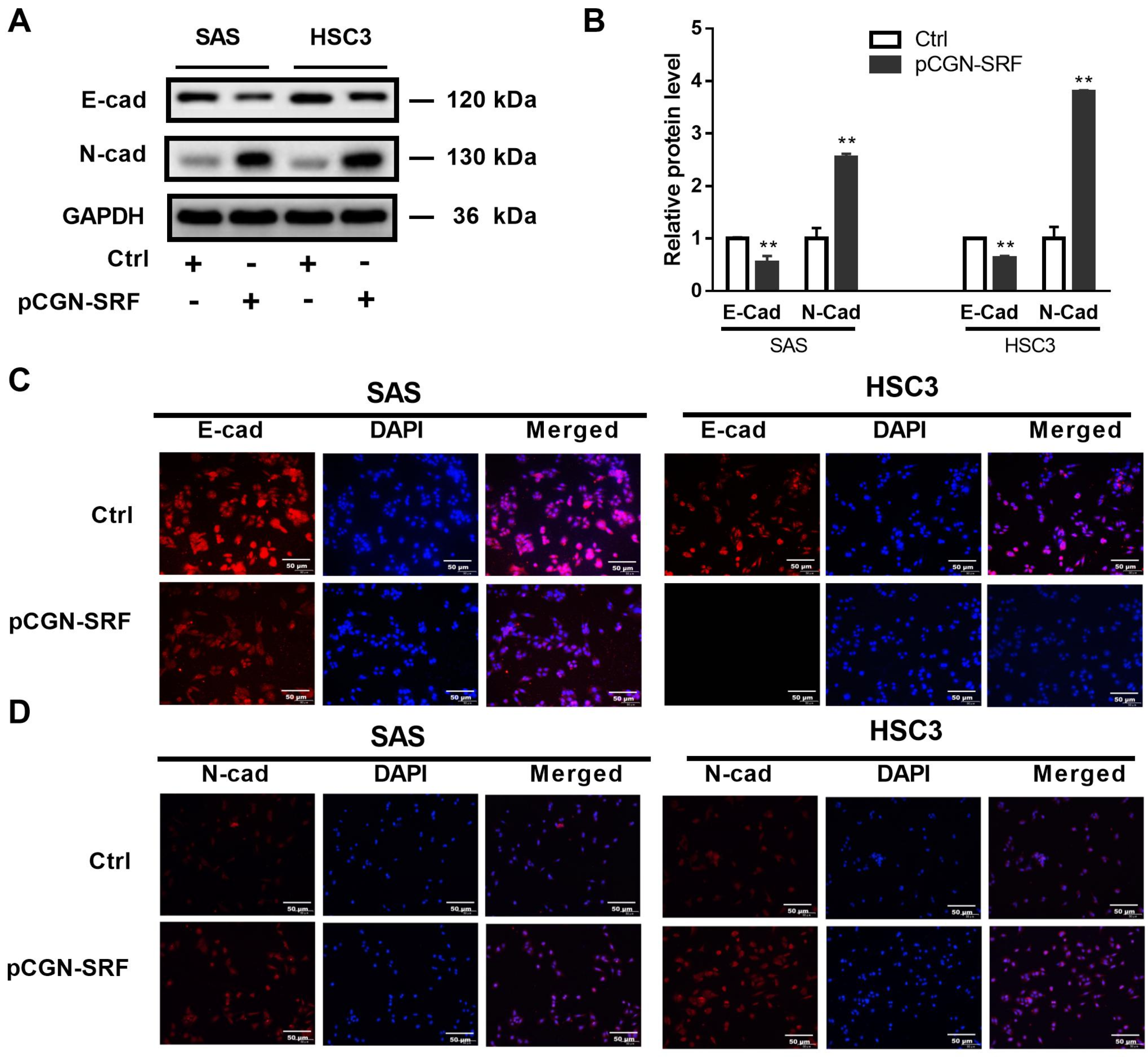
3.3. Overexpression of SRF Increases EMT of OSCC Cells
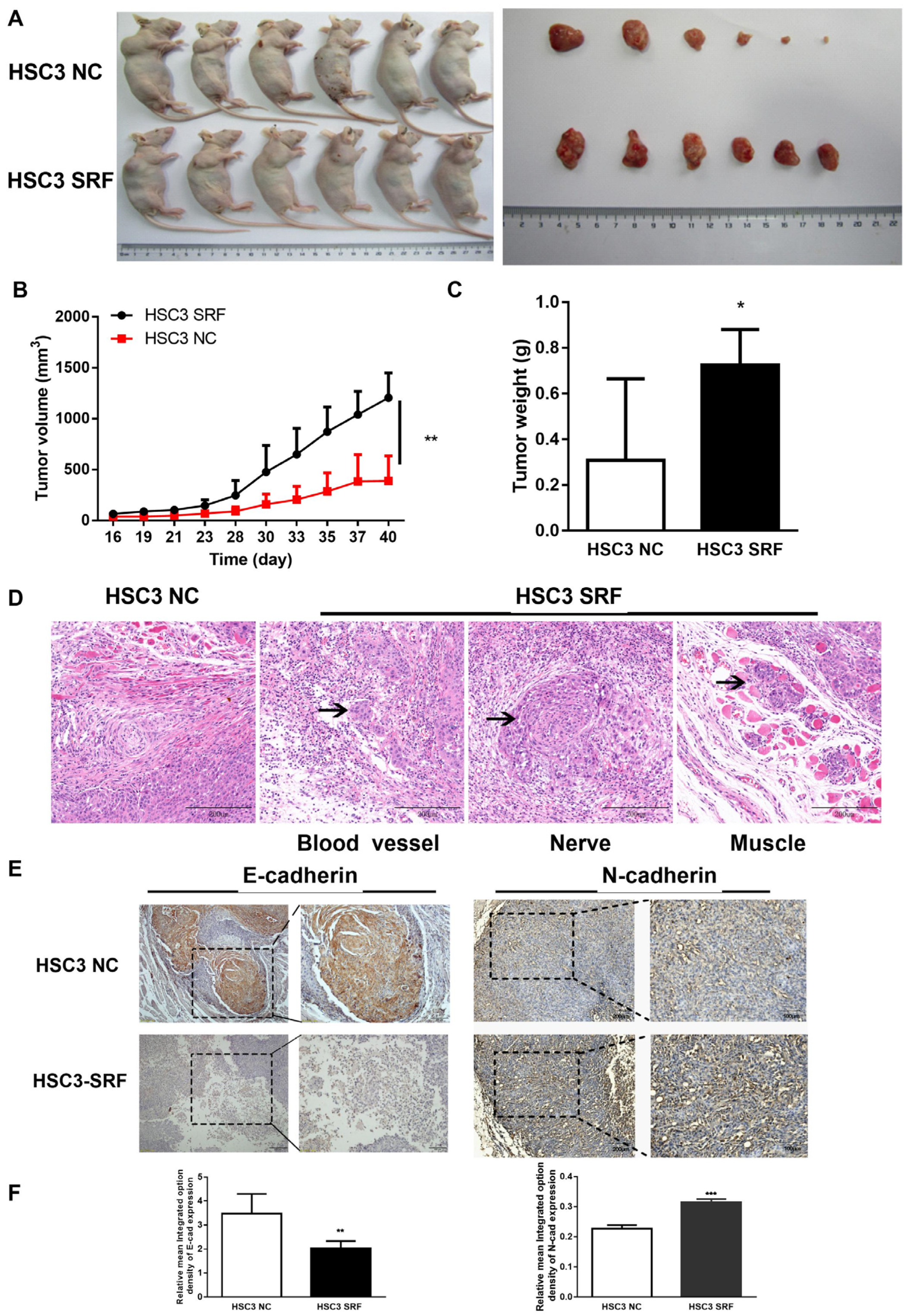
3.4. Overexpression of SRF Promotes OSCC Tumorigenesis and EMT In Vivo
3.5. SRF Activates the IDO1/Kyn-AhR Signaling Pathway in OSCC Cells
3.6. Migration and Invasion of OSCC Is Facilitated by SRF through IDO1 Transcriptional Upregulation
3.7. Aryl Hydrocarbon Receptors Are Involved in SRF-promoted OSCC Cell Migration, Invasion, and EMT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, C.M.; Chu, T.H.; Chou, C.C.; Chien, C.Y.; Wang, J.S.; Huang, C.C. Exosome-derived microRNAs in oral squamous cell carcinomas impact disease prognosis. Oral Oncol. 2021, 120, 105402. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Sasahira, T.; Kirita, T.; Kuniyasu, H. Update of molecular pathobiology in oral cancer: A review. Int. J. Clin. Oncol. 2014, 19, 431–436. [Google Scholar] [CrossRef]
- Chai, A.W.Y.; Lim, K.P.; Cheong, S.C. Translational genomics and recent advances in oral squamous cell carcinoma. Semin. Cancer Biol. 2020, 61, 71–83. [Google Scholar] [CrossRef]
- Kim, D.; Li, R. Contemporary Treatment of Locally Advanced Oral Cancer. Curr. Treat. Options. Oncol. 2019, 20, 32. [Google Scholar] [CrossRef]
- Gharat, S.A.; Momin, M.; Bhavsar, C. Oral Squamous Cell Carcinoma: Current Treatment Strategies and Nanotechnology-Based Approaches for Prevention and Therapy. Crit. Rev. Ther. Drug Carrier Syst. 2016, 33, 363–400. [Google Scholar] [CrossRef]
- Ling, Z.; Cheng, B.; Tao, X. Epithelial-to-mesenchymal transition in oral squamous cell carcinoma: Challenges and opportunities. Int. J. Cancer 2021, 148, 1548–1561. [Google Scholar] [CrossRef]
- Loh, C.Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef]
- Aiello, N.M.; Kang, Y. Context-dependent EMT programs in cancer metastasis. J. Exp. Med. 2019, 216, 1016–1026. [Google Scholar] [CrossRef]
- Ota, I.; Masui, T.; Kurihara, M.; Yook, J.I.; Mikami, S.; Kimura, T.; Shimada, K.; Konishi, N.; Yane, K.; Yamanaka, T.; et al. Snail-induced EMT promotes cancer stem cell-like properties in head and neck cancer cells. Oncol. Rep. 2016, 35, 261–266. [Google Scholar] [CrossRef]
- de Freitas Silva, B.S.; Yamamoto-Silva, F.P.; Pontes, H.A.; Pinto Junior Ddos, S. E-cadherin downregulation and Twist overexpression since early stages of oral carcinogenesis. J. Oral Pathol. Med. 2014, 43, 125–131. [Google Scholar] [CrossRef]
- Yao, X.; Sun, S.; Zhou, X.; Zhang, Q.; Guo, W.; Zhang, L. Clinicopathological significance of ZEB-1 and E-cadherin proteins in patients with oral cavity squamous cell carcinoma. Onco. Targets Ther. 2017, 10, 781–790. [Google Scholar] [CrossRef]
- Remadevi, V.; Muraleedharan, P.; Sreeja, S. FOXO1: A pivotal pioneer factor in oral squamous cell carcinoma. Am. J. Cancer Res. 2021, 11, 4700–4710. [Google Scholar]
- Modak, C.; Chai, J. Serum response factor: Look into the gut. World J. Gastroenterol. 2010, 16, 2195–2201. [Google Scholar] [CrossRef]
- Onuh, J.O.; Qiu, H. Serum response factor-cofactor interactions and their implications in disease. FEBS J. 2021, 288, 3120–3134. [Google Scholar] [CrossRef]
- Ohrnberger, S.; Thavamani, A.; Braeuning, A.; Lipka, D.B.; Kirilov, M.; Geffers, R.; Autenrieth, S.E.; Romer, M.; Zell, A.; Bonin, M.; et al. Dysregulated serum response factor triggers formation of hepatocellular carcinoma. Hepatology 2015, 61, 979–989. [Google Scholar] [CrossRef]
- Watson, R.W.; Azam, H.; Aura, C.; Russell, N.; McCormack, J.; Corey, E.; Morrissey, C.; Crown, J.; Gallagher, W.M.; Prencipe, M. Inhibition of serum response factor improves response to enzalutamide in prostate cancer. Cancers 2020, 12, 3540. [Google Scholar] [CrossRef]
- Yin, J.; Lv, X.; Hu, S.; Zhao, X.; Liu, Q.; Xie, H. Overexpression of serum response factor is correlated with poor prognosis in patients with gastric cancer. Hum. Pathol. 2019, 85, 10–17. [Google Scholar] [CrossRef]
- Ma, L.; Yu, Y.; Qu, X. Suppressing serum response factor inhibits invasion in cervical cancer cell lines via regulating Egr-1 and epithelial-mesenchymal transition. Int. J. Mol. Med. 2019, 43, 614–620. [Google Scholar] [CrossRef]
- Platten, M.; von Knebel Doeberitz, N.; Oezen, I.; Wick, W.; Ochs, K. Cancer immunotherapy by targeting IDO1/TDO and their downstream effectors. Front. Immunol. 2014, 5, 673. [Google Scholar] [CrossRef]
- Amani, H.; Shahbazi, M.A.; D'Amico, C.; Fontana, F.; Abbaszadeh, S.; Santos, H.A. Microneedles for painless transdermal immunotherapeutic applications. J. Control. Release Off. J. Control. Release Soc. 2021, 330, 185–217. [Google Scholar] [CrossRef]
- Litzenburger, U.M.; Opitz, C.A.; Sahm, F.; Rauschenbach, K.J.; Trump, S.; Winter, M.; Ott, M.; Ochs, K.; Lutz, C.; Liu, X.; et al. Constitutive IDO expression in human cancer is sustained by an autocrine signaling loop involving IL-6, STAT3 and the AHR. Oncotarget 2014, 5, 1038–1051. [Google Scholar] [CrossRef]
- Lin, D.J.; Ng, J.C.K.; Huang, L.; Robinson, M.; O’Hara, J.; Wilson, J.A.; Mellor, A.L. The immunotherapeutic role of indoleamine 2,3-dioxygenase in head and neck squamous cell carcinoma: A systematic review. Clin. Otolaryngol. 2021, 46, 919–934. [Google Scholar] [CrossRef]
- Vered, M.; Wright, J.M. Update from the 5th Edition of the World Health Organization Classification of Head and Neck Tumors: Odontogenic and Maxillofacial Bone Tumours. Head Neck Pathol. 2022, 16, 63–75. [Google Scholar] [CrossRef]
- Mittal, V. Epithelial mesenchymal transition in tumor metastasis. Annu. Rev. Pathol. 2018, 13, 395–412. [Google Scholar] [CrossRef]
- Anzai, H.; Yoshimoto, S.; Okamura, K.; Hiraki, A.; Hashimoto, S. IDO1-mediated Trp-kynurenine-AhR signal activation induces stemness and tumor dormancy in oral squamous cell carcinomas. Oral Sci. Int. 2022, 19, 31–43. [Google Scholar] [CrossRef]
- Azam, H.; Pierro, L.; Reina, M.; Gallagher, W.M.; Prencipe, M. Emerging role for the Serum Response Factor (SRF) as a potential therapeutic target in cancer. Expert. Opin. Ther. Targets 2022, 26, 155–169. [Google Scholar] [CrossRef]
- Leon, X.; Pujals, G.; Sauter, B.; Neumann, E.; Pujol, A.; Quer, M. Differential characteristics of patients with squamous cell carcinoma of the head and neck with no history of tobacco or alcohol use. Acta Otorrinolaringol. Esp. (Engl. Ed.) 2023. [Google Scholar] [CrossRef]
- Song, Z.; Liu, Z.; Sun, J.; Sun, F.L.; Li, C.Z.; Sun, J.Z.; Xu, L.Y. The MRTF-A/B function as oncogenes in pancreatic cancer. Oncol. Rep. 2016, 35, 127–138. [Google Scholar] [CrossRef]
- Lu, J.; Shenoy, A.K. Epithelial-to-Pericyte Transition in Cancer. Cancers 2017, 9, 77. [Google Scholar] [CrossRef]
- Bae, J.S.; Noh, S.J.; Kim, K.M.; Jang, K.Y.; Chung, M.J.; Kim, D.G.; Moon, W.S. Serum response factor induces epithelial to mesenchymal transition with resistance to sorafenib in hepatocellular carcinoma. Int. J. Oncol. 2014, 44, 129–136. [Google Scholar] [CrossRef]
- Choi, H.N.; Kim, K.R.; Lee, J.H.; Park, H.S.; Jang, K.Y.; Chung, M.J.; Hwang, S.E.; Yu, H.C.; Moon, W.S. Serum response factor enhances liver metastasis of colorectal carcinoma via alteration of the E-cadherin/beta-catenin complex. Oncol. Rep. 2009, 21, 57–63. [Google Scholar]
- Zhao, X.; He, L.; Li, T.; Lu, Y.; Miao, Y.; Liang, S.; Guo, H.; Bai, M.; Xie, H.; Luo, G.; et al. SRF expedites metastasis and modulates the epithelial to mesenchymal transition by regulating miR-199a-5p expression in human gastric cancer. Cell Death Differ. 2014, 21, 1900–1913. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Si, Q.; Qi, R.; Liu, W.; Li, M.; Guo, M.; Wei, L.; Yao, Z. Indoleamine 2,3-Dioxygenase 1: A Promising Therapeutic Target in Malignant Tumor. Front. Immunol. 2021, 12, 800630. [Google Scholar] [CrossRef]
- Jiao, R.; Zheng, X.; Sun, Y.; Feng, Z.; Song, S.; Ge, H. IDO1 Expression Increased After Neoadjuvant Therapy Predicts Poor Pathologic Response and Prognosis in Esophageal Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 1099. [Google Scholar] [CrossRef]
- Hacking, S.; Vitkovski, T.; Jain, S.; Jin, C.; Chavarria, H.; Wu, D.; Nasim, M. Clinical Significance of Program Death Ligand-1 and Indoleamine-2,3-Dioxygenase Expression in Colorectal Carcinoma. Appl. Immunohistochem. Mol. Morphol. 2021, 29, 201–208. [Google Scholar] [CrossRef]
- Rosenbaum, M.W.; Gigliotti, B.J.; Pai, S.I.; Parangi, S.; Wachtel, H.; Mino-Kenudson, M.; Gunda, V.; Faquin, W.C. PD-L1 and IDO1 Are Expressed in Poorly Differentiated Thyroid Carcinoma. Endocr. Pathol. 2018, 29, 59–67. [Google Scholar] [CrossRef]
- Mandarano, M.; Bellezza, G.; Belladonna, M.L.; Vannucci, J.; Gili, A.; Ferri, I.; Lupi, C.; Ludovini, V.; Falabella, G.; Metro, G.; et al. Indoleamine 2,3-Dioxygenase 2 Immunohistochemical Expression in Resected Human Non-small Cell Lung Cancer: A Potential New Prognostic Tool. Front. Immunol. 2020, 11, 839. [Google Scholar] [CrossRef]
- Laimer, K.; Troester, B.; Kloss, F.; Schafer, G.; Obrist, P.; Perathoner, A.; Laimer, J.; Brandacher, G.; Rasse, M.; Margreiter, R.; et al. Expression and prognostic impact of indoleamine 2,3-dioxygenase in oral squamous cell carcinomas. Oral Oncol. 2011, 47, 352–357. [Google Scholar] [CrossRef]
- Moretti, S.; Nucci, N.; Menicali, E.; Morelli, S.; Bini, V.; Colella, R.; Mandarano, M.; Sidoni, A.; Puxeddu, E. The Aryl Hydrocarbon Receptor Is Expressed in Thyroid Carcinoma and Appears to Mediate Epithelial-Mesenchymal-Transition. Cancers 2020, 12, 145. [Google Scholar] [CrossRef]

| Clinical Parameters | SRF Expression | p-Value 1 | ||
|---|---|---|---|---|
| All Cases | High | Low | ||
| n = 70 | n = 56 | n = 14 | ||
| Sex | 0.393 | |||
| Male | 42 | 35 | 7 | |
| Female | 28 | 21 | 7 | |
| Age (y) | ||||
| <60 | 37 | 29 | 8 | 0.719 |
| ≥60 | 33 | 27 | 6 | |
| Depth of invasion | 0.008 * | |||
| T1/T2 | 51 | 37 | 14 | |
| T3/T4 | 19 | 19 | — | |
| Lymph node metastasis | 0.005 * | |||
| Negative | 42 | 29 | 13 | |
| Positive | 28 | 27 | 1 | |
| Differentiation | 0.546 | |||
| Well | 40 | 31 | 9 | |
| Moderate/poor | 30 | 25 | 5 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, M.; Zhu, F.; Yin, Q.; Yin, H.; Fang, S.; Luo, G.; Huang, J.; Huang, W.; Liu, F.; Zhong, M.; et al. Serum Response Factor-Regulated IDO1/Kyn-Ahr Pathway Promotes Tumorigenesis of Oral Squamous Cell Carcinoma. Cancers 2023, 15, 1319. https://doi.org/10.3390/cancers15041319
Xu M, Zhu F, Yin Q, Yin H, Fang S, Luo G, Huang J, Huang W, Liu F, Zhong M, et al. Serum Response Factor-Regulated IDO1/Kyn-Ahr Pathway Promotes Tumorigenesis of Oral Squamous Cell Carcinoma. Cancers. 2023; 15(4):1319. https://doi.org/10.3390/cancers15041319
Chicago/Turabian StyleXu, Mingyan, Feixiang Zhu, Qi Yin, Hao Yin, Shaobin Fang, Gongwei Luo, Jie Huang, Wenxia Huang, Fan Liu, Ming Zhong, and et al. 2023. "Serum Response Factor-Regulated IDO1/Kyn-Ahr Pathway Promotes Tumorigenesis of Oral Squamous Cell Carcinoma" Cancers 15, no. 4: 1319. https://doi.org/10.3390/cancers15041319
APA StyleXu, M., Zhu, F., Yin, Q., Yin, H., Fang, S., Luo, G., Huang, J., Huang, W., Liu, F., Zhong, M., & Deng, X. (2023). Serum Response Factor-Regulated IDO1/Kyn-Ahr Pathway Promotes Tumorigenesis of Oral Squamous Cell Carcinoma. Cancers, 15(4), 1319. https://doi.org/10.3390/cancers15041319






