Simple Summary
Glioblastoma is a complex and aggressive primary brain tumour that is rapidly fatal. Timely and accurate diagnosis is therefore crucial. Here, we explore the newly emerging field of epitranscriptomics to understand the modifications that occur on RNA molecules in the healthy and diseased brain, focusing on glioblastoma. RNA modifications are modulated by various regulators and are diverse, specific, reversible, and involved in many aspects of brain tumour biology. Epitranscriptomic biomarkers may therefore be ideal candidates for clinical diagnostic workflows. This review summarises the current understanding of epitranscriptomics and its clinical relevance in brain cancer diagnostics.
Abstract
RNA modifications are diverse, dynamic, and reversible transcript alterations rapidly gaining attention due to their newly defined RNA regulatory roles in cellular pathways and pathogenic mechanisms. The exciting emerging field of ‘epitranscriptomics’ is predominantly centred on studying the most abundant mRNA modification, N6-methyladenine (m6A). The m6A mark, similar to many other RNA modifications, is strictly regulated by so-called ‘writer’, ‘reader’, and ‘eraser’ protein species. The abundance of genes coding for the expression of these regulator proteins and m6A levels shows great potential as diagnostic and predictive tools across several cancer fields. This review explores our current understanding of RNA modifications in glioma biology and the potential of epitranscriptomics to develop new diagnostic and predictive classification tools that can stratify these highly complex and heterogeneous brain tumours.
1. Introduction
Analogous to the epigenome, the epitranscriptome comprises a vast, dense, and complex web of chemical modifications on RNA molecules that facilitate the fine-tuning of gene expression regulation [1,2]. RNA modifications are reversible and highly controlled by protein regulators. These protein regulators include ‘writers’ that deposit chemical modifications on RNA molecules, ‘readers’ that recognise different RNA modifications, and ‘erasers’, which oppose the writers’ function by removing changes from RNA molecules [3,4]. There have been more than 100 RNA modifications identified to date, with the n6-methyladenine (m6A) modification being the most well studied, perhaps owing to its highest frequency of detection [5]. The m6A mRNA modifications are typically identified on 6–7 nucleotides per 3000-nucleotide-long mRNA and are presumed to have crucial regulatory roles [6,7]. While the exact downstream impacts of epitranscriptomic modifications on gene expression and biological function are not well understood, epitranscriptomic alterations are known to influence the nuclear transport of RNA molecules, the decay and translation of mRNA [8,9,10,11,12], and mRNA processing mechanisms such as splicing [13] and 3′ end processing, as well as miRNA processing [14,15,16,17].
Alterations in the genome, epigenome, and/or epitranscriptome are understood to underpin cancer molecular pathology, with implications for all aspects of cancer biology, including tumourigenesis and disease progression [18,19]. Glioblastoma (GBM), the most common and deadliest brain tumour in adults, is no exception [18,20,21]. While genomic and epigenomic alterations have been extensively investigated in GBM, the epitranscriptome remains largely unexplored.
Although RNA modifications in mammalian cells were first described almost 50 years ago [7], it is only until the recent emergence of new state-of-the-art technologies that large-scale explorations of epitranscriptomic modifications have been facilitated. It is now recognised that RNA modifications are diverse, dynamic, and of reversible nature, and this facilitates their role as fine tuners of RNA structure and function. There is also a growing appreciation of their regulation by various environmental factors. Therefore, a more thorough understanding of the epitranscriptome and its intricacies, particularly in cancer, will likely lead to innovative diagnostic tools and adjuncts for personalised therapeutics. In this review, we will discuss epitranscriptomics with a focus on m6A regulators and their roles, m6A modifications in the healthy and cancerous brain, and the current and future perspectives of epitranscriptomics in neurological disease and GBM diagnostics.
2. Epitranscriptomic Diversity and the Cellular Roles of RNA Modifications
There is great diversity among the RNA modification types, with more than 100 different chemical modifications reported in RNA species [22], which are precisely regulated by RNA-binding proteins. The most well-known RNA modifications, their epitranscriptomic regulators, and their roles are presented in Figure 1 and summarised below. Epitranscriptomic regulators and RNA modification types vary enormously, rendering biological pathways both complex and well regulated.
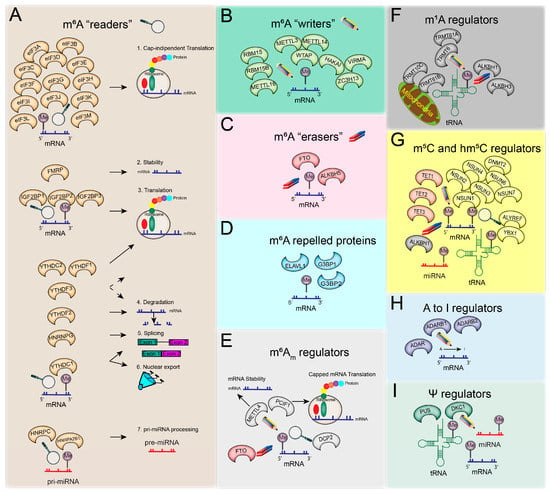
Figure 1.
RNA modifications are mediated by known epitranscriptomic regulators: readers (magnifying glass), writers (pencil), and erasers (rubber). The panels depict (A) m6A reader members with their roles in different aspects of RNA biology; (B) m6A writers; (C) m6A erasers; (D) proteins repelled by m6A methylation; (E) m6Am regulators and their known roles; (F) m1A regulators and their main role during tRNA and mitochondrial tRNA life; (G) m5C and hm5C regulators; (H) known adenosine-to-inosine (A–I) regulators; (I) Ψ regulators.
2.1. N6-Methyladenosine (m6A) RNA Modification
The methylation of adenosine (A) at the N6 position generates N6-methyladenosine (m6A), the most abundant and reversible epitranscriptomic modification in mammalian mRNA and noncoding RNAs (ncRNA). There are usually 1–3 m6A sites per mRNA molecule, which comprises approximately 0.2% of the overall transcript [7,23]. The m6A modification is preferentially found at RRACH positions (R = guanine (G) or adenosine (A); H = adenosine (A), cytosine (C), or uracil (U)) of consensus sequences [24], although this modification is not defined strictly by the presence of these sequences [25]. The distribution of m6A modifications occurs preferentially in different regions along the mRNA length, including around stop codons, long internal exons, and the 3′UTR end. m6A modifications at the 5′UTR end are less frequent; when present, m6A acts to promote mRNA translation at the 5′UTR [11,24,26]. Nevertheless, m6A distribution patterns along the mRNA sequence can change with environmental stressors, such as UV or heat shock [11].
2.1.1. m6A Writers
The m6A methylation mark (in eukaryotes) is deposited by RNA methylation regulators (protein complexes with methyltransferase function) termed ‘writers’ (Figure 1B) during transcription in the nucleus. The m6A ‘writer’ complex comprises three main subunits—(1) methyltransferase-like 3 (METTL3), the catalytic subunit; (2) methyltransferase-like 14 (METTL14), the noncatalytic subunit that has a role in maintaining the integrity of this complex and ensuring RNA-binding [27]; and (3) Wilms’ tumour 1-associating protein (WTAP), which acts to recruit the methyltransferase complex to a target RNA by specifically binding to the Rm6ACH motif of mRNA molecules. The methyltransferase complex formed by METTL3, METTL14, and WTAP regulators preferentially targets internal adenosine residues and is termed WTAP-dependent methylation [15,28]; however, there are specific WTAP-independent methylation sites present around the RNA cap structure [28]. In addition to these subunits, there are proteins that interact with the m6A ‘writer’ complex and appear to be essential for methylation, such as KIAA1429. However, KIAA1429 knockdown experiments yielded a smaller reduction in methylation than WTAP-silencing experiments [28]. This knockdown investigation revealed the relationship between gene levels and methylation densities. Low-expression genes are more likely to be methylated than highly expressed genes; typically, ‘housekeeping’ genes lack methylation sites. WTAP-dependent mediated methylation is inversely correlated with mRNA stability [28], and through methyltransferase complex regulation, WTAP and METTL3 are implicated in the expression and alternative splicing of genes associated with transcription and RNA processing. Unexpected new evidence shows that METTL3 can also bind to non-m6A-modified RNA and enhance the translation of epigenetic factors [29].
In various coding and noncoding RNAs, i.e., pre-mRNAs and small nuclear RNA (snRNA), methyltransferase-like 16 (METTL16) was found to be a cytoplasmic RNA-binding ‘writer’ protein but also promoted translation in an m6A-independent manner [30,31,32,33]. In mouse embryonic stem cells (mESC), additional components of the ‘writer’ methyltransferase complex were identified, such as Zc3h13 and Hakai. These two components were found to interact with WTAP and Virilizer (the mouse homolog of KIAA1429, also named VIRMA in humans), suggesting the involvement of these components in the m6A methylation process [34]. In human cells, it was also shown that VIRMA interacts with WTAP/HAKAI/ZC3H13 to recruit METTL3 and METTL14 to the methyltransferase complex [35]. The interaction between HAKAI and WTAP is implicated in cell cycle regulation [36]. Two other possible interactor components of the ‘writer’ complex include the RNA-binding motif protein 15 (RBM15) and RBM15B, identified around the methylated DRACH motif. This suggests a role in recruiting additional writer proteins to specific sites in X-inactive specific transcript (XIST) RNA [37]. The m6A ‘writer’ complex is mainly comprised of METTL3, METTL14, WTAP, HAKAI, ZC3H13, VIRMA, RBM15, and RBM15B, while other diverse proteins (e.g., METTL16) with specific functions can have ‘writer’ role, as described in the literature to date.
2.1.2. m6A Readers
The YTH protein family members contain the YTH domain that specifically recognises or ‘reads’ the m6A mark on mRNA molecules compared to unmethylated adenosine residues [38]. With affinity proteomic experiments, it was shown that three members of the YTH ‘reader’ protein family (YTHDF1, YTHDF2, and YTHDF3) selectively bind to m6A sites, suggesting a possible physical interaction between writers and readers [28]. YTHDF1 has binding sites around stop codons, which are regions enriched in m6A modification sites. The function of YTHDF1 in RNA translation was elucidated by knockdown in HeLa cells showing reduced translation efficiency and the number of ribosome-bound reads of the target genes. The YTHDF1 and YTHDF2 have targets that are regulated separately, of which 50% are in common, with YTHDF1 responsible for translating methylated mRNA while YTHDF2 mediates mRNA degradation [9,39]. Another ‘reader’ from the same family, YTHDF3, interacts with YTHDF1 to facilitate protein synthesis and with YTHDF2 to degrade methylated RNA. While these three members of the ‘readers’ (YTHDF1, YTHDF2, and YTHDF3) perform highly dynamic and reversible modulations of mRNA in the cytoplasm, another ‘reader’, YTHDC1, oversees methylated RNA transport from the nucleus to the cytoplasm [10,40]. YTHDC1 is also involved in mRNA splicing by promoting exon inclusion through the SRSF3 splicing factor recruitment and blocking the SRSF10 binding to mRNA [41]. The YTHDC2 is another ‘reader’ of the YTH protein family that contains RNA helicase domains and regulates its targets through a 3’→5’ RNA helicase activity [42]. YTHDC2 also facilitates the translation efficiency of m6A-marked targets [43]. In genetic disorders such as fragile X syndrome, the fragile X mental retardation protein (FMRP) has been found to bind to m6A sites and interact with YTHDF2 in the cerebral cortex of mice. The FMRP ensures RNA stability, while YTHDF2 promotes RNA decay. This suggests that FMRP has a ‘reader’ role and, through its interaction with YTHDF2, contributes to the fragile X syndrome phenotype, a common inherited intellectual disability known to be driven by the loss of functional FMRP [44].
In addition to the YTH protein family members, numerous other m6A readers have been characterised (Figure 1A). The eukaryotic initiation factor 3 (eIF3) is a specific ‘reader’ of m6A in the 5′UTR region, where it directly binds m6A to promote cap-independent translation [11]. Several heterogeneous nuclear ribonucleoproteins (hnRNPs) family members are also known as ‘readers’. One such m6A reader, HNRNPA2B1, has an essential role in primary microRNA (pri-miRNA) processing [17]. HNRNPA2B1 directly interacts with DGCR8, a miRNA processing complex (Microprocessor) member, to facilitate pri-miRNA processing into the precursor (pre)-miRNA form [17]. Interestingly, pri-miRNAs that have m6A marks deposited by METTL3 are more efficiently processed by the Microprocessor complex than those that are unmethylated [16]. Another RNA-binding protein, HNRNPC, is also a ‘reader’ involved in pre-mRNA processing, where HNRNPC binding is facilitated by m6A modifications to adjacent mRNAs and long noncoding RNA (lncRNA) structures [45]. Similarly, it was found that m6A modifications provide better accessibility for HNRNPG binding in a study that identified more than 10,000 m6A sites regulating this RNA-HNRNPG interaction, suggesting possible effects on expression changes and splicing [46]. RNA-binding insulin-like growth factor 2 (IGF2BP) protein family members (IGF2BP1/2/3) are also novel m6A methylation ‘readers’, which protect RNA from degradation by enabling translation and RNA stability. Some IGF2BPs target, stabilise, and translate oncogenic mRNAs, such as MYC [47], highlighting the role of m6A methylation and regulation in cancer cell biology.
2.1.3. m6A ‘Erasers’
The m6A methylation mark can be removed by m6A demethylase enzymes, termed ‘erasers’ (Figure 1C). The obesity-associated protein (FTO) was one of the first m6A regulators to be identified for its essential role in demethylation; FTO knockdown was shown to increase m6A levels, whereas FTO overexpression decreased the relative m6A methylation in HeLa and 293FT cells [48]. Furthermore, FTO was found to mediate m6A demethylation on internal nucleotides along the RNA, as well as other demethylation activities in the cap region (m6Am) and in small nuclear RNA (snRNA; m6A and m6Am) and transfer RNA (tRNA; m1A). Interestingly, the FTO-mediated m6A demethylation significantly influences the levels of target transcripts that have internal m6A and a combination of both internal and cap region methylation but does not influence transcripts with only cap m6Am modifications. FTO was shown to target nuclear and cytoplasmic m6A sites; however, it preferentially demethylates m6Am modifications [49], thus reducing the stability of transcripts methylated in the 5′ cap region (m6Am) [50]. Another m6A demethylase, ALKBH5, was also found to remove m6A oxidatively and affect RNA metabolism and RNA export to the cytoplasm [8].
2.1.4. Repelled Proteins
Some RNA-binding proteins are repelled by the m6A mark and instead preferentially bind nonmethylated adenosine, influencing RNA stability and degradation (Figure 1D). The two most well-known m6A-repelled proteins are the stress granule proteins, G3BP1 and G3BP2, that regulate mRNA stability and are involved in embryonic development [51]. ELAV-like RNA-binding protein 1 (ELAVL1), also known as human antigen R (HuR), is another RNA-binding protein repelled by m6A. It is a well-known RNA stabiliser protein. If an m6A mark is proximate to the ELAVL1 binding site on an mRNA molecule, it promotes the binding of ELAVL1, leading to RNA stabilisation. However, if the m6A mark is further from the ELAVL1-binding site, the spatial distance will not allow ELAVL1-binding and will result in RNA degradation [52,53].
2.2. Other RNA Modifications
2.2.1. N6,2-O-Dimethyladenosine: m6Am Modifications
N6, 2′-Odimethyladenosine (m6Am) methylation is abundant in human mRNA (92%) and occurs at the transcription start site just after the N7-methylguanosine (m7G) mRNA cap. M6Am methylation is mediated by phosphorylated CTD interacting factor 1 (PCIF1), a cap-specific adenosine-N6-methyltransferase (CAPAM), and facilitates the translation of capped mRNAs [54,55] (Figure 1E). Where m6A modification is associated with RNA degradation, m6Am modifications are important for RNA stabilisation. This methylation mark can also be catalysed by methyltransferase 4, N6-Adenosine (METTL4), a protein encoded by the METTL4 gene. METTL4 is well characterised in GBM as harbouring a high frequency of missense mutations [19,56]. The m6Am mark can be removed by the m6A ‘eraser’ FTO and can be ‘read’ by decapping MRNA 2 (DCP2) [19].
2.2.2. N1-Methyladenosine: m1A Modifications
M1A modifications play a critical role in tRNA stability and have been detected in mitochondrial transcripts, mRNA, and ribosomal RNA (rRNA). M1A modifications are added to tRNA by specific m1A writers, tRNA methyltransferase 61A (TRMT61A), and tRNA methyltransferase 6 noncatalytic subunits (TRMT6), and to mitochondrial tRNAs by tRNA methyltransferase 10C, mitochondrial RNase P subunit (TRMT10C), and tRNA methyltransferase 61B (TRMT61B) (Figure 1F). The erasers for this mark are alkB homolog 1, histone H2A dioxygenase (ALKBH1) and alkB homolog 3, and alpha-ketoglutarate dependent dioxygenase (ALKBH3) [57].
2.2.3. 5-Methylcytosine (m5C) and 5-Hydroxymethylcytidine (hm5C) Modifications
DNA methylation is one of the most well-known epigenetic modifications that mainly occurs on cytosine residues (m5C) [58]. While this modification is less known in RNA, several recent studies describe the importance of m5C in diverse functions of mRNA, ncRNA, rRNA, and tRNA. m5C is enriched in the 5′UTR and 3′UTR regions, and it is regulated by writers (including NOP2/Sun RNA methyltransferase 1–7 (NSUN1–7) and tRNA aspartic acid methyltransferase (DNMT2)), readers (Aly/REF export factor (ALYREF) and Y-box binding protein 1 (YBX1)), and erasers (tet methylcytosine dioxygenase 1–3 (TET1–3) and ALKBH1) (Figure 1G) [59,60]. Similar to DNA, the m5C can be oxidized in RNA, resulting in 5-hydroxymethylcytidine (hm5C) [61]. M5C ‘erasers’ from the TET family have a dual role as hm5C ‘writers’.
2.2.4. Adenosine-to-Inosine (A-to-I) Modifications
RNA sequences can undergo specific editing, resulting in the synthesis of protein isoforms that are different to what is encoded by the original genomic sequences [62]. Such editing events include base modifications through deamination reactions, i.e., uridine (U) substitution of cytidine (C) or adenosine (A) converted to inosine (I) [63]. The adenosine deaminase RNA specific (ADAR) family members perform adenosine-to-inosine RNA editing and are also known to have an epitranscriptomic ‘writer’ role; the I ‘writer’ proteins are ADAR, ADARB1, and ADARB2 (Figure 1H).
2.2.5. Pseudouridine (Ψ) Modifications
Pseudouridine (Ψ), termed the fifth RNA nucleotide [19], is the 5-ribosyl isomer of uridine and was the first RNA modification to be discovered. Ψ is the most abundant RNA modification and can be found in diverse RNA types, including mRNA, rRNA, tRNA, small nucleolar RNA (snoRNA), and snRNA [64,65]. The Ψ modification is catalysed in eukaryotes by several pseudouridine synthases (Pus enzymes), and its most well-known writer is dyskerin pseudouridine synthase 1 (DKC1) (Figure 1I).
3. Epitranscriptomics in Neurobiology and Brain Cancer
Although the abundance and roles of some RNA modifications are still in dispute, the epitranscriptome is thought to have specific relevance to stem cell biology and neurobiology. Human genetic analysis and animal model studies have revealed that alterations in RNA regulatory programs have critical roles in the aetiology of neurogenerative disorders, intellectual disability, mental disorders, and brain cancers [59,66,67,68,69]. To date, m6A in fragile X syndrome is the most well-characterised interaction, and a growing number of studies have linked mutations in epitranscriptomic enzymes with intellectual disability [44]. m6A modifications are highly abundant in the brain and play essential roles in embryonic stem cell differentiation, neural development, and neurodevelopmental disorders [70]. The roles of m6A regulators in neural development, glioma stem cells (GSCs), glioma progression, and treatment are expanded on below.
3.1. m6A Regulators in Neural Development and Physiology
In mammals, the brain shows the highest abundance of m6A methylation in the body, where it is linked to several dynamic developmental and physiological processes such as neurogenesis, axonal growth, synaptic plasticity, circadian rhythm, cognitive function, and stress response [71]. Several studies show that m6A methylation is developmentally altered. Temporal-specific m6A-methylation features were identified in lncRNA in the developing cortex [72], and levels of m6A in mRNA were observed to remain low throughout embryogenesis but dramatically increase by adulthood [11,24,26]. A transcriptome-wide m6A methylation study revealed differences in the distribution of this epitranscriptomic modification, where m6A levels were twice as high in the mouse cerebellum compared to the cortex [73]. Moreover, the temporal regulation of the m6A methylation regulators METTL3, METTL14, WTAP, ALKBH5, and FTO is essential for the precise control of postnatal cerebellar development [74]. m6A methylation is also biased toward neuronal transcripts relative to glial cells, which are localised in axons and dendritic shafts, indicating their role in neuronal-activity-dependent gene regulation [71,75,76]. In addition to spatiotemporal and cell type preferences, m6A methylation is essential in governing direct reprogramming into neuronal cells [77] (Figure 2A).
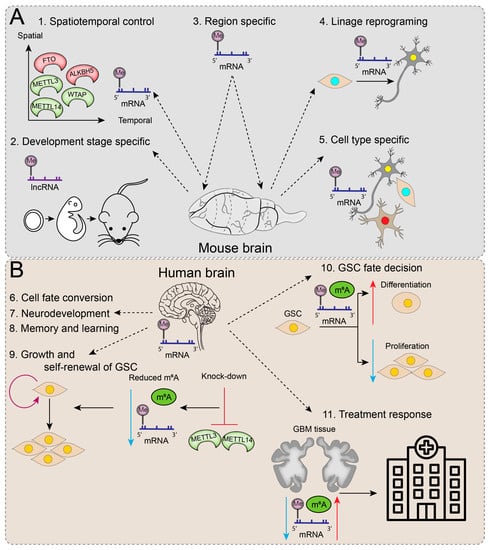
Figure 2.
Functions of epitranscriptomic regulators and RNA modifications in the brain. (A) In an animal model (mouse), RNA methylation is predominantly related to spatiotemporal control and is specific to developmental stage, region, lineage, and cell type. (B) The main functions attributed to RNA methylations in the human brain are associated with cell fate conversion, neurodevelopment, memory, and learning. The knockdown of m6A ‘writers’ (METTL3 and METTL4) causes a decrease in m6A methylation, which is thought to play a role in GSC growth and cell renewal. The m6A methylation levels can influence the GSC fate decision towards differentiation (increase; red upward arrow) or proliferation (decrease; blue downward arrow). RNA modifications detected in GBM tissue reflect treatment-related responses, which could have great importance for clinical monitoring of GBM.
During gliogenesis, when neural stem cell (NSC) populations are replaced by glial, astrocyte, and oligodendrocyte precursor cells (GPC, APC, and OPCs, respectively) [78], several m6A methylation-related changes take place. This was evidenced by Mettl3 and Mettl14 knockouts in embryonic mice brains, leading to ablation of RNA m6A levels and prolonged radial glia cell cycles, with the extension of cortical neurogenesis into postnatal stages [79]. Similarly, m6A signalling was also demonstrated to regulate neurogenesis in human forebrain organoids, where human ‘brain-disorder risk genes’ were observed to be enriched in m6A-tagged transcripts [79]. Dynamic RNA methylation also plays important regulatory roles in oligodendrocyte development and CNS myelination [80]. Conditional inactivation of Mettl14 in mice disrupted oligodendroglial maturation with the selective depletion of oligodendrocytes where OPC numbers were within normal limits, with distinct effects on OPC and oligodendrocyte transcriptomes [80]. Oligodendrocyte specification and myelination are also influenced by epitranscriptomic regulators, such as m6A ‘reader’ Prrc2a (Proline rich coiled-coil 2 A) [81]. While key regulators of m6A methylation have been implicated in various neurological, neurodevelopmental, and neuropsychiatric disorders, m6A has specific roles in glioblastoma.
3.2. m6A Regulators in Glioma Stem-like Cells (GSCs) and Tumourigenicity
Glioblastomas are highly heterogeneous primary brain tumours consisting of diverse cellular populations, including glioma stem-like cells (GSCs). GSCs are a subset of distinct aberrant neural stem cells that possess glioma self-renewal potential and are thought to be responsible for driving GBM initiation, progression, treatment resistance, and recurrence. Interestingly, m6A mRNA methylation machinery was demonstrated to play a vital role in GSC self-renewal and tumourigenesis (Figure 2B) [82]. In knockdown experiments of m6A ‘writers’, METTL3 and METTL14 reduced m6A levels and dramatically enhanced GSC growth and self-renewal. Conversely, METLL3 overexpression or inhibition of the m6A ‘eraser’, FTO, increased m6A methylation levels and suppressed GSC self-renewal, proliferation, and tumour progression, resulting in prolonged survival of GSC-grafted mice [82]. To extrapolate, m6A methylation in GSCs may direct cell fate decisions, with higher m6A levels promoting GSC differentiation, whereas reduced m6A levels enhance GSC proliferation and tumourigenesis.
Indeed, GSCs and differentiated GBM cells have distinct patterns of m6A methylation [83]. However, a set of common transcripts from primary GSC spheroid cultures that were observed to lose RNA methylation during cell differentiation correlated with increased translation rates during the GSC transition to differentiated GBM cells. Moreover, miRNA-binding sites were observed to overlap with areas rich in RRACH motifs, i.e., m6A binding sites, which led to the identification of specific tumour-suppressive RRACH binding miRNAs that facilitate FTO-dependent, transcript-specific demethylation [83]. Taken together, this study showed that specific miRNAs may directly influence mRNA stability without engaging in expected miRNA mediated transcript downregulation, facilitating a fast and accurate adaptation to meet the cellular requirements of GSC differentiation [83].
The other m6A ‘eraser’, ALKBH5, has also been linked to GSC self-renewal and tumourigenesis [84] through its regulation of FOXM1 expression, a pivotal cell cycle regulation transcription factor that functions to maintain GSC properties [85] and is overexpressed and associated with poor survival in GBM [86]. ALKBH5 cooperates with a nuclear lncRNA, FOXM1-AS, to demethylate FOXM1 nascent transcripts and stimulate FOXM1 expression in GSCs, with the inhibition of ALKBH5 repressing GSC proliferation. In this process, ELAVL1, a nuclear RNA-binding protein that preferentially binds nonmethylated RNA, plays an important role in regulating FOXM1 expression by recruiting ALKBH5 to bind to the 3′UTR of unmethylated FOXM1 transcripts and promoting FOXM1 expression, contributing to GSC tumourigenicity [84].
Contrary to reports demonstrating positive associations between reduced m6A methylation and increased GSC tumourigenicity [82,84,87,88,89], several studies implicate an oncogenic role for METTL3 in cancer stem cells [90,91,92,93]. One such study purported that high levels of METTL3 and METTL3-dependent m6A modification are essential for GSC maintenance and dedifferentiation, with the preservation of a stem-like phenotype mediated by SOX2 mRNA stabilisation via the recruitment of ELAV1 to m6A-modified SOX2 [91]. The function of SOX2 as a transcription factor essential for embryonic and neural stem cell maintenance is well established. This study also implicates METTL3 in DNA repair efficiency, radiation resistance of GSCs, and survival outcomes in a GBM orthotopic mouse model.
Discrepancies between different studies might be explained by the diversity of cell types and m6A-modified RNA species analysed, as well as the intra- and intertumoural heterogeneity of GBM. Future experimental work must be context- and compartment-dependent to explain these inconsistencies and further our understanding of m6A RNA methylation and the interplay among regulator proteins in GSCs [91].
3.3. m6A Modifications and Regulators in Glioblastoma with Potential Therapeutic Implications for Improving Treatment Response
RNA modifications play increasingly important roles in the tumourigenesis and development of GBM; several ‘writers’, ‘erasers’, and even ‘readers’ of RNA modifications are proposed as potential diagnostic biomarkers and novel targets for treatment [19]. Yet, there are striking inconsistencies in m6A RNA methylation levels and m6A regulator expression reported in GBM [4,19]. In paediatric medulloblastoma, high METTL3 expression is associated with increased m6A levels and low survival, and a specific inhibitor of METTL3 slowed tumour progression in a medulloblastoma mouse model [94]. In line with the observed inverse correlation between GSC tumourigenicity and the general levels of m6A RNA methylation explored above [82], Li et al. reported that glioma tumours have lower global m6A RNA methylation levels relative to normal brain tissue, with decreased METTL3 and increased FTO levels in glioma tissues offered as a causative link [87]. Here, METTL3 overexpression in an established GBM cell line increased m6A levels and reduced cell proliferation and migration in vitro, perhaps related to a disruption in HSP90 chaperone activity and enhanced apoptosis [87]. Other studies corroborated the association between high METTL3 expression and reduced glioma proliferation; however, METTL3 silencing was shown to suppress vasculogenic mimicry, important for tumour angiogenesis [95,96]. Meanwhile, overexpression of another m6A writer, METTL4, was shown to promote glioma cell proliferation and migration [95,97]. Recently, FTO knockdown or inhibition was shown to suppress GBM proliferation via an m6A-dependent depletion of MYC levels [98]. Moreover, targeted FTO inhibition enhanced the efficiency of temozolomide (TMZ), the standard-of-care chemotherapy, in killing GBM cells [98]. Promisingly, FTO inhibition also shows antitumoural effects within tumour microenvironments, enhancing T cell-mediated cytotoxicity and reducing immune evasion by suppressing immune checkpoint genes [99]. While seemingly a promising novel therapeutic target, there are conflicting reports, including reduced FTO expression levels in more aggressive glioma tumour subtypes, with low FTO expression levels significantly associated with poor survival outcomes [100,101]. Similarly, specific inhibitors of another m6A ‘eraser’, ALKBH5, have been tested and show efficient inhibition of GBM cell proliferation [102] enhanced radiosensitivity and reduced invasiveness of GSCs [103], and improved anti-PD1 treatment efficiency in a preclinical glioma model [104]. While ALKBH5 was reported as a significant negative prognostic factor for GBM patients [105,106], reported ALKBH5 expression levels in GBM tissue are inconsistent [100,101,105,107].
Another approach to sensitise glioma cells to treatment was explored by modulating m6A ‘reader’ and member of the YTH protein family, YTHDF1. Relative to normal brain tissue, YTHDF1 has the highest expression in GBM, and is stabilised by musashi RNA-binding protein 1 (MSI1). GBM cell proliferation and migration are enhanced by the MSI1 stabilisation of YTHDF1, and high expression of these genes in GBM is associated with reduced survival [108]. Notably, YTHDF1 knockdown increased the sensitivity of glioma cells to the antiproliferative effects of TMZ, demonstrating its potential as a synergistic target for improved treatment response in GBM. Another YTH family member, YTHDF2, is also a potential target for glioma treatment, with studies linking YTHDF2 expression to glioma progression [97,109]. Increased in GSCs, YTHDF2 plays a vital role in stem maintenance, and it enhances tumour growth in mice through stabilising MYC (YTHDF2-MYC) and targeting IGFBP3. Interestingly, YTHDF2 expressing GSC have an increased sensitivity to cell killing by linsitinib, an IGF1/IGF1R inhibitor. As NSC cell viability is unaffected by linsitinib treatment, this drug is proposed as a potentially specific anti-GBM treatment [110].
Epitranscriptomic regulators and RNA modifications are extensively involved in essential functions of the healthy brain and play various roles in brain disease. This is well shown in glioblastoma, where m6A can also have a role in treatment response. Unfortunately, there are several inconsistencies in the literature regarding the expression levels of the regulators and global m6A levels, and more in-depth studies are required to decipher the intricate web of RNA modifications in glioblastoma.
3.4. Other RNA Modifications Implicated in Glioblastoma Biology and Treatment
Beyond m6A methylation, other forms of RNA methylation also play important roles in GBM biology. For instance, the downregulation of m6Am writer, PCIF1, promotes glioma cell proliferation and tumour growth in mice [111]. m5C methylated mature miRNAs are linked to poor GBM patient outcomes through a mechanism where this modification inhibits miRNA gene silencing [112]. hm5C methylation was found to have a significant role in GBM formation through a mechanism involving ‘eraser’ TET1, while TET2 and TET3 downregulation is linked to GBM tumourigenesis [113,114,115]. Transcript levels of I ‘writers’ ADAR, ADARB1, and ADARB2 are reduced in different grades and types of brain tumours [116]; ADAR2 was found to inhibit GBM cell growth, and an abnormally expressed splice variant supressed adenosine-to-inosine RNA editing [117,118]. Upregulation of the Ψ writer, DKC1, was also shown to promote glioma cell proliferation, migration, and invasion [119,120]. Likewise, a Ψ regulator, pseudouridine synthase 7 (PUS7), is highly expressed in glioma tissue, associated with poor survival, and PUS7 inhibitors could suppress tumour growth in mice [121].
4. Epitranscriptomics in Diagnosis
4.1. RNA Methylation Detection Methods
The field of epitranscriptomics is rapidly expanding and is anticipated to impact clinical diagnostics. The search for rapid, simple, and reliable methods to explore and detect new disease classification and therapeutic targets are essential steps for precision medicine. Currently, several RNA methylation detection methods exist, with diverse sensitivity, sample input requirements, and data analytical processes. Epitranscriptomics platforms may be antibody-based or employ mass spectrometry, polymerase chain reaction (PCR), next-generation sequencing (NGS), or nanopore direct RNA sequencing [122].
Probably, the simplest, most accessible, and affordable methods are antibody-based, i.e., m6A or m5C enzyme-linked immunoassay (ELISA). ELISA kits are useful for assessing global m6A/m5C methylation transcriptome-wide but are unable to distinguish m6A from m6Am or detect a methylation mark at single-nucleotide or even at gene resolution. The detection of modifications at a single RNA species level can be measured by Dot-blot, another antibody-based method. This is also a relatively inexpensive method, with 100 ng to 1 μg required starting material, and can be used to detect RNA modifications in diverse RNA species [123,124].
Mass-spectrometry-based methods, i.e., liquid chromatography–coupled tandem mass spectrometry (LC–MS/MS), are more sensitive than antibody-based approaches. LC–MS/MS allows the absolute quantification of modified nucleosides, and, when coupled with nucleic acid isotope labelling (NAIL–MS), can assess the temporal dynamics of RNA modifications [125]. A drawback of LC–MS/MS is the high starting material needed (i.e., more than 1 μg of purified RNA), which may be unfeasible for some experimental setups [122]. Another method that uses isotope labelling in its workflow for RNA modification detection is 2D thin-layer chromatography (2D-TLC) [126]. This method is inexpensive and requires less starting material (50–200 ng) than LC–MS/MS; however, 2D-TLC can be adversely impacted by RNA digestion and labelling efficiencies [122].
PCR-based approaches are locus-specific RNA modification detection methods based on the premise that the queried modification impedes reverse transcription. These methods have high sensitivity and specificity; thus, they can be used for various RNA species. Some of the most well-known PCR-based approaches are the m6A-reverse transcription (RT)-quantitative PCR, single-base-elongation- and ligation-based PCR amplification method (SELECT), and primer-extension-based method [122,127].
NGS-based detection methods are diverse and frequently used for whole transcriptome profiling at single-nucleotide resolution. Some approaches require antibody coupling, such as m6A-seq or methylated RNA immunoprecipitation sequencing (MeRIP-seq) [26]. Other standard NGS-based detection methods are m6A individual-nucleotide resolution crosslinking and immunoprecipitation sequencing (miCLIP-seq), m6A-label-seq, m6A-sensitive RNA-endoribonuclease-facilitated sequencing (m6A-REF-seq), MazF RNase-assisted sequencing (MAZTER-seq), and deamination adjacent to RNA modification targets (DART-seq) [128,129,130,131]. More recent approaches include evolved TadA-assisted N6-methyladenosine sequencing (eTAM-seq), glyoxal- and nitrite-mediated deamination of unmethylated adenosines (GLORI), and m6A-selective allyl chemical labelling and sequencing (m6A-SAC-seq) [131,132,133,134,135]. m6A-SAC-seq requires very little starting material (2 ng of poly A+ RNA) but is a particularly laborious process.
One of the most promising epitranscriptomic approaches is Oxford Nanopore Technologies (ONT) sequencing, which can detect modifications in naïve RNA, such as m6A, inosine, or pseudouridine [136,137]. Compared to other methods, this method can detect various modifications in parallel from the same sample, directly from the RNA, without biases that can be introduced through reverse transcription required by NGS-based methods. The technology is based on the specific ionic current changes induced by single RNA molecules while passing through the nanopore. The current fluctuations differ between modified and nonmodified RNA molecules and can be decoded computationally [138]. Algorithms that can identify and analyse RNA modifications are fast evolving. Epitranscriptomic bioinformatic tools of note include—‘Explore the epitranscriptional landscape inferring from glitches of ONT signals’ (ELIGOS) that can predict known types of RNA modification sites, ‘ModPhred’ for user-friendly analysis of DNA and RNA modifications detected by ONT, and ‘MasterOfPores’, a processing pipeline for analysing direct RNA sequencing reads [139,140,141].
Beyond the m6A mark, other detection and data analytical methods facilitate the exploration of m5C, m7G, inosine, pseudouridine, and many other modifications on RNA. Although RNA modification detection methodology and bioinformatics tools are advancing rapidly, these techniques will require further adaptation before their implementation in diagnostic clinical service laboratories where there are real sample quantity and time constraints for analytical processes and data interpretation.
4.2. Epitranscriptomics in Current Diagnostic Approaches
The potential use of epitranscriptomic regulators as biomarkers has been explored across multiple cancer types; however, only a handful of studies have attempted to measure RNA modifications in body fluids directly. Of note, high global m6A levels are reported as potential diagnostic markers in the peripheral blood of small-cell lung (NSCLC) and breast and gastric cancer patients relative to healthy and/or benign cancer controls [142,143,144]. Here, m6A levels correlated to tumour-staging parameters were depleted following surgery, and accompanying upregulations in ‘writer’ and/or downregulations in ‘eraser’ genes were observed [142,143,144]. Likewise, m6A levels in rheumatoid arthritis (RA) patients’ and ischemic stroke patients’ blood were elevated relative to healthy controls, along with decreased ‘eraser’ ALKBH5 and FTO levels and increased YTHDF2 ‘reader’ levels in RA samples [145,146]. In contrast, m6A levels in the peripheral blood of individuals with Type 2 diabetes mellitus (T2DM) are lower than in controls, with concomitantly high m6A ‘eraser’ FTO gene expression [147]. Similar trends are observed in ageing, COVID-19 infection, and smoking [148,149,150]. A summary of studies investigating global m6A methylation status and gene expression of m6A regulators as diagnostic tools across a range of pathologies are presented in Table 1.

Table 1.
m6A regulators expression direction and global m6A methylation status with biomarker value detected from blood. Increased and decreased m6A levels are depicted by up and down arrows, respectively.
In recent epitranscriptomics studies exploring new diagnostic classification markers, investigations often stem from in silico analyses of public sequencing datasets (i.e., TCGA, CCGA) to identify the predictive and prognostic significance of the diverse regulators of RNA modifications. For instance, in lung adenocarcinoma, high HNRNPC and IGF2BP3 levels correlate with reduced overall survival [156,157], and a six-gene risk signature (KIAA1429, ALKBH5, METTL3, HNRNPC, YTHDC2, and YTHDF1) strongly correlated with various clinical and pathological features [158]. A further study also identified METTL3, YTHDF1, and YTHDF2 as prognostic lung adenocarcinoma biomarkers [159]. In lung squamous cell carcinoma, reduced KIAA1429, ALKBH5, METTL3, and HNRNPC expressions were predictive of better responses to chemotherapy and immunotherapy. Identified m6A regulators with significant prognostic value in a diverse range of cancer types are presented in Table 2. There is no clear consensus for a pan-cancer epitranscriptomic regulation profile but rather a range of epitranscriptomic players that are differentially modulated across conditions, suggesting that the expression of epitranscriptomic patterns is cancer subtype-specific.

Table 2.
Expression of epitranscriptomic regulators and their observed roles in diverse conditions.
4.3. Epitranscriptomics in Glioma Diagnostics
Like other cancer types, the value of epitranscriptomics as a tool for brain cancer diagnostics is still in its early days. The WHO classification system for primary brain tumours has integrated molecular diagnostics with traditional histopathology [197]. While the identification of molecular alterations, such as isocitrate dehydrogenase IDH1/2 mutations, 1p/19q codeletion, and MGMT promoter methylation, has improved the accuracy of diagnostics, prognostication, and prediction of treatment response for glioma patients, there has been no tangible improvement in clinical management and patient outcomes. Epitranscriptomics of known glioma sub-entities may yield further advances in tumour stratification and help guide treatment selection for precision medicine, as was shown in preliminary studies of colon cancer [152]
Several studies explored m6A RNA methylation regulators in public primary brain cancer sequencing datasets, and these key findings are summarised in Table 3. Many of these studies are not based on integrated brain tumour diagnostic definitions (WHO 2021 classification), and tumours are often referred to in broad terms (GBM, glioma, astrocytoma, or low-grade glioma, LGG). In this review, we use the diagnostic terms used by the original studies; however, caution must be taken when interpreting RNA modification changes relevant to specific glioma subtypes.
Key findings in glioma patients in relation to m6A ‘readers’ include YTHDF2 overexpression, induced through the EGFR/SRC/ERK pathway to promote tumourigenesis, invasiveness, and cell proliferation independently associated with poor prognosis [198]. Similarly, expression of the YTHDF1 paralog is negatively associated with survival and promotes glioma cell proliferation and growth in vitro [100]. A systematic meta-analysis of m6A ‘reader’ eIF3 was performed in independent glioma cohorts as several eIF3 subunits are localised to chromosomes 1p and 19q [199,200], regions co-deleted in IDH-mutant oligodendroglioma [201]. All 13 eIF3 subunits were significantly differentially expressed between the glioma subgroups in both TCGA and CGGA datasets [202]. Among other findings, eIF3i expression was shown to be an independent prognostic factor in IDH-mutant LGG and could also predict the 1p/19q codeletion status [202]. m6A regulators IGF2BP2, IGF2BP3, HNRNPC, and YTHDF2 and m5C-related long noncoding RNAs CIRBP-AS1, GDNF-AS1, LINC00265, and ZBTB20-AS4 have prognostic significance in LGG [203,204].
Expression of the m6A ‘writer’ METTL3 was positively associated with a higher malignant grade and poorer prognosis of IDH-wildtype but not IDH-mutant gliomas [205]. Another study identified significant fluctuations in m6A regulator expression when comparing glioma cohorts comprising different IDH mutational and 1p/19q codeletion states. While these cohorts represent different glioma sub-entities with distinct molecular pathophysiology, the average survival times are vastly different, particularly between oligodendroglioma (IDH-mutant, 1p/19q co-deleted) and GBM (IDH-wildtype, 1p19q non-co-deleted), and thus divergent epitranscriptomic regulator expression patterns may relate to the very different biology of these distinct pathological entities [101]. In this study, two glioma subgroups were identified based on the expression of m6A regulators, and a risk signature (ALKBH5, IGF2BP3, KIAA1429, and YTHDF2) was significantly associated with prognosis, the immune microenvironment and treatment efficacy [101]. A similar risk signature (ALKBH5, IGF2BP2, IGF2BP3, HNRNPA2B1, YTHDF1, YTHDF2, RBM15, and WTAP) was shown to be prognostic for glioma patients, and the expression of IGF2BP2 and IGF2BP3 was linked to tumour occurrence, development, and progression [206]. A recent study also reported independent prognostic significance of ALKBH5, which is upregulated in glioma and associated with immune signalling, and inflammatory and metabolic pathways genes [207]. Another study calculated significant prognostic values for four hub genes (EMP3, PDPN, TAGLN2, and TIMP1) related to m6A regulators, all with high expression in high-grade glioma, and further correlation to IDH status and transcriptome subtype [208]. Links between clinical outcomes and m6A regulators function in the tumour microenvironment, tumour cell stemness, and infiltration have also been established [209]. Likewise, epitranscriptomic regulation of lncRNAs are also associated with glioma patient prognosis [210]. Further studies highlighting the close link between m6A RNA methylation regulators and clinicopathological features and treatment sensitivity of gliomas are outlined in Table 3.

Table 3.
m6A regulators identified as biomarkers in glioma.
Table 3.
m6A regulators identified as biomarkers in glioma.
| Increased m6A Regulator(s) | Decreased m6A Regulator(s) | Data Used | Observations/Role | Ref. |
|---|---|---|---|---|
| YTHDF2 | TCGA, REMBRANDT French, Kawaguchi, Paugh | YTHDF2 is linked to glioma malignancy and invasiveness. | [198] | |
| YTHDF2, YTHDF1, METTL3, RBM15, HNRNPC | ALKBH5, WTAP, YTHDC2, ZC3H13 METTL14, FTO | TCGA | YTHDF1 overexpression correlates with the advanced stage of disease. YTHDF1 contributes to glioma progression. | [100] |
| eIF3e | Oncomine, TCGA | eIF3 subunits show varied expression in distinct regions of GBM tumours. eIF3e proteins expression correlates with glioma grade, highest expression in GBM, and increases in recurrences. eIF3e upregulation in recurrences may have a role in treatment resistance. | [211] | |
| eIF3b, eIF3i, eIF3k and eIF3m (poor OS) | eIF3a and eIF3l (better OS) | CGGA, TCGA | Expression of eIF3d, eIF3e, eIF3f, eIF3h, and eIF3l correlates with the IDH-mutant status of gliomas. eIF3i and eIF3k expressions increase with tumour grade and are associated with poor OS. eIF3i is an independent prognostic factor in IDH-mutant LGG and can predict the 1p/19q codeletion status in IDH-mutant LGG. High eIF3i expression correlates with cell proliferation, mRNA processing, translation, T-cell receptor signalling, NF-kB signalling, and many others. | [202] |
| METTL3 | CGGA, TCGA | METTL3 promotes the malignant progression of gliomas in vitro and in vivo. METTL3 correlates with poor OS in IDH-wildtype but not in IDH-mutant gliomas. | [205] | |
| eIF3A, FMR1, FTO, METTL14, METTL16, METTL3, RBMX, YTHDC, YTHDF3 and ZC3H13 (IDH-mutant vs. IDH-wildtype) | ALKBH5, IGF2BP2, IGF2BP3, RBM15, WTAP and YTHDF1 (IDH-mutant vs. IDH-wildtype) | CGGA, TCGA, REMBRANDT | Expression of m6A regulators is associated with Prognosis, grade, IDH, and 1p/19q status. Lower m6A regulators expression (except FTO) is associated with longer OS. A prognostic risk signature—ALKBH5, IGF2BP3, KIAA1429, and YTHDF2. | [101] |
| ALBKH5, RBM15, YTHDF and WTAP (increased tumour grade) | FTO | CGGA, TCGA | RBM15, METTL3, METTL14, ALKBH5, FTO, YTHDC1, and YTHDF2 are significantly differentially expressed between IDH-mutant and IDH-wildtype LGG. METTL3, FTO, and YTHDC1 are significantly differentially expressed between IDH-mutant and IDH-wildtype GBM. The risk signature comprises RBM15, WTAP, ALBKH5, FTO, YTHDC1, YTHDF1, and YTHDF2, all of which are independent prognostic markers and predictive of clinicopathological features and treatment sensitivity. | [105] |
| RBM15, RBM15B, METTL3, METTL14, WTAP, HNRNPA2B1, HNRNPC, YTHDF1, YTHDF2, YTHDF3, and YTHDC2 (gliomas vs. control) ALKBH5, RBM15, WTAP and YTHDF2 (in GBM vs. LGG) | FTO and ZC3H13 (gliomas vs. control) FTO, KIAA1429, METTL3, ZC3H13, HNRNPC, and YTHDC2 (in GBM vs. LGG) | CGGA | Four m6A-related lncRNAs that have prognostic values: LINC00900 and MIR155HG, increased in higher-grade tumours, while MIR9-3HG and LINC00515 have lower expression in HGG vs. LGG. | [107] |
| IGF2BP3 | YTHDC2 | TCGA, CGGA cBioportal | IGF2BP3 expression increases with tumour grade and correlates with shorter OS. YTHDC2 and IGF2BP3 are negative and positive prognostic factors for OS. | [212] |
| HNRNPC, WTAP, YTHDF2 and, YTHDF1 | TCGA | Defined prognostic risk signature: HNRNPC, ZC3H13, and YTHDF2. HNRNPC plays an important role in malignancy and contributes to the development of gliomas. High expression of HNRNPC correlates with a favourable prognosis. | [213] | |
| LINC00265 | C6orf3, GDNF-AS1, LINC00925, LINC00237 | TCGA, CGGA | Twenty-four prognostic m6A-related lncRNAs were identified as prognostic lncRNAs. m6A-related lncRNA prognostic signature (m6A-LPS). | [107] |
| METTL14, IGF2BP2, IGF2BP3, HNRNPA2B1, YTHDF1, YTHDF3,HNRNPC, RBMX, WTAP, YTHDF2, and IGF2BP1 | TCGA, CGGA | Defined prognostic risk signature: ALKBH5, IGF2BP2, IGF2BP3, HNRNPA2B1, YTHDF1, YTHDF2, RBM15, and WTAP. | [206] |
Again, there are several conflicting reports of m6A regulator expression levels among the different brain tumour entities and non-tumour brain tissue, including ALKBH5, WTAP, METTL14, and YTHDC2 [100,101,105,107,213]. The contradictory findings of m6A RNA methylation regulating gene expression levels may help to explain the opposing reports of high and low global m6A in brain tumour tissue relative to ‘normal’ brains [4,87]. There is some consensus, however, for the relative expressions of several m6A regulator genes, which are summarised in Table 4. Of these, one of the most discussed is m6A ‘erasers’ FTO, which is frequently observed to be downregulated in GBM relative to other tumours and control tissue [100,101]. If taken to be true, this suggests that GBM tumours have increased global m6A levels.

Table 4.
Diverse m6A regulators and the direction of their expression in different glioma grades with a possible role in diagnostics (these regulators do not have contradictory expressions in the studies presented in Table 3). Regulators in bold were detected in more than one study; relative increases and decreases in m6A regulator levels are indicated by up and down arrows, respectively.
5. Conclusions and Future Perspectives
Epitranscriptome profiling studies, particularly in cancer research, are becoming more common. Studies investigating global m6A, m5C, and other RNA methylation profiles have helped to identify epitranscriptomic regulators and their involvement in cancer biology. While the direct assessments of RNA modifications in diagnostic laboratory settings are technologically challenging, there is a wealth of evidence supporting the significant relationships between epitranscriptomic regulator gene expressions and clinicopathological parameters in cancer, including their use as diagnostic, prognostic and/or predictive biomarkers. With distinct roles defined in stem cell biology and neural development along with high levels detected in brain tissue, RNA modifications are, theoretically, highly relevant to multiple aspects of gliomagenesis and biology. Indeed, multiple studies have reported epitranscriptome regulator signatures that are highly predictive of glioma pathology and patient outcomes.
Further technological advances are needed to adapt epitranscriptomic approaches for clinical diagnostics and will likely uncover entirely novel targets for further stratifying tumour molecular phenotypes, directing treatment(s) and guiding patient management. In the interim, the detection of regulator genes and global m6A methylation levels in highly annotated cohorts of discrete tumour types as well as biofluids are warranted.
Author Contributions
Conceptualization, Á.T. and K.L.A.; investigation Á.T. and K.L.A.; resources, M.E.B. and K.L.A.; writing—original draft preparation, Á.T., S.H., L.S., K.L.A. and M.E.B.; writing—review and editing, Á.T., S.H., L.S., M.E.B. and K.L.A.; visualisation, Á.T.; supervision, K.L.A.; project administration, K.L.A.; funding acquisition, K.L.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by BrainStorm, a brain cancer research charity of the Sydney Local Health District, the BF Foundation, James N. Kirby Foundation, Mark Hughes Foundation, and Cure My Brain.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Goldberg, A.D.; Allis, C.D.; Bernstein, E. Epigenetics: A landscape takes shape. Cell 2007, 128, 635–638. [Google Scholar] [CrossRef]
- Livneh, I.; Moshitch-Moshkovitz, S.; Amariglio, N.; Rechavi, G.; Dominissini, D. The m(6)A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2020, 21, 36–51. [Google Scholar] [CrossRef] [PubMed]
- He, C. Grand challenge commentary: RNA epigenetics? Nat. Chem. Biol. 2010, 6, 863–865. [Google Scholar] [CrossRef] [PubMed]
- Galardi, S.; Michienzi, A.; Ciafrè, S.A. Insights into the Regulatory Role of m(6)A Epitranscriptome in Glioblastoma. Int. J. Mol. Sci. 2020, 21, 2816. [Google Scholar] [CrossRef]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, K.P. RNA synthesis and processing reactions in a subcellular system from mouse L cells. Hoppe Seylers Z. Physiol. Chem. 1982, 363, 33–43. [Google Scholar] [CrossRef]
- Perry, R.; Kelley, D.J.C. Existence of Methylated Messenger RNA in Mouse L Cells. Cell 1974, 1, 37–42. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef]
- Louloupi, A.; Ntini, E.; Conrad, T.; Ørom, U.A.V. Transient N-6-Methyladenosine Transcriptome Sequencing Reveals a Regulatory Role of m6A in Splicing Efficiency. Cell Rep. 2018, 23, 3429–3437. [Google Scholar] [CrossRef]
- Ke, S.; Alemu, E.A.; Mertens, C.; Gantman, E.C.; Fak, J.J.; Mele, A.; Haripal, B.; Zucker-Scharff, I.; Moore, M.J.; Park, C.Y.; et al. A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes Dev. 2015, 29, 2037–2053. [Google Scholar] [CrossRef] [PubMed]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Alarcon, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef]
- Kim, T.Y.; Zhong, S.; Fields, C.R.; Kim, J.H.; Robertson, K.D. Epigenomic profiling reveals novel and frequent targets of aberrant DNA methylation-mediated silencing in malignant glioma. Cancer Res. 2006, 66, 7490–7501. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Cui, H. The Emerging Roles of RNA Modifications in Glioblastoma. Cancers 2020, 12, 736. [Google Scholar] [CrossRef]
- Das, K.K.; Kumar, R. Pediatric Glioblastoma. In Glioblastoma; De Vleeschouwer, S., Ed.; Codon Publications: Brisbane, Austrilia, 2017. [Google Scholar]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Wei, C.M.; Gershowitz, A.; Moss, B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 1975, 4, 379–386. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W.V.; Bell, T.A.; Schaening, C. Messenger RNA modifications: Form, distribution, and function. Science 2016, 352, 1408–1412. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Śledź, P.; Jinek, M. Structural insights into the molecular mechanism of the m(6)A writer complex. eLife 2016, 5, e18434. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef]
- Wei, X.; Huo, Y.; Pi, J.; Gao, Y.; Rao, S.; He, M.; Wei, Q.; Song, P.; Chen, Y.; Lu, D.; et al. METTL3 preferentially enhances non-m(6)A translation of epigenetic factors and promotes tumourigenesis. Nat. Cell Biol. 2022, 24, 1278–1290. [Google Scholar] [CrossRef] [PubMed]
- Nance, D.J.; Satterwhite, E.R.; Bhaskar, B.; Misra, S.; Carraway, K.R.; Mansfield, K.D. Characterization of METTL16 as a cytoplasmic RNA binding protein. PloS ONE 2020, 15, e0227647. [Google Scholar] [CrossRef]
- Warda, A.S.; Kretschmer, J.; Hackert, P.; Lenz, C.; Urlaub, H.; Höbartner, C.; Sloan, K.E.; Bohnsack, M.T. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017, 18, 2004–2014. [Google Scholar] [CrossRef]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017, 169, 824–835.e814. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Dong, L.; Li, Y.; Gao, M.; He, P.C.; Liu, W.; Wei, J.; Zhao, Z.; Gao, L.; Han, L.; et al. METTL16 exerts an m(6)A-independent function to facilitate translation and tumorigenesis. Nat. Cell Biol. 2022, 24, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 2018, 69, 1028–1038.e1026. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef]
- Horiuchi, K.; Kawamura, T.; Iwanari, H.; Ohashi, R.; Naito, M.; Kodama, T.; Hamakubo, T. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 2013, 288, 33292–33302. [Google Scholar] [CrossRef] [PubMed]
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef]
- Liu, J.; Harada, B.T.; He, C. Regulation of Gene Expression by N(6)-methyladenosine in Cancer. Trends Cell Biol. 2019, 29, 487–499. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Luo, G.Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. eLife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Roundtree, I.A.; He, C. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Trends Genet. 2016, 32, 320–321. [Google Scholar] [CrossRef]
- Wojtas, M.N.; Pandey, R.R.; Mendel, M.; Homolka, D.; Sachidanandam, R.; Pillai, R.S. Regulation of m(6)A Transcripts by the 3′→5′ RNA Helicase YTHDC2 Is Essential for a Successful Meiotic Program in the Mammalian Germline. Mol. Cell 2017, 68, 374–387.e312. [Google Scholar] [CrossRef]
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Kang, Y.; Wang, M.; Li, Y.; Xu, T.; Yang, W.; Song, H.; Wu, H.; Shu, Q.; Jin, P. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 2018, 27, 3936–3950. [Google Scholar] [CrossRef]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef]
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Liu, F.; Lu, Z.; Fei, Q.; Ai, Y.; He, P.C.; Shi, H.; Cui, X.; Su, R.; Klungland, A.; et al. Differential m(6)A, m(6)A(m), and m(1)A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 2018, 71, 973–985.e975. [Google Scholar] [CrossRef]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.J.; Chen, Q.; et al. Reversible methylation of m(6)A(m) in the 5’ cap controls mRNA stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef]
- Edupuganti, R.R.; Geiger, S.; Lindeboom, R.G.H.; Shi, H.; Hsu, P.J.; Lu, Z.; Wang, S.Y.; Baltissen, M.P.A.; Jansen, P.; Rossa, M.; et al. N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 2017, 24, 870–878. [Google Scholar] [CrossRef]
- Lee, Y.; Choe, J.; Park, O.H.; Kim, Y.K. Molecular Mechanisms Driving mRNA Degradation by m(6)A Modification. Trends Genet. 2020, 36, 177–188. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, K.; Cai, J.; Zhang, M.; Zhang, X.; Xiong, X.; Meng, H.; Xu, X.; Huang, Z.; Peng, J.; et al. Landscape and Regulation of m(6)A and m(6)Am Methylome across Human and Mouse Tissues. Mol. Cell 2020, 77, 426–440.e426. [Google Scholar] [CrossRef] [PubMed]
- Akichika, S.; Hirano, S.; Shichino, Y.; Suzuki, T.; Nishimasu, H.; Ishitani, R.; Sugita, A.; Hirose, Y.; Iwasaki, S.; Nureki, O.; et al. Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 2019, 363. [Google Scholar] [CrossRef]
- Asif, S.; Fatima, R.; Krc, R.; Bennett, J.; Raza, S. Comparative proteogenomic characterization of glioblastoma. CNS Oncol. 2019, 8, Cns37. [Google Scholar] [CrossRef]
- Zhang, C.; Jia, G. Reversible RNA Modification N(1)-methyladenosine (m(1)A) in mRNA and tRNA. Genom. Proteom. Bioinform. 2018, 16, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Portela, A.; Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010, 28, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Yang, W.L.; Zhao, Y.L.; Yang, Y.G. Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip. Rev. RNA 2021, 12, e1639. [Google Scholar] [CrossRef]
- Yang, X.; Yang, Y.; Sun, B.F.; Chen, Y.S.; Xu, J.W.; Lai, W.Y.; Li, A.; Wang, X.; Bhattarai, D.P.; Xiao, W.; et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017, 27, 606–625. [Google Scholar] [CrossRef]
- Fu, L.; Guerrero, C.R.; Zhong, N.; Amato, N.J.; Liu, Y.; Liu, S.; Cai, Q.; Ji, D.; Jin, S.G.; Niedernhofer, L.J.; et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J. Am. Chem. Soc. 2014, 136, 11582–11585. [Google Scholar] [CrossRef]
- Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002, 71, 817–846. [Google Scholar] [CrossRef] [PubMed]
- Gerber, A.P.; Keller, W. RNA editing by base deamination: More enzymes, more targets, new mysteries. Trends Biochem. Sci. 2001, 26, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Charette, M.; Gray, M.W. Pseudouridine in RNA: What, where, how, and why. IUBMB Life 2000, 49, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef]
- Blanco, S.; Dietmann, S.; Flores, J.V.; Hussain, S.; Kutter, C.; Humphreys, P.; Lukk, M.; Lombard, P.; Treps, L.; Popis, M.; et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. Embo J. 2014, 33, 2020–2039. [Google Scholar] [CrossRef]
- Park, C.W.; Lee, S.M.; Yoon, K.J. Epitranscriptomic regulation of transcriptome plasticity in development and diseases of the brain. BMB Rep. 2020, 53, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Pupak, A.; Singh, A.; Sancho-Balsells, A.; Alcalá-Vida, R.; Espina, M.; Giralt, A.; Martí, E.; Ørom, U.A.V.; Ginés, S.; Brito, V. Altered m6A RNA methylation contributes to hippocampal memory deficits in Huntington’s disease mice. Cell. Mol. Life Sci. CMLS 2022, 79, 416. [Google Scholar] [CrossRef] [PubMed]
- Ming, Y.; Deng, Z.; Tian, X.; Jia, Y.; Ning, M.; Cheng, S. m6A Methyltransferase METTL3 Reduces Hippocampal Neuron Apoptosis in a Mouse Model of Autism Through the MALAT1/SFRP2/Wnt/β-catenin Axis. Psychiatry Investig. 2022, 19, 771–787. [Google Scholar] [CrossRef] [PubMed]
- Vissers, C.; Sinha, A.; Ming, G.L.; Song, H. The epitranscriptome in stem cell biology and neural development. Neurobiol. Dis. 2020, 146, 105139. [Google Scholar] [CrossRef]
- Chokkalla, A.K.; Mehta, S.L.; Vemuganti, R. Epitranscriptomic regulation by m(6)A RNA methylation in brain development and diseases. J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab. 2020, 40, 2331–2349. [Google Scholar] [CrossRef]
- Nie, Y.; Tian, G.G.; Zhang, L.; Lee, T.; Zhang, Z.; Li, J.; Sun, T. Identifying cortical specific long noncoding RNAs modified by m(6)A RNA methylation in mouse brains. Epigenetics 2021, 16, 1260–1276. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Lv, H.; Zhang, W.; Ma, C.; He, X.; Zhao, S.; Zhang, Z.W.; Zeng, Y.X.; Song, S.; Niu, Y.; et al. Region-specific RNA m(6)A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol. 2017, 7, 170166. [Google Scholar] [CrossRef]
- Ma, C.; Chang, M.; Lv, H.; Zhang, Z.W.; Zhang, W.; He, X.; Wu, G.; Zhao, S.; Zhang, Y.; Wang, D.; et al. RNA m(6)A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018, 19, 68. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chen, M.; Huang, H.; Zhu, J.; Song, H.; Zhu, J.; Park, J.; Ji, S.J. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 2018, 46, 1412–1423. [Google Scholar] [CrossRef] [PubMed]
- Merkurjev, D.; Hong, W.T.; Iida, K.; Oomoto, I.; Goldie, B.J.; Yamaguti, H.; Ohara, T.; Kawaguchi, S.Y.; Hirano, T.; Martin, K.C.; et al. Synaptic N(6)-methyladenosine (m(6)A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 2018, 21, 1004–1014. [Google Scholar] [CrossRef]
- Choi, H.; Baek, S.; Cho, B.; Kim, S.; Kim, J.; Chang, Y.; Shin, J.; Kim, J. Epitranscriptomic N(6)-Methyladenosine Modification Is Required for Direct Lineage Reprogramming into Neurons. ACS Chem. Biol. 2020, 15, 2087–2097. [Google Scholar] [CrossRef]
- Laug, D.; Glasgow, S.M.; Deneen, B. A glial blueprint for gliomagenesis. Nat. Rev. Neurosci. 2018, 19, 393–403. [Google Scholar] [CrossRef]
- Yoon, K.J.; Ringeling, F.R.; Vissers, C.; Jacob, F.; Pokrass, M.; Jimenez-Cyrus, D.; Su, Y.; Kim, N.S.; Zhu, Y.; Zheng, L.; et al. Temporal Control of Mammalian Cortical Neurogenesis by m(6)A Methylation. Cell 2017, 171, 877–889.e817. [Google Scholar] [CrossRef]
- Xu, H.; Dzhashiashvili, Y.; Shah, A.; Kunjamma, R.B.; Weng, Y.L.; Elbaz, B.; Fei, Q.; Jones, J.S.; Li, Y.I.; Zhuang, X.; et al. m(6)A mRNA Methylation Is Essential for Oligodendrocyte Maturation and CNS Myelination. Neuron 2020, 105, 293–309.e295. [Google Scholar] [CrossRef]
- Wu, R.; Li, A.; Sun, B.; Sun, J.G.; Zhang, J.; Zhang, T.; Chen, Y.; Xiao, Y.; Gao, Y.; Zhang, Q.; et al. A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019, 29, 23–41. [Google Scholar] [CrossRef]
- Cui, Q.; Shi, H.; Ye, P.; Li, L.; Qu, Q.; Sun, G.; Sun, G.; Lu, Z.; Huang, Y.; Yang, C.G.; et al. m(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017, 18, 2622–2634. [Google Scholar] [CrossRef]
- Zepecki, J.P.; Karambizi, D.; Fajardo, J.E.; Snyder, K.M.; Guetta-Terrier, C.; Tang, O.Y.; Chen, J.S.; Sarkar, A.; Fiser, A.; Toms, S.A.; et al. miRNA-mediated loss of m6A increases nascent translation in glioblastoma. PLoS Genet. 2021, 17, e1009086. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, B.S.; Zhou, A.; Lin, K.; Zheng, S.; Lu, Z.; Chen, Y.; Sulman, E.P.; Xie, K.; Bögler, O.; et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017, 31, 591–606.e596. [Google Scholar] [CrossRef] [PubMed]
- Gong, A.H.; Wei, P.; Zhang, S.; Yao, J.; Yuan, Y.; Zhou, A.D.; Lang, F.F.; Heimberger, A.B.; Rao, G.; Huang, S. FoxM1 Drives a Feed-Forward STAT3-Activation Signaling Loop That Promotes the Self-Renewal and Tumorigenicity of Glioblastoma Stem-like Cells. Cancer Res. 2015, 75, 2337–2348. [Google Scholar] [CrossRef]
- Liu, M.; Dai, B.; Kang, S.H.; Ban, K.; Huang, F.J.; Lang, F.F.; Aldape, K.D.; Xie, T.X.; Pelloski, C.E.; Xie, K.; et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006, 66, 3593–3602. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhang, C.; Zhang, G. m6A RNA Methylation Controls Proliferation of Human Glioma Cells by Influencing Cell Apoptosis. Cytogenet. Genome Res. 2019, 159, 119–125. [Google Scholar] [CrossRef]
- Malacrida, A.; Rivara, M.; Di Domizio, A.; Cislaghi, G.; Miloso, M.; Zuliani, V.; Nicolini, G. 3D proteome-wide scale screening and activity evaluation of a new ALKBH5 inhibitor in U87 glioblastoma cell line. Bioorg. Med. Chem. 2020, 28, 115300. [Google Scholar] [CrossRef]
- Su, R.; Dong, L.; Li, C.; Nachtergaele, S.; Wunderlich, M.; Qing, Y.; Deng, X.; Wang, Y.; Weng, X.; Hu, C.; et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell 2018, 172, 90–105.e123. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Xue, Y.; Zheng, J.; Liu, X.; Ma, J.; Liu, Y. WTAP Expression Predicts Poor Prognosis in Malignant Glioma Patients. J. Mol. Neurosci. 2016, 60, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Visvanathan, A.; Patil, V.; Arora, A.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Somasundaram, K. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 2018, 37, 522–533. [Google Scholar] [CrossRef]
- Visvanathan, A.; Patil, V.; Abdulla, S.; Hoheisel, J.D.; Somasundaram, K. N⁶-Methyladenosine Landscape of Glioma Stem-Like Cells: METTL3 Is Essential for the Expression of Actively Transcribed Genes and Sustenance of the Oncogenic Signaling. Genes 2019, 10, 141. [Google Scholar] [CrossRef]
- Li, F.; Yi, Y.; Miao, Y.; Long, W.; Long, T.; Chen, S.; Cheng, W.; Zou, C.; Zheng, Y.; Wu, X.; et al. N(6)-Methyladenosine Modulates Nonsense-Mediated mRNA Decay in Human Glioblastoma. Cancer Res. 2019, 79, 5785–5798. [Google Scholar] [CrossRef]
- Zhang, Z.W.; Teng, X.; Zhao, F.; Ma, C.; Zhang, J.; Xiao, L.F.; Wang, Y.; Chang, M.; Tian, Y.; Li, C.; et al. METTL3 regulates m(6)A methylation of PTCH1 and GLI2 in Sonic hedgehog signaling to promote tumor progression in SHH-medulloblastoma. Cell Rep. 2022, 41, 111530. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yin, X.; Zhao, W.; Xu, W.; Chen, L. Molecular mechanism of m(6)A methylation of circDLC1 mediated by RNA methyltransferase METTL3 in the malignant proliferation of glioma cells. Cell Death Discov. 2022, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Lin, Y.; Wei, N. N6-methyladenosine-modified HOTAIRM1 promotes vasculogenic mimicry formation in glioma. Cancer Sci. 2023, 114, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.Q.; Chen, W.; Zhou, J.; Shen, Q.; Sun, Y.; Li, T.; Wang, S.C. N(6)-adenosine-methyltransferase-14 promotes glioma tumorigenesis by repressing argininosuccinate synthase 1 expression in an m6A-dependent manner. Bioengineered 2022, 13, 1858–1871. [Google Scholar] [CrossRef]
- Xiao, L.; Li, X.; Mu, Z.; Zhou, J.; Zhou, P.; Xie, C.; Jiang, S. FTO Inhibition Enhances the Antitumor Effect of Temozolomide by Targeting MYC-miR-155/23a Cluster-MXI1 Feedback Circuit in Glioma. Cancer Res. 2020, 80, 3945–3958. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Dong, L.; Li, Y.; Gao, M.; Han, L.; Wunderlich, M.; Deng, X.; Li, H.; Huang, Y.; Gao, L.; et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell 2020, 38, 79–96.e11. [Google Scholar] [CrossRef]
- Xu, C.; Yuan, B.; He, T.; Ding, B.; Li, S. Prognostic values of YTHDF1 regulated negatively by mir-3436 in Glioma. J. Cell. Mol. Med. 2020, 24, 7538–7549. [Google Scholar] [CrossRef]
- Xu, S.; Tang, L.; Dai, G.; Luo, C.; Liu, Z. Expression of m6A Regulators Correlated with Immune Microenvironment Predicts Therapeutic Efficacy and Prognosis in Gliomas. Front. Cell Dev. Biol. 2020, 8, 594112. [Google Scholar] [CrossRef]
- Takahashi, H.; Hase, H.; Yoshida, T.; Tashiro, J.; Hirade, Y.; Kitae, K.; Tsujikawa, K. Discovery of two novel ALKBH5 selective inhibitors that exhibit uncompetitive or competitive type and suppress the growth activity of glioblastoma multiforme. Chem. Biol. Drug Des. 2022, 100, 1–12. [Google Scholar] [CrossRef]
- Kowalski-Chauvel, A.; Lacore, M.G.; Arnauduc, F.; Delmas, C.; Toulas, C.; Cohen-Jonathan-Moyal, E.; Seva, C. The m6A RNA Demethylase ALKBH5 Promotes Radioresistance and Invasion Capability of Glioma Stem Cells. Cancers 2020, 13, 40. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Xu, N.; Zhou, J.; He, Z.; Lenahan, C.; Wang, C.; Ji, H.; Liu, B.; Zou, Y.; Zeng, H.; et al. ALKBH5 promotes PD-L1-mediated immune escape through m6A modification of ZDHHC3 in glioma. Cell Death Discov. 2022, 8, 497. [Google Scholar] [CrossRef]
- Chai, R.C.; Wu, F.; Wang, Q.X.; Zhang, S.; Zhang, K.N.; Liu, Y.Q.; Zhao, Z.; Jiang, T.; Wang, Y.Z.; Kang, C.S. m(6)A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging 2019, 11, 1204–1225. [Google Scholar] [CrossRef]
- Tao, M.; Li, X.; He, L.; Rong, X.; Wang, H.; Pan, J.; Lu, Z.; Zhang, X.; Peng, Y. Decreased RNA m(6)A methylation enhances the process of the epithelial mesenchymal transition and vasculogenic mimicry in glioblastoma. Am. J. Cancer Res. 2022, 12, 893–906. [Google Scholar]
- Wang, W.; Li, J.; Lin, F.; Guo, J.; Zhao, J. Identification of N(6)-methyladenosine-related lncRNAs for patients with primary glioblastoma. Neurosurg. Rev. 2021, 44, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Yarmishyn, A.A.; Yang, Y.P.; Lu, K.H.; Chen, Y.C.; Chien, Y.; Chou, S.J.; Tsai, P.H.; Ma, H.I.; Chien, C.S.; Chen, M.T.; et al. Musashi-1 promotes cancer stem cell properties of glioblastoma cells via upregulation of YTHDF1. Cancer Cell Int. 2020, 20, 597. [Google Scholar] [CrossRef]
- Chai, R.C.; Chang, Y.Z.; Chang, X.; Pang, B.; An, S.Y.; Zhang, K.N.; Chang, Y.H.; Jiang, T.; Wang, Y.Z. YTHDF2 facilitates UBXN1 mRNA decay by recognizing METTL3-mediated m(6)A modification to activate NF-κB and promote the malignant progression of glioma. J. Hematol. Oncol. 2021, 14, 109. [Google Scholar] [CrossRef] [PubMed]
- Dixit, D.; Prager, B.C.; Gimple, R.C.; Poh, H.X.; Wang, Y.; Wu, Q.; Qiu, Z.; Kidwell, R.L.; Kim, L.J.Y.; Xie, Q.; et al. The RNA m6A Reader YTHDF2 Maintains Oncogene Expression and Is a Targetable Dependency in Glioblastoma Stem Cells. Cancer Discov. 2021, 11, 480–499. [Google Scholar] [CrossRef]
- Gao, S.; Zhou, J.; Hu, Z.; Zhang, S.; Wu, Y.; Musunuru, P.P.; Zhang, T.; Yang, L.; Luo, X.; Bai, J.; et al. Effects of the m6Am methyltransferase PCIF1 on cell proliferation and survival in gliomas. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166498. [Google Scholar] [CrossRef]
- Cheray, M.; Etcheverry, A.; Jacques, C.; Pacaud, R.; Bougras-Cartron, G.; Aubry, M.; Denoual, F.; Peterlongo, P.; Nadaradjane, A.; Briand, J.; et al. Cytosine methylation of mature microRNAs inhibits their functions and is associated with poor prognosis in glioblastoma multiforme. Mol. Cancer 2020, 19, 36. [Google Scholar] [CrossRef]
- Carella, A.; Tejedor, J.R.; García, M.G.; Urdinguio, R.G.; Bayón, G.F.; Sierra, M.; López, V.; García-Toraño, E.; Santamarina-Ojeda, P.; Pérez, R.F.; et al. Epigenetic downregulation of TET3 reduces genome-wide 5hmC levels and promotes glioblastoma tumorigenesis. Int. J. Cancer 2020, 146, 373–387. [Google Scholar] [CrossRef] [PubMed]
- García, M.G.; Carella, A.; Urdinguio, R.G.; Bayón, G.F.; Lopez, V.; Tejedor, J.R.; Sierra, M.I.; García-Toraño, E.; Santamarina, P.; Perez, R.F.; et al. Epigenetic dysregulation of TET2 in human glioblastoma. Oncotarget 2018, 9, 25922–25934. [Google Scholar] [CrossRef]
- Takai, H.; Masuda, K.; Sato, T.; Sakaguchi, Y.; Suzuki, T.; Suzuki, T.; Koyama-Nasu, R.; Nasu-Nishimura, Y.; Katou, Y.; Ogawa, H.; et al. 5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 2014, 9, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Paz, N.; Levanon, E.Y.; Amariglio, N.; Heimberger, A.B.; Ram, Z.; Constantini, S.; Barbash, Z.S.; Adamsky, K.; Safran, M.; Hirschberg, A.; et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007, 17, 1586–1595. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Li, Z.; Du, C.; Qi, B.; Zhao, X.; Wang, L.; Bi, L.; Wang, G.; Zhang, X.; Su, X.; et al. Abnormal expression of an ADAR2 alternative splicing variant in gliomas downregulates adenosine-to-inosine RNA editing. Acta Neurochir. 2014, 156, 1135–1142. [Google Scholar] [CrossRef]
- Galeano, F.; Rossetti, C.; Tomaselli, S.; Cifaldi, L.; Lezzerini, M.; Pezzullo, M.; Boldrini, R.; Massimi, L.; Di Rocco, C.M.; Locatelli, F.; et al. ADAR2-editing activity inhibits glioblastoma growth through the modulation of the CDC14B/Skp2/p21/p27 axis. Oncogene 2013, 32, 998–1009. [Google Scholar] [CrossRef]
- Rintala-Dempsey, A.C.; Kothe, U. Eukaryotic stand-alone pseudouridine synthases - RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 2017, 14, 1185–1196. [Google Scholar] [CrossRef]
- Miao, F.A.; Chu, K.; Chen, H.R.; Zhang, M.; Shi, P.C.; Bai, J.; You, Y.P. Increased DKC1 expression in glioma and its significance in tumor cell proliferation, migration and invasion. Invest. New Drugs 2019, 37, 1177–1186. [Google Scholar] [CrossRef]
- Cui, Q.; Yin, K.; Zhang, X.; Ye, P.; Chen, X.; Chao, J.; Meng, H.; Wei, J.; Roeth, D.; Li, L.; et al. Targeting PUS7 suppresses tRNA pseudouridylation and glioblastoma tumorigenesis. Nat. Cancer 2021, 2, 932–949. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, L.; Li, X. Detection technologies for RNA modifications. Exp. Mol. Med. 2022, 54, 1601–1616. [Google Scholar] [CrossRef]
- Du, Y.; Xia, M.; Hu, Z. Analysis of N 6-methyladenosine RNA Modification Levels by Dot Blotting. Bio-Protoc. 2022, 12, e4565. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Liang, Z.; Yu, H. Dot Blot Analysis of N6-methyladenosine RNA Modification Levels. Bio-Protoc. 2017, 7, e2095. [Google Scholar] [CrossRef]
- Heiss, M.; Borland, K.; Yoluç, Y.; Kellner, S. Quantification of Modified Nucleosides in the Context of NAIL-MS. Methods Mol. Biol. 2021, 2298, 279–306. [Google Scholar] [CrossRef]
- Keith, G. Mobilities of modified ribonucleotides on two-dimensional cellulose thin-layer chromatography. Biochimie 1995, 77, 142–144. [Google Scholar] [CrossRef] [PubMed]
- Olazagoitia-Garmendia, A.; Castellanos-Rubio, A. Relative Quantification of Residue-Specific m(6)A RNA Methylation Using m(6)A-RT-QPCR. Methods Mol. Biol. 2021, 2298, 185–195. [Google Scholar] [CrossRef]
- Shu, X.; Cao, J.; Cheng, M.; Xiang, S.; Gao, M.; Li, T.; Ying, X.; Wang, F.; Yue, Y.; Lu, Z.; et al. A metabolic labeling method detects m(6)A transcriptome-wide at single base resolution. Nat. Chem. Biol. 2020, 16, 887–895. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, L.Q.; Zhao, Y.L.; Yang, C.G.; Roundtree, I.A.; Zhang, Z.; Ren, J.; Xie, W.; He, C.; Luo, G.Z. Single-base mapping of m(6)A by an antibody-independent method. Sci. Adv. 2019, 5, eaax0250. [Google Scholar] [CrossRef]
- Garcia-Campos, M.A.; Edelheit, S.; Toth, U.; Safra, M.; Shachar, R.; Viukov, S.; Winkler, R.; Nir, R.; Lasman, L.; Brandis, A.; et al. Deciphering the “m(6)A Code” via Antibody-Independent Quantitative Profiling. Cell 2019, 178, 731–747.e716. [Google Scholar] [CrossRef]
- Meyer, K.D. DART-seq: An antibody-free method for global m(6)A detection. Nat. Methods 2019, 16, 1275–1280. [Google Scholar] [CrossRef] [PubMed]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef]
- Ge, R.; Ye, C.; Peng, Y.; Dai, Q.; Zhao, Y.; Liu, S.; Wang, P.; Hu, L.; He, C. m(6)A-SAC-seq for quantitative whole transcriptome m(6)A profiling. Nat. Protoc. 2023, 8, 626–657. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.L.; Liu, S.; Ge, R.; Wu, Y.; He, C.; Chen, M.; Tang, W. Transcriptome-wide profiling and quantification of N(6)-methyladenosine by enzyme-assisted adenosine deamination. Nat. Biotechnol. 2023, Online ahead of print. [Google Scholar] [CrossRef]
- Liu, C.; Sun, H.; Yi, Y.; Shen, W.; Li, K.; Xiao, Y.; Li, F.; Li, Y.; Hou, Y.; Lu, B.; et al. Absolute quantification of single-base m(6)A methylation in the mammalian transcriptome using GLORI. Nat. Biotechnol. 2022, Online ahead of print. [Google Scholar] [CrossRef]
- Liu, H.; Begik, O.; Lucas, M.C.; Ramirez, J.M.; Mason, C.E.; Wiener, D.; Schwartz, S.; Mattick, J.S.; Smith, M.A.; Novoa, E.M. Accurate detection of m(6)A RNA modifications in native RNA sequences. Nat. Commun. 2019, 10, 4079. [Google Scholar] [CrossRef]
- Begik, O.; Mattick, J.S.; Novoa, E.M. Exploring the epitranscriptome by native RNA sequencing. RNA 2022, 28, 1430–1439. [Google Scholar] [CrossRef] [PubMed]
- Leger, A.; Amaral, P.P.; Pandolfini, L.; Capitanchik, C.; Capraro, F.; Miano, V.; Migliori, V.; Toolan-Kerr, P.; Sideri, T.; Enright, A.J.; et al. RNA modifications detection by comparative Nanopore direct RNA sequencing. Nat. Commun. 2021, 12, 7198. [Google Scholar] [CrossRef] [PubMed]
- Jenjaroenpun, P.; Wongsurawat, T.; Wadley, T.D.; Wassenaar, T.M.; Liu, J.; Dai, Q.; Wanchai, V.; Akel, N.S.; Jamshidi-Parsian, A.; Franco, A.T.; et al. Decoding the epitranscriptional landscape from native RNA sequences. Nucleic Acids Res. 2021, 49, e7. [Google Scholar] [CrossRef]
- Pryszcz, L.P.; Novoa, E.M. ModPhred: An integrative toolkit for the analysis and storage of nanopore sequencing DNA and RNA modification data. Bioinformatics 2021, 38, 257–260. [Google Scholar] [CrossRef]
- Cozzuto, L.; Liu, H.; Pryszcz, L.P.; Pulido, T.H.; Delgado-Tejedor, A.; Ponomarenko, J.; Novoa, E.M. MasterOfPores: A Workflow for the Analysis of Oxford Nanopore Direct RNA Sequencing Datasets. Front. Genet. 2020, 11, 211. [Google Scholar] [CrossRef]
- Pei, Y.; Lou, X.; Li, K.; Xu, X.; Guo, Y.; Xu, D.; Yang, Z.; Xu, D.; Cui, W.; Zhang, D. Peripheral Blood Leukocyte N6-methyladenosine is a Noninvasive Biomarker for Non-small-cell Lung Carcinoma. OncoTargets Ther. 2020, 13, 11913–11921. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Fan, X.; Zhang, R.; Wu, G. Upregulated N6-Methyladenosine RNA in Peripheral Blood: Potential Diagnostic Biomarker for Breast Cancer. Cancer Res. Treat. 2021, 53, 399–408. [Google Scholar] [CrossRef]
- Ge, L.; Zhang, N.; Chen, Z.; Song, J.; Wu, Y.; Li, Z.; Chen, F.; Wu, J.; Li, D.; Li, J.; et al. Level of N6-Methyladenosine in Peripheral Blood RNA: A Novel Predictive Biomarker for Gastric Cancer. Clin. Chem. 2020, 66, 342–351. [Google Scholar] [CrossRef]
- Luo, Q.; Gao, Y.; Zhang, L.; Rao, J.; Guo, Y.; Huang, Z.; Li, J. Decreased ALKBH5, FTO, and YTHDF2 in Peripheral Blood Are as Risk Factors for Rheumatoid Arthritis. BioMed Res. Int. 2020, 2020, 5735279. [Google Scholar] [CrossRef]
- Zhu, L.; Liu, S.; Liao, F.; Yang, J.; Liang, T.; Yang, Y.; Huang, X.; Gu, L.; Su, L. Comprehensive Analysis of Blood-Based m6A Methylation in Human Ischemic Stroke. Mol. Neurobiol. 2023, 60, 431–446. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Huang, W.; Huang, J.T.; Xiong, J.; Yang, Y.; Wu, K.; Jia, G.F.; Chen, J.; Feng, Y.Q.; Yuan, B.F.; et al. Decreased N(6)-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J. Clin. Endocrinol. Metab. 2015, 100, E148–E154. [Google Scholar] [CrossRef]
- Min, K.W.; Zealy, R.W.; Davila, S.; Fomin, M.; Cummings, J.C.; Makowsky, D.; McDowell, C.H.; Thigpen, H.; Hafner, M.; Kwon, S.H.; et al. Profiling of m6A RNA modifications identified an age-associated regulation of AGO2 mRNA stability. Aging Cell 2018, 17, e12753. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Xie, Z.; Liao, Y.; Jiang, J.; Dong, W.; Yin, F.; Li, W.X.; Ye, L.; Lin, J.; Liang, H. Systematic analysis of clinical relevance and molecular characterization of m(6)A in COVID-19 patients. Genes Dis. 2022, 9, 1170–1173. [Google Scholar] [CrossRef]
- Kupsco, A.; Gonzalez, G.; Baker, B.H.; Knox, J.M.; Zheng, Y.; Wang, S.; Chang, D.; Schwartz, J.; Hou, L.; Wang, Y.; et al. Associations of smoking and air pollution with peripheral blood RNA N(6)-methyladenosine in the Beijing truck driver air pollution study. Environ. Int. 2020, 144, 106021. [Google Scholar] [CrossRef] [PubMed]
- Naren, D.; Yan, T.; Gong, Y.; Huang, J.; Zhang, D.; Sang, L.; Zheng, X.; Li, Y. High Wilms’ tumor 1 associating protein expression predicts poor prognosis in acute myeloid leukemia and regulates m(6)A methylation of MYC mRNA. J. Cancer Res. Clin. Oncol. 2021, 147, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Huang, Z.; Jiang, P.; Wu, R.; Jiang, H.; Luo, C.; Hong, H.; Yin, H. Elevated N6-Methyladenosine RNA Levels in Peripheral Blood Immune Cells: A Novel Predictive Biomarker and Therapeutic Target for Colorectal Cancer. Front. Immunol 2021, 12, 760747. [Google Scholar] [CrossRef]
- Wang, S.; Xie, X.; Li, C.; Jia, J.; Chen, C. Integrative network analysis of N(6) methylation-related genes reveal potential therapeutic targets for spinal cord injury. Math. Biosci. Eng. 2021, 18, 8174–8187. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Rao, J.; Zhang, L.; Fu, B.; Guo, Y.; Huang, Z.; Li, J. The study of METTL14, ALKBH5, and YTHDF2 in peripheral blood mononuclear cells from systemic lupus erythematosus. Mol. Genet. Genom. Med. 2020, 8, e1298. [Google Scholar] [CrossRef]
- Chang, J.S.; Lin, Z.X.; Liu, Y.J.; Yang, S.M.; Zhang, Y.; Yu, X.Y. Ultra performance liquid chromatography-tandem mass spectrometry assay for the quantification of RNA and DNA methylation. J. Pharm. Biomed. Anal. 2021, 197, 113969. [Google Scholar] [CrossRef]
- Guo, W.; Huai, Q.; Zhang, G.; Guo, L.; Song, P.; Xue, X.; Tan, F.; Xue, Q.; Gao, S.; He, J. Elevated Heterogeneous Nuclear Ribonucleoprotein C Expression Correlates With Poor Prognosis in Patients With Surgically Resected Lung Adenocarcinoma. Front. Oncol. 2020, 10, 598437. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Huai, Q.; Wan, H.; Guo, L.; Song, P.; Gao, S.; He, J. Prognostic Impact of IGF2BP3 Expression in Patients with Surgically Resected Lung Adenocarcinoma. DNA Cell Biol. 2021, 40, 316–331. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, M.; Hu, D. Deciphering N(6)-Methyladenosine-Related Genes Signature to Predict Survival in Lung Adenocarcinoma. BioMed Res. Int. 2020, 2020, 2514230. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.; Liu, L.; Li, J.; Hu, Q.; Sun, R. Expression and Prognostic Significance of m6A-Related Genes in Lung Adenocarcinoma. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020, 26, e919644. [Google Scholar] [CrossRef]
- Luo, Q.; Fu, B.; Zhang, L.; Guo, Y.; Huang, Z.; Li, J. Decreased Peripheral Blood ALKBH5 Correlates with Markers of Autoimmune Response in Systemic Lupus Erythematosus. Dis. Markers 2020, 2020, 8193895. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Gan, L.; Sun, J. Identification and Validation of Three m6A Regulators: FTO, HNRNPC, and HNRNPA2B1 as Potential Biomarkers for Endometriosis. Genes 2022, 14, 86. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.C.; Sun, Q.; Su, W.J.; Li, J.C.; Liu, Y.S.; Zhang, K.; Yang, L.Q. Analysis of m(6)A modulator-mediated methylation modification patterns and the tumor microenvironment in lung adenocarcinoma. Sci. Rep. 2022, 12, 20684. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Guo, X.; Zhao, M.; Ao, H.; Leng, X.; Liu, M.; Wu, C.; Ma, J.; Zhu, J. Contributions and prognostic values of m(6) A RNA methylation regulators in non-small-cell lung cancer. J. Cell. Physiol. 2020, 235, 6043–6057. [Google Scholar] [CrossRef]
- Gu, Z.; Yang, Y.; Ma, Q.; Wang, H.; Zhao, S.; Qi, Y.; Li, Y. HNRNPC, a predictor of prognosis and immunotherapy response based on bioinformatics analysis, is related to proliferation and invasion of NSCLC cells. Respir. Res. 2022, 23, 362. [Google Scholar] [CrossRef]
- Xu, F.; Zhang, H.; Chen, J.; Lin, L.; Chen, Y. Immune signature of T follicular helper cells predicts clinical prognostic and therapeutic impact in lung squamous cell carcinoma. Int. Immunopharmacol. 2020, 81, 105932. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, Y.; Chen, J.; Peng, C.; Zhang, Y.; Tong, R.; Cheng, Q.; Yang, B.; Feng, X.; Lu, Y.; et al. ALKBH5 suppresses malignancy of hepatocellular carcinoma via m(6)A-guided epigenetic inhibition of LYPD1. Mol. Cancer 2020, 19, 123. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Chen, Y.; Mao, Q.; Jiang, X.; Jiang, W.; Chen, J.; Xu, W.; Zhong, L.; Sun, X. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. Sect. A Dis. Markers 2018, 21, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Qu, N.; Qin, S.; Zhang, X.; Bo, X.; Liu, Z.; Tan, C.; Wen, G.; Jiang, H. Multiple m(6)A RNA methylation modulators promote the malignant progression of hepatocellular carcinoma and affect its clinical prognosis. BMC Cancer 2020, 20, 165. [Google Scholar] [CrossRef]
- Li, Z.; Li, F.; Peng, Y.; Fang, J.; Zhou, J. Identification of three m6A-related mRNAs signature and risk score for the prognostication of hepatocellular carcinoma. Cancer Med. 2020, 9, 1877–1889. [Google Scholar] [CrossRef]
- Lu, J.; Wang, H.; Cao, W.; Chen, D.; He, Z.; Xu, J. Construction of a m5C-related long non-coding RNA signature for the prognosis of hepatocellular carcinoma. Hum. Cell 2022, Online ahead of print. [Google Scholar] [CrossRef]
- Liu, T.; Yang, S.; Cheng, Y.P.; Kong, X.L.; Du, D.D.; Wang, X.; Bai, Y.F.; Yin, L.H.; Pu, Y.P.; Liang, G.Y. The N6-Methyladenosine (m6A) Methylation Gene YTHDF1 Reveals a Potential Diagnostic Role for Gastric Cancer. Cancer Manag. Res. 2020, 12, 11953–11964. [Google Scholar] [CrossRef]
- Zhang, B.; Wu, Q.; Li, B.; Wang, D.; Wang, L.; Zhou, Y.L. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol. Cancer 2020, 19, 53. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chen, C.; Ding, Q.; Zhao, Y.; Wang, Z.; Chen, J.; Jiang, Z.; Zhang, Y.; Xu, G.; Zhang, J.; et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 2020, 69, 1193–1205. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, D.; Wang, F.; Xu, Y.; Yu, H.; Zhang, H. Expression of Demethylase Genes, FTO and ALKBH1, Is Associated with Prognosis of Gastric Cancer. Dig. Dis. Sci. 2019, 64, 1503–1513. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Feng, S.; Sun, Y.; Jiang, Y.; Tang, C.; Sun, D. Identification of a five m6A-relevant mRNAs signature and risk score for the prognostication of gastric cancer. J. Gastrointest. Oncol 2022, 13, 2234–2248. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Wang, Z.; Li, H.; Zhang, H.; Luo, L. Gene Signature and Identification of Clinical Trait-Related m(6) A Regulators in Pancreatic Cancer. Front. Genet. 2020, 11, 522. [Google Scholar] [CrossRef]
- Zhao, Z.; Ju, Q.; Ji, J.; Li, Y.; Zhao, Y. N6-Methyladenosine Methylation Regulator RBM15 is a Potential Prognostic Biomarker and Promotes Cell Proliferation in Pancreatic Adenocarcinoma. Front. Mol. Biosci. 2022, 9, 842833. [Google Scholar] [CrossRef]
- Xu, D.; Shao, J.; Song, H.; Wang, J. The YTH Domain Family of N6-Methyladenosine “Readers” in the Diagnosis and Prognosis of Colonic Adenocarcinoma. BioMed Res. Int. 2020, 2020, 9502560. [Google Scholar] [CrossRef]
- Yang, P.; Wang, Q.; Liu, A.; Zhu, J.; Feng, J. ALKBH5 Holds Prognostic Values and Inhibits the Metastasis of Colon Cancer. Pathol. Oncol. Res. POR 2020, 26, 1615–1623. [Google Scholar] [CrossRef]
- Liao, X.; Chen, L.; Liu, J.; Hu, H.; Hou, D.; You, R.; Wang, X.; Huang, H. m(6)A RNA methylation regulators predict prognosis and indicate characteristics of tumour microenvironment infiltration in acute myeloid leukaemia. Epigenetics 2022, 1–20, Online ahead of print. [Google Scholar] [CrossRef]
- Wang, Z.; Cheng, H.; Xu, H.; Yu, X.; Sui, D. A five-gene signature derived from m6A regulators to improve prognosis prediction of neuroblastoma. Cancer Biomark. Sect. A Dis. Markers 2020, 28, 275–284. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, J.N.; Wang, E.H.; Gong, C.J.; Lan, K.F.; Ding, X. The m6A reader protein YTHDC2 is a potential biomarker and associated with immune infiltration in head and neck squamous cell carcinoma. PeerJ 2020, 8, e10385. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, J.; Liu, M.; Feng, Q.; Peng, H. IGF2BP2 serves as a core m6A regulator in head and neck squamous cell carcinoma. Biosci. Rep. 2022, 42, BSR20221311. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wan, Q.; Lu, J. The prognostic values of m6A RNA methylation regulators in uveal melanoma. BMC Cancer 2020, 20, 674. [Google Scholar] [CrossRef]
- Li, T.; Gu, M.; Deng, A.; Qian, C. Increased expression of YTHDF1 and HNRNPA2B1 as potent biomarkers for melanoma: A systematic analysis. Cancer Cell Int. 2020, 20, 239. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Bai, J.; Zhang, Y.; Xue, Y.; Peng, Q. N(6)-Methyladenosine regulator RBM15B acts as an independent prognostic biomarker and its clinical significance in uveal melanoma. Front. Immunol. 2022, 13, 918522. [Google Scholar] [CrossRef]
- Li, J.; Rao, B.; Yang, J.; Liu, L.; Huang, M.; Liu, X.; Cui, G.; Li, C.; Han, Q.; Yang, H.; et al. Dysregulated m6A-Related Regulators Are Associated with Tumor Metastasis and Poor Prognosis in Osteosarcoma. Front. Oncol. 2020, 10, 769. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Shan, H.; Zhang, Y.; Fan, Y.; Wu, B. m(6)A RNA methylation regulators have prognostic value in papillary thyroid carcinoma. Am. J. Otolaryngol. 2020, 41, 102547. [Google Scholar] [CrossRef]
- Li, K.; Luo, H.; Luo, H.; Zhu, X. Clinical and prognostic pan-cancer analysis of m6A RNA methylation regulators in four types of endocrine system tumors. Aging 2020, 12, 23931–23944. [Google Scholar] [CrossRef]
- Fu, Y.; Sun, S.; Bi, J.; Kong, C.; Yin, L. Expression patterns and prognostic value of m6A RNA methylation regulators in adrenocortical carcinoma. Medicine 2021, 100, e25031. [Google Scholar] [CrossRef]
- Zhang, Q.J.; Luan, J.C.; Song, L.B.; Cong, R.; Ji, C.J.; Zhou, X.; Xia, J.D.; Song, N.H. m6A RNA methylation regulators correlate with malignant progression and have potential predictive values in clear cell renal cell carcinoma. Exp. Cell Res. 2020, 392, 112015. [Google Scholar] [CrossRef]
- Zheng, Z.; Mao, S.; Guo, Y.; Zhang, W.; Liu, J.; Li, C.; Yao, X. N6-methyladenosine RNA methylation regulators participate in malignant progression and have prognostic value in clear cell renal cell carcinoma. Oncol. Rep. 2020, 43, 1591–1605. [Google Scholar] [CrossRef]
- Strick, A.; von Hagen, F.; Gundert, L.; Klümper, N.; Tolkach, Y.; Schmidt, D.; Kristiansen, G.; Toma, M.; Ritter, M.; Ellinger, J. The N(6) -methyladenosine (m(6) A) erasers alkylation repair homologue 5 (ALKBH5) and fat mass and obesity-associated protein (FTO) are prognostic biomarkers in patients with clear cell renal carcinoma. BJU Int. 2020, 125, 617–624. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, H.; Chen, Q.; Wan, Z.; Gao, X.; Qian, W. Identification of METTL14 in Kidney Renal Clear Cell Carcinoma Using Bioinformatics Analysis. Dis. Markers 2019, 2019, 5648783. [Google Scholar] [CrossRef]
- Wu, J.; Wang, X.; Xu, H.; Tian, J.; Ji, H.; Zhu, J.; Guo, H.; Chen, Z. Bioinformatics analysis of the correlation between m6A RNA methylation regulators and the immune infiltration and prognosis of bladder cancer. Ann. Transl. Med. 2022, 10, 1386. [Google Scholar] [CrossRef]
- Chen, M.; Nie, Z.Y.; Wen, X.H.; Gao, Y.H.; Cao, H.; Zhang, S.F. m6A RNA methylation regulators can contribute to malignant progression and impact the prognosis of bladder cancer. Biosci. Rep. 2019, 39, BSR20192892. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Fang, R.; Chen, X.; Zhang, S.; Shi, H.; Ye, Y.; Shi, H.; Zou, Z.; Li, P.; Guo, Q.; Ma, L.; et al. EGFR/SRC/ERK-stabilized YTHDF2 promotes cholesterol dysregulation and invasive growth of glioblastoma. Nat. Commun. 2021, 12, 177. [Google Scholar] [CrossRef] [PubMed]
- Hershey, J.W. The role of eIF3 and its individual subunits in cancer. Biochim. Biophys. Acta 2015, 1849, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhang, Y.; Yao, S.; Shi, H.; Huang, X.; Li, Y.; Wei, Y.; Lin, S. The translation initiation factor eIF3i up-regulates vascular endothelial growth factor A, accelerates cell proliferation, and promotes angiogenesis in embryonic development and tumorigenesis. J. Biol. Chem. 2014, 289, 28310–28323. [Google Scholar] [CrossRef]
- Hu, X.; Martinez-Ledesma, E.; Zheng, S.; Kim, H.; Barthel, F.; Jiang, T.; Hess, K.R.; Verhaak, R.G.W. Multigene signature for predicting prognosis of patients with 1p19q co-deletion diffuse glioma. Neuro Oncol. 2017, 19, 786–795. [Google Scholar] [CrossRef] [PubMed]
- Chai, R.C.; Wang, N.; Chang, Y.Z.; Zhang, K.N.; Li, J.J.; Niu, J.J.; Wu, F.; Liu, Y.Q.; Wang, Y.Z. Systematically profiling the expression of eIF3 subunits in glioma reveals the expression of eIF3i has prognostic value in IDH-mutant lower grade glioma. Cancer Cell Int. 2019, 19, 155. [Google Scholar] [CrossRef]
- Bai, Z.; Wang, X.; Zhang, Z. Establishment and Validation of a 5 m6A RNA Methylation Regulatory Gene Prognostic Model in Low-Grade Glioma. Front. Genet. 2022, 13, 655169. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, N.; Wu, J.; Gao, X.; Zhao, H.; Liu, Z.; Yan, X.; Dong, J.; Wang, F.; Ba, Y.; et al. 5-Methylcytosine Related LncRNAs Reveal Immune Characteristics, Predict Prognosis and Oncology Treatment Outcome in Lower-Grade Gliomas. Front. Immunol. 2022, 13, 844778. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.Z.; Chai, R.C.; Pang, B.; Chang, X.; An, S.Y.; Zhang, K.N.; Jiang, T.; Wang, Y.Z. METTL3 enhances the stability of MALAT1 with the assistance of HuR via m6A modification and activates NF-κB to promote the malignant progression of IDH-wildtype glioma. Cancer Lett. 2021, 511, 36–46. [Google Scholar] [CrossRef]
- Guan, S.; He, Y.; Su, Y.; Zhou, L. A Risk Signature Consisting of Eight m(6)A Methylation Regulators Predicts the Prognosis of Glioma. Cell. Mol. Neurobiol. 2022, 42, 2733–2743. [Google Scholar] [CrossRef]
- Wei, C.; Wang, B.; Peng, D.; Zhang, X.; Li, Z.; Luo, L.; He, Y.; Liang, H.; Du, X.; Li, S.; et al. Pan-Cancer Analysis Shows That ALKBH5 Is a Potential Prognostic and Immunotherapeutic Biomarker for Multiple Cancer Types Including Gliomas. Front. Immunol. 2022, 13, 849592. [Google Scholar] [CrossRef]
- Lin, S.; Xu, H.; Zhang, A.; Ni, Y.; Xu, Y.; Meng, T.; Wang, M.; Lou, M. Prognosis Analysis and Validation of m(6)A Signature and Tumor Immune Microenvironment in Glioma. Front. Oncol. 2020, 10, 541401. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cao, H.; Zhong, Y.; Ji, P.; Chen, F. The Role of m6A Regulator-Mediated Methylation Modification and Tumor Microenvironment Infiltration in Glioblastoma Multiforme. Front. Cell Dev. Biol. 2022, 10, 842835. [Google Scholar] [CrossRef] [PubMed]
- Shao, D.; Li, Y.; Wu, J.; Zhang, B.; Xie, S.; Zheng, X.; Jiang, Z. An m6A/m5C/m1A/m7G-Related Long Non-coding RNA Signature to Predict Prognosis and Immune Features of Glioma. Front. Genet. 2022, 13, 903117. [Google Scholar] [CrossRef]
- Bertorello, J.; Sesen, J.; Gilhodes, J.; Evrard, S.; Courtade-Saïdi, M.; Augustus, M.; Uro-Coste, E.; Toulas, C.; Moyal, E.C.; Seva, C.; et al. Translation reprogramming by eIF3 linked to glioblastoma resistance. NAR Cancer 2020, 2, zcaa020. [Google Scholar] [CrossRef]
- Sun, C.; Zheng, X.; Sun, Y.; Yu, J.; Sheng, M.; Yan, S.; Zhu, Q.; Lan, Q. Identification of IGF2BP3 as an Adverse Prognostic Biomarker of Gliomas. Front. Genet. 2021, 12, 743738. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.C.; Chen, S.H.; Shen, X.L.; Li, D.C.; Liu, H.Y.; Ji, Y.L.; Li, M.; Yu, K.; Yang, H.; Chen, J.J.; et al. M6A RNA Methylation Regulator HNRNPC Contributes to Tumorigenesis and Predicts Prognosis in Glioblastoma Multiforme. Front. Oncol. 2020, 10, 536875. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.; Shi, L.; Ye, Y.; Shi, H.; Zeng, L.; Tiwary, S.; Huse, J.T.; Huo, L.; Ma, L.; Ma, Y.; et al. YTHDF3 Induces the Translation of m(6)A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell 2020, 38, 857–871.e857. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).