Body Composition and Metabolic Dysfunction Really Matter for the Achievement of Better Outcomes in High-Grade Serous Ovarian Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Data Collection
2.2. Genetic and Clinical Assessment of Nutritional Status and Lipid-Metabolism-Related Disorders
2.3. Co-Morbidities and mFI-5
2.4. Statistical Analyses
3. Results
3.1. Cohort Characteristics and Clinical Parameters Associated with Disease Recurrence and Survival
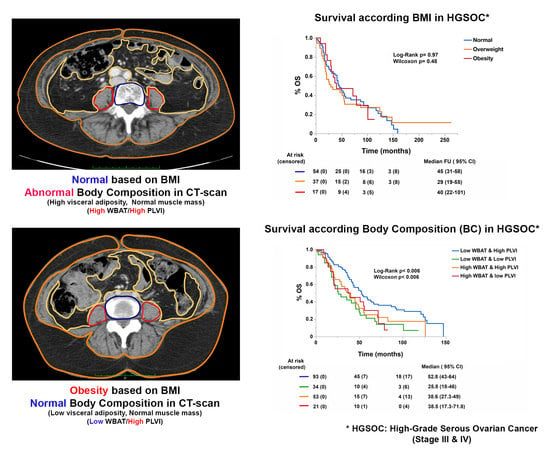
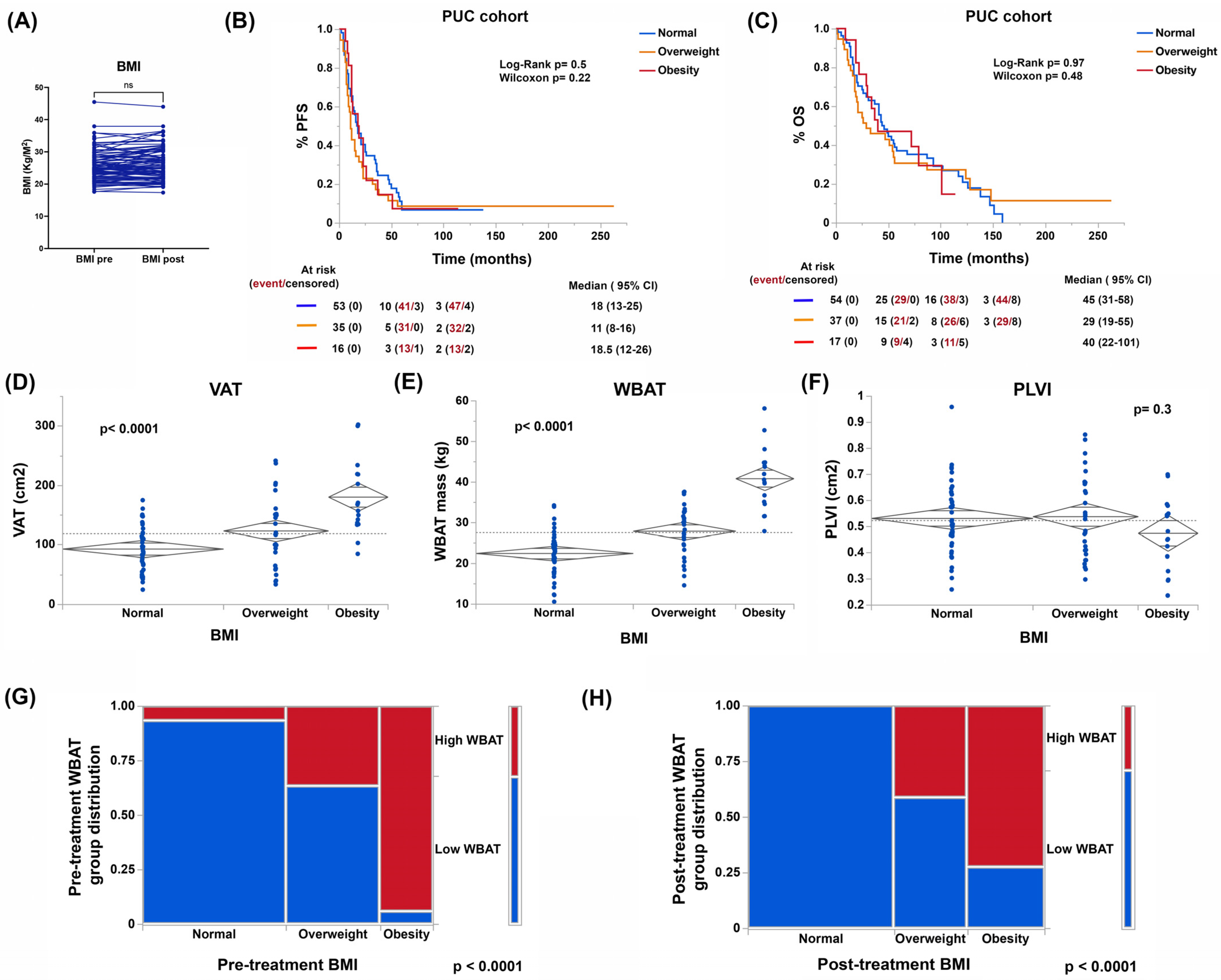
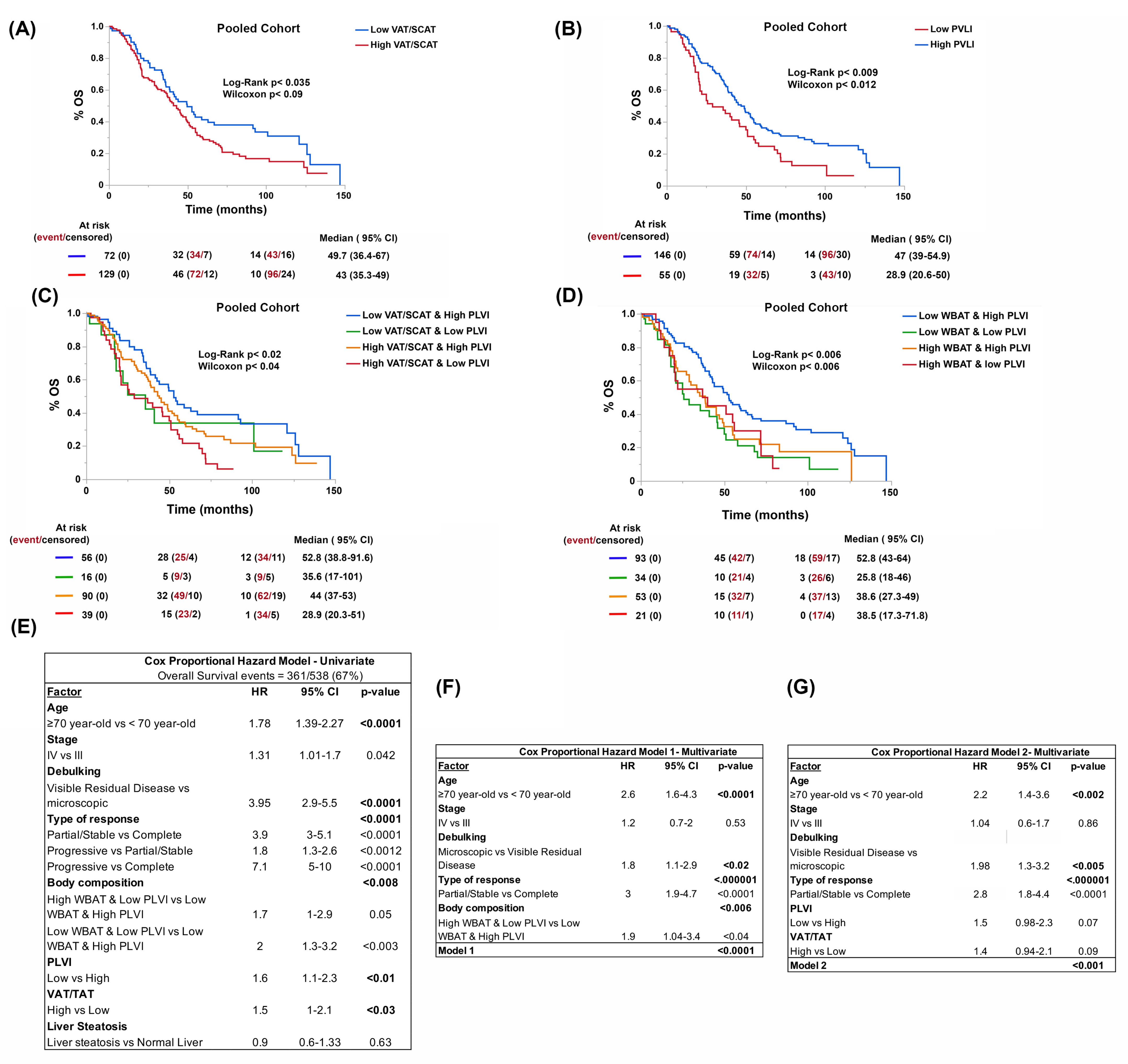
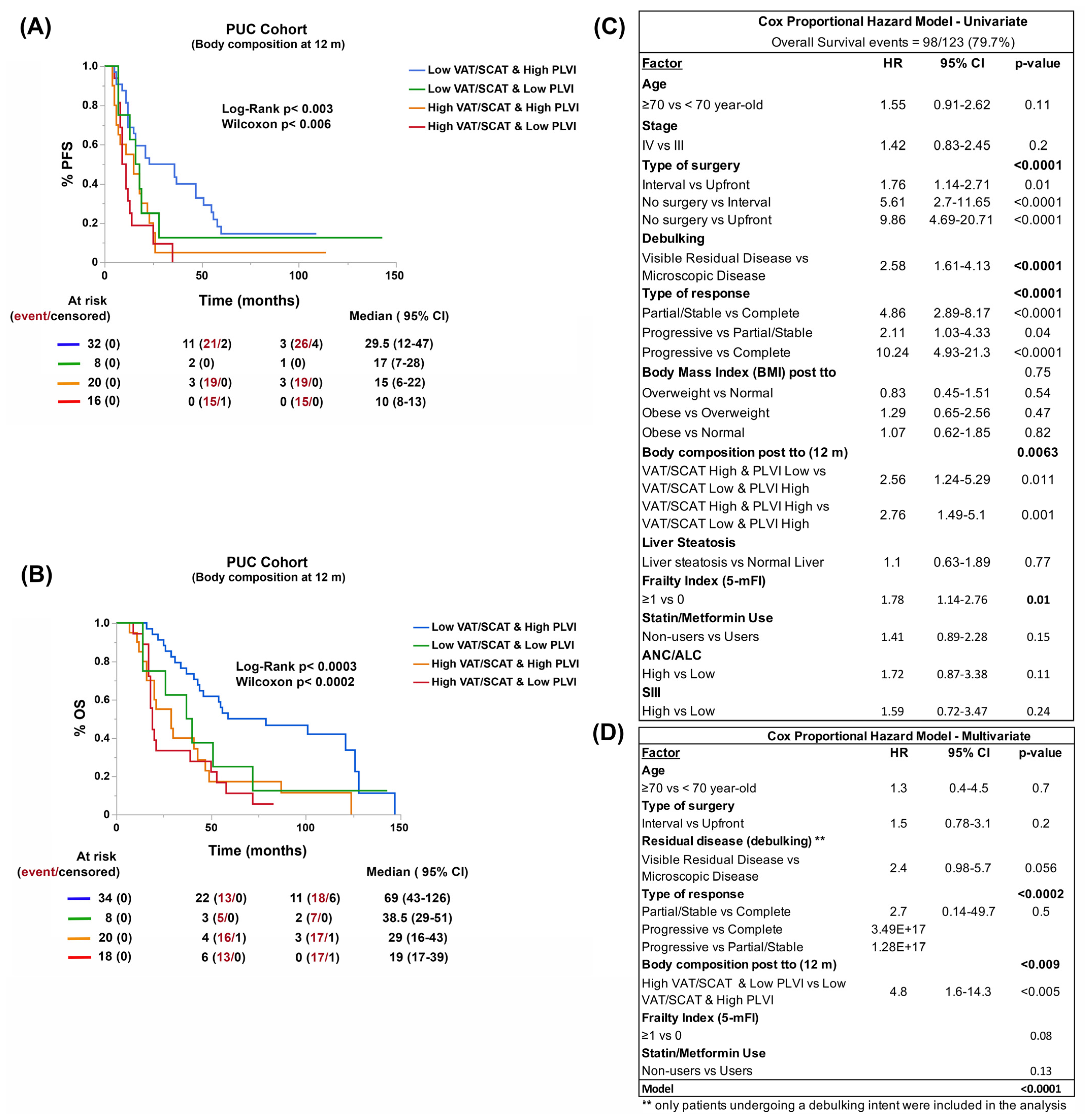
3.2. Body Composition, and Not BMI, Is Associated with Patient Outcomes
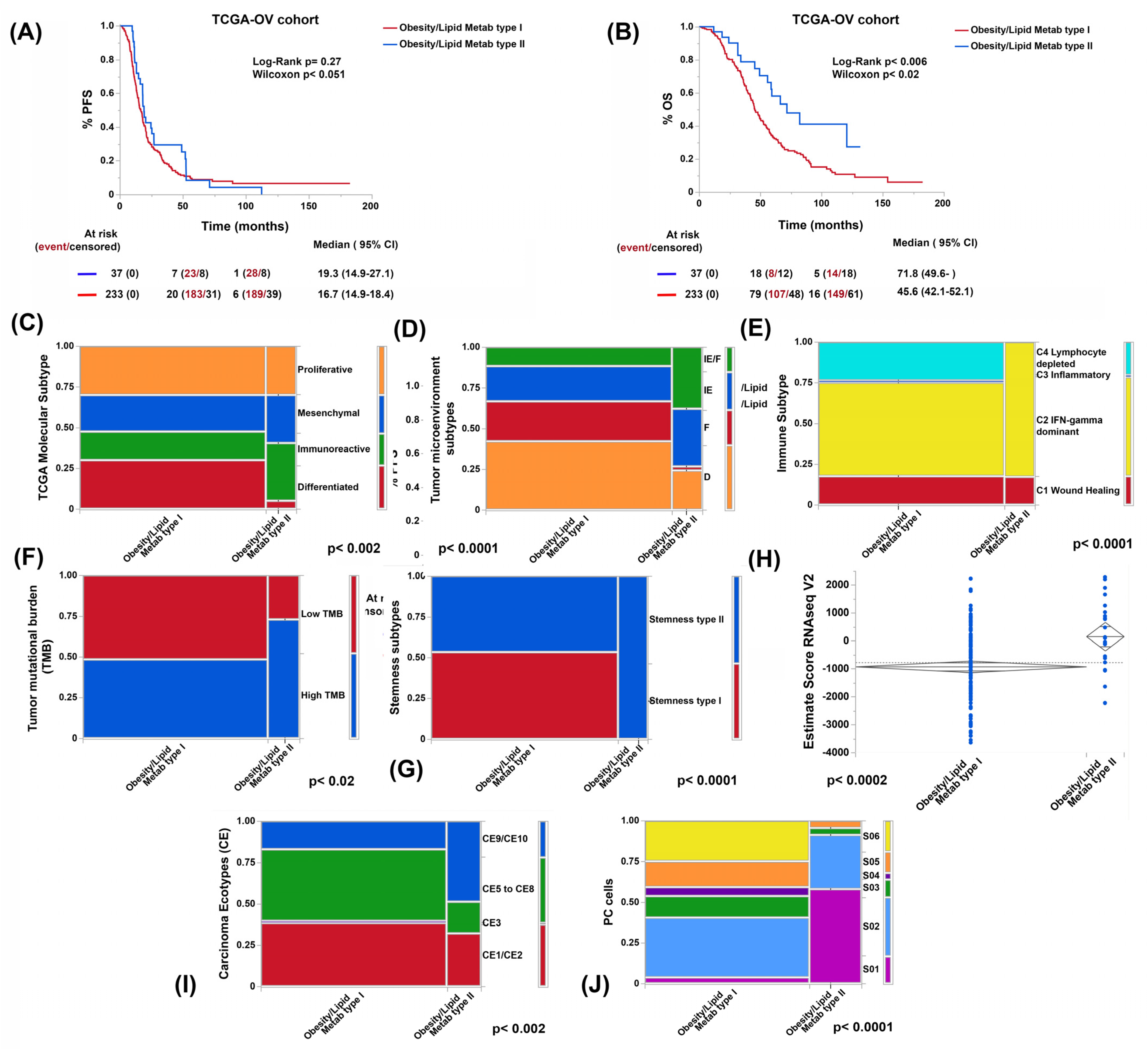
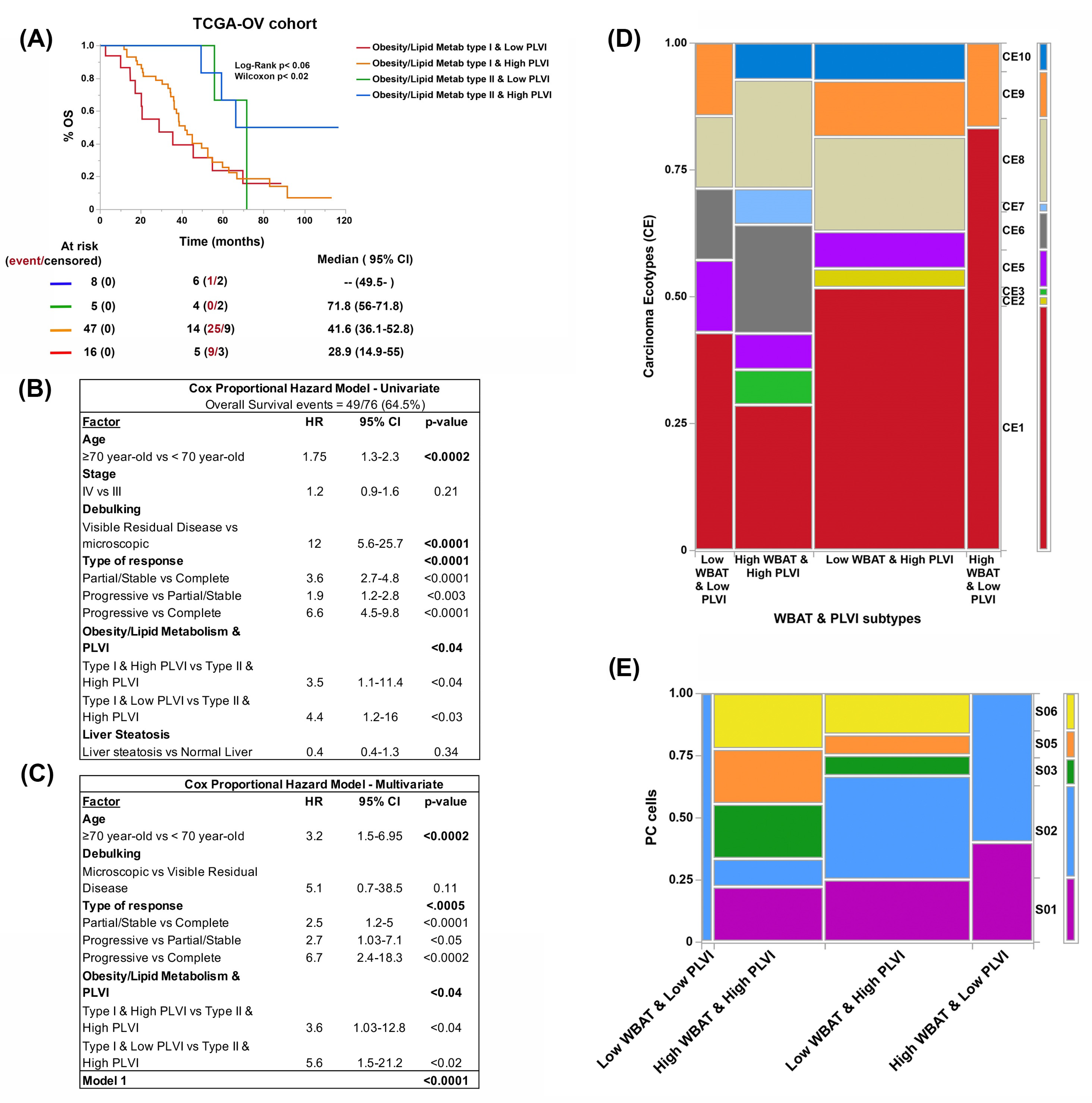
3.3. Genes Related to Obesity and Lipid Metabolism Distinguish Two Clusters of Patients with Marked Differences in Survival in the TCGA-OV Cohort
3.4. Obesity- and Lipid-Metabolism-Related Clusters Associate with Molecular Features Predictive of ICB Response
3.5. Both Obesity and Lipid Metabolism Clusters and BC Types Have Different Compositions of Immune Cell Types and States
3.6. Reduction of Visceral Adiposity, Increase of Muscle Mass, and Use of Metformin and Statins Improve Patient Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berek, J.S.; Renz, M.; Kehoe, S.; Kumar, L.; Friedlander, M. Cancer of the ovary, fallopian tube, and peritoneum: 2021 update. Int. J. Gynaecol. Obstet. 2021, 155 (Suppl. S1), 61–85. [Google Scholar] [CrossRef] [PubMed]
- Kurnit, K.C.; Fleming, G.F.; Lengyel, E. Updates and New Options in Advanced Epithelial Ovarian Cancer Treatment. Obstet. Gynecol. 2021, 137, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.E.; Tripcony, L.; Sanday, K.; Robertson, J.; Perrin, L.; Chetty, N.; Land, R.; Garrett, A.; Obermair, A.; Nascimento, M.; et al. Survival outcomes after delayed cytoreduction surgery following neoadjuvant chemotherapy in advanced epithelial ovarian cancer. Int. J. Gynecol. Cancer 2020, 30, 1935–1942. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Nag, S.; Aggarwal, S.; Rauthan, A.; Warrier, N. Maintenance therapy for recurrent epithelial ovarian cancer: Current therapies and future perspectives-a review. J. Ovarian. Res. 2019, 12, 103. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, E.; Minn, I.; Chambers, J.E.; Searson, P.C. In vitro characterization of pralidoxime transport and acetylcholinesterase reactivation across MDCK cells and stem cell-derived human brain microvascular endothelial cells (BC1-hBMECs). Fluids Barriers CNS 2016, 13, 10. [Google Scholar] [CrossRef]
- Gaitskell, K.; Hermon, C.; Barnes, I.; Pirie, K.; Floud, S.; Green, J.; Beral, V.; Reeves, G.K.; Million Women Study, C. Ovarian cancer survival by stage, histotype, and pre-diagnostic lifestyle factors, in the prospective UK Million Women Study. Cancer Epidemiol. 2022, 76, 102074. [Google Scholar] [CrossRef]
- Cheng, E.; Kirley, J.; Cespedes Feliciano, E.M.; Caan, B.J. Adiposity and cancer survival: A systematic review and meta-analysis. Cancer Causes Control 2022, 33, 1219–1246. [Google Scholar] [CrossRef]
- De Lorenzo, A.; Soldati, L.; Sarlo, F.; Calvani, M.; Di Lorenzo, N.; Di Renzo, L. New obesity classification criteria as a tool for bariatric surgery indication. World J. Gastroenterol. 2016, 22, 681–703. [Google Scholar] [CrossRef]
- Clarke, C.L.; Kushi, L.H.; Chubak, J.; Pawloski, P.A.; Bulkley, J.E.; Epstein, M.M.; Burnett-Hartman, A.N.; Powell, B.; Pearce, C.L.; Spencer Feigelson, H. Predictors of Long-Term Survival among High-Grade Serous Ovarian Cancer Patients. Cancer Epidemiol. Biomarkers Prev. 2019, 28, 996–999. [Google Scholar] [CrossRef]
- Marchetti, C.; D’Indinosante, M.; Bottoni, C.; Di Ilio, C.; Di Berardino, S.; Costantini, B.; Minucci, A.; Vertechy, L.; Scambia, G.; Fagotti, A. NLR and BRCA mutational status in patients with high grade serous advanced ovarian cancer. Sci. Rep. 2021, 11, 11125. [Google Scholar] [CrossRef]
- Pinto, M.P.; Balmaceda, C.; Bravo, M.L.; Kato, S.; Villarroel, A.; Owen, G.I.; Roa, J.C.; Cuello, M.A.; Ibanez, C. Patient inflammatory status and CD4+/CD8+ intraepithelial tumor lymphocyte infiltration are predictors of outcomes in high-grade serous ovarian cancer. Gynecol. Oncol. 2018, 151, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Susok, L.; Said, S.; Reinert, D.; Mansour, R.; Scheel, C.H.; Becker, J.C.; Gambichler, T. The pan-immune-inflammation value and systemic immune-inflammation index in advanced melanoma patients under immunotherapy. J. Cancer Res. Clin. Oncol. 2022, 11, 3103–3108. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hong, N.; Robertson, M.; Wang, C.; Jiang, G. Preoperative red cell distribution width and neutrophil-to-lymphocyte ratio predict survival in patients with epithelial ovarian cancer. Sci. Rep. 2017, 7, 43001. [Google Scholar] [CrossRef] [PubMed]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic. Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef] [PubMed]
- Jagannadham, J.; Jaiswal, H.K.; Agrawal, S.; Rawal, K. Comprehensive Map of Molecules Implicated in Obesity. PLoS ONE 2016, 11, e0146759. [Google Scholar] [CrossRef]
- Cuello, M.A.; Kato, S.; Liberona, F. The impact on high-grade serous ovarian cancer of obesity and lipid metabolism-related gene expression patterns: The underestimated driving force affecting prognosis. J. Cell Mol. Med. 2018, 22, 1805–1815. [Google Scholar] [CrossRef]
- Brunet, J.P.; Tamayo, P.; Golub, T.R.; Mesirov, J.P. Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. USA 2004, 101, 4164–4169. [Google Scholar] [CrossRef]
- Reich, M.; Liefeld, T.; Gould, J.; Lerner, J.; Tamayo, P.; Mesirov, J.P. GenePattern 2.0. Nat. Genet. 2006, 38, 500–501. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Luca, B.A.; Steen, C.B.; Matusiak, M.; Azizi, A.; Varma, S.; Zhu, C.; Przybyl, J.; Espin-Perez, A.; Diehn, M.; Alizadeh, A.A.; et al. Atlas of clinically distinct cell states and ecosystems across human solid tumors. Cell 2021, 184, 5482–5496.e5428. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Yu, Q.; Pang, J.; Chen, Y.; Sheng, M.; Tang, W. The Value of the Stemness Index in Ovarian Cancer Prognosis. Genes 2022, 13, 993. [Google Scholar] [CrossRef] [PubMed]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e814. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, W.; Li, S.; Sun, R.; Cheng, W. Multi-omics analysis of tumor mutational burden combined with prognostic assessment in epithelial ovarian cancer based on TCGA database. Int. J. Med. Sci. 2020, 17, 3200–3213. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Bagaev, A.; Kotlov, N.; Nomie, K.; Svekolkin, V.; Gafurov, A.; Isaeva, O.; Osokin, N.; Kozlov, I.; Frenkel, F.; Gancharova, O.; et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell 2021, 39, 845–865.e847. [Google Scholar] [CrossRef] [PubMed]
- Ebbeling, L.; Grabo, D.J.; Shashaty, M.; Dua, R.; Sonnad, S.S.; Sims, C.A.; Pascual, J.L.; Schwab, C.W.; Holena, D.N. Psoas:lumbar vertebra index: Central sarcopenia independently predicts morbidity in elderly trauma patients. Eur. J. Trauma Emerg Surg. 2014, 40, 57–65. [Google Scholar] [CrossRef]
- Kim, E.H.; Kim, H.K.; Lee, M.J.; Bae, S.J.; Choe, J.; Jung, C.H.; Kim, C.H.; Park, J.Y.; Lee, W.J. Sex Differences of Visceral Fat Area and Visceral-to-Subcutaneous Fat Ratio for the Risk of Incident Type 2 Diabetes Mellitus. Diabetes Metab. J. 2022, 46, 486–498. [Google Scholar] [CrossRef]
- Lacoste Jeanson, A.; Dupej, J.; Villa, C.; Bruzek, J. Body composition estimation from selected slices: Equations computed from a new semi-automatic thresholding method developed on whole-body CT scans. Peer J. 2017, 5, e3302. [Google Scholar] [CrossRef]
- Jawahar, A.; Gonzalez, B.; Balasubramanian, N.; Adams, W.; Goldberg, A. Comparison of computed tomography hepatic steatosis criteria for identification of abnormal liver function and clinical risk factors, in incidentally noted fatty liver. Eur. J. Gastroenterol. Hepatol. 2020, 32, 216–221. [Google Scholar] [CrossRef]
- Subramaniam, S.; Aalberg, J.J.; Soriano, R.P.; Divino, C.M. New 5-Factor Modified Frailty Index Using American College of Surgeons NSQIP Data. J. Am. Coll Surg. 2018, 226, 173–181.e178. [Google Scholar] [CrossRef] [PubMed]
- George, E.M.; Burke, W.M.; Hou, J.Y.; Tergas, A.I.; Chen, L.; Neugut, A.I.; Ananth, C.V.; Hershman, D.L.; Wright, J.D. Measurement and validation of frailty as a predictor of outcomes in women undergoing major gynaecological surgery. BJOG 2016, 123, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Di Donato, V.; Di Pinto, A.; Giannini, A.; Caruso, G.; D’Oria, O.; Tomao, F.; Fischetti, M.; Perniola, G.; Palaia, I.; Muzii, L.; et al. Modified fragility index and surgical complexity score are able to predict postoperative morbidity and mortality after cytoreductive surgery for advanced ovarian cancer. Gynecol. Oncol. 2021, 161, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.Z.I.; Turin, T.C. Variable selection strategies and its importance in clinical prediction modelling. Fam. Med. Commun. Health 2020, 8, e000262. [Google Scholar] [CrossRef] [PubMed]
- Torres, D.; Wang, C.; Kumar, A.; Bakkum-Gamez, J.N.; Weaver, A.L.; McGree, M.E.; Konecny, G.E.; Goode, E.L.; Cliby, W.A. Factors that influence survival in high-grade serous ovarian cancer: A complex relationship between molecular subtype, disease dissemination, and operability. Gynecol. Oncol. 2018, 150, 227–232. [Google Scholar] [CrossRef]
- Fabbro, M.; Colombo, P.E.; Leaha, C.M.; Rouanet, P.; Carrere, S.; Quenet, F.; Gutowski, M.; Mourregot, A.; D’Hondt, V.; Coupier, I.; et al. Conditional Probability of Survival and Prognostic Factors in Long-Term Survivors of High-Grade Serous Ovarian Cancer. Cancers 2020, 12, 2184. [Google Scholar] [CrossRef]
- Kato, S.; Liberona, M.F.; Cerda-Infante, J.; Sanchez, M.; Henriquez, J.; Bizama, C.; Bravo, M.L.; Gonzalez, P.; Gejman, R.; Branes, J.; et al. Simvastatin interferes with cancer ‘stem-cell’ plasticity reducing metastasis in ovarian cancer. Endocr. Relat. Cancer 2018, 25, 821–836. [Google Scholar] [CrossRef]
- Wang, H.; Liu, J.; Yang, J.; Wang, Z.; Zhang, Z.; Peng, J.; Wang, Y.; Hong, L. A novel tumor mutational burden-based risk model predicts prognosis and correlates with immune infiltration in ovarian cancer. Front. Immunol. 2022, 13, 943389. [Google Scholar] [CrossRef]
- Jardim, D.L.; Goodman, A.; de Melo Gagliato, D.; Kurzrock, R. The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer Cell 2021, 39, 154–173. [Google Scholar] [CrossRef]
- Shen, X.; Gu, X.; Ma, R.; Li, X.; Wang, J. Identification of the Immune Signatures for Ovarian Cancer Based on the Tumor Immune Microenvironment Genes. Front. Cell Dev. Biol. 2022, 10, 772701. [Google Scholar] [CrossRef]
- Bersanelli, M.; Cortellini, A.; Buti, S. The interplay between cholesterol (and other metabolic conditions) and immune-checkpoint immunotherapy: Shifting the concept from the “inflamed tumor” to the “inflamed patient”. Hum. Vaccin Immunother. 2021, 17, 1930–1934. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yi, Z. Obesity paradox and aging: Visceral Adiposity Index and all-cause mortality in older individuals: A prospective cohort study. Front. Endocrinol. (Lausanne) 2022, 13, 975209. [Google Scholar] [CrossRef] [PubMed]
- Strulov Shachar, S.; Williams, G.R. The Obesity Paradox in Cancer-Moving Beyond BMI. Cancer Epidemiol. Biomarkers Prev. 2017, 26, 13–16. [Google Scholar] [CrossRef] [PubMed]
- McSharry, V.; Mullee, A.; McCann, L.; Rogers, A.C.; McKiernan, M.; Brennan, D.J. The Impact of Sarcopenia and Low Muscle Attenuation on Overall Survival in Epithelial Ovarian Cancer: A Systematic Review and Meta-analysis. Ann. Surg. Oncol. 2020, 27, 3553–3564. [Google Scholar] [CrossRef] [PubMed]
- Pfister, D.; Nunez, N.G.; Pinyol, R.; Govaere, O.; Pinter, M.; Szydlowska, M.; Gupta, R.; Qiu, M.; Deczkowska, A.; Weiner, A.; et al. NASH limits anti-tumour surveillance in immunotherapy-treated HCC. Nature 2021, 592, 450–456. [Google Scholar] [CrossRef]
- Cataldi, M.; Manco, F.; Tarantino, G. Steatosis, Steatohepatitis and Cancer Immunotherapy: An Intricate Story. Int. J. Mol. Sci. 2021, 22, 12947. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, C.; Cao, F.; Wang, C.B.; Dong, J.N.; Wang, Z.H. Nomogram of Combining CT-Based Body Composition Analyses and Prognostic Inflammation Score: Prediction of Survival in Advanced Epithelial Ovarian Cancer Patients. Acad. Radiol. 2022, 29, 1394–1403. [Google Scholar] [CrossRef]
- Zumsteg, D.M.; Chu, C.E.; Midwinter, M.J. Radiographic assessment of sarcopenia in the trauma setting: A systematic review. Trauma Surg. Acute Care Open 2020, 5, e000414. [Google Scholar] [CrossRef]
- Wang, Y.; Thai, T.; Moore, K.; Ding, K.; McMeekin, S.; Liu, H.; Zheng, B. Quantitative measurement of adiposity using CT images to predict the benefit of bevacizumab-based chemotherapy in epithelial ovarian cancer patients. Oncol. Lett. 2016, 12, 680–686. [Google Scholar] [CrossRef]
- Kroeger, D.R.; Milne, K.; Nelson, B.H. Tumor-Infiltrating Plasma Cells Are Associated with Tertiary Lymphoid Structures, Cytolytic T-Cell Responses, and Superior Prognosis in Ovarian Cancer. Clin. Cancer Res. 2016, 22, 3005–3015. [Google Scholar] [CrossRef]
- Shalapour, S.; Font-Burgada, J.; Di Caro, G.; Zhong, Z.; Sanchez-Lopez, E.; Dhar, D.; Willimsky, G.; Ammirante, M.; Strasner, A.; Hansel, D.E.; et al. Immunosuppressive plasma cells impede T-cell-dependent immunogenic chemotherapy. Nature 2015, 521, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Crombe, A.; Kind, M.; Toulmonde, M.; Italiano, A.; Cousin, S. Impact of CT-based body composition parameters at baseline, their early changes and response in metastatic cancer patients treated with immune checkpoint inhibitors. Eur. J. Radiol. 2020, 133, 109340. [Google Scholar] [CrossRef] [PubMed]
- Trestini, I.; Caldart, A.; Dodi, A.; Avancini, A.; Tregnago, D.; Sartori, G.; Belluomini, L.; Milella, M.; Pilotto, S. Body composition as a modulator of response to immunotherapy in lung cancer: Time to deal with it. ESMO Open 2021, 6, 100095. [Google Scholar] [CrossRef] [PubMed]
- Wichmann, I.A.; Cuello, M.A. Obesity and gynecological cancers: A toxic relationship. Int. J. Gynaecol. Obstet. 2021, 155 (Suppl. S1), 123–134. [Google Scholar] [CrossRef]
- Mao, W.; Cai, Y.; Chen, D.; Jiang, G.; Xu, Y.; Chen, R.; Wang, F.; Wang, X.; Zheng, M.; Zhao, X.; et al. Statin shapes inflamed tumor microenvironment and enhances immune checkpoint blockade in non-small cell lung cancer. JCI Insight 2022, 7, e161940. [Google Scholar] [CrossRef]
- McSharry, V.; Glennon, K.; Mullee, A.; Brennan, D. The impact of body composition on treatment in ovarian cancer: A current insight. Expert Rev. Clin. Pharmacol. 2021, 14, 1065–1074. [Google Scholar] [CrossRef]

| Variable | PUC Cohort | TCGA-OV Cohort | p-Value |
|---|---|---|---|
| n sample (n with imaging) | 123 (104) | n = 415 (97) | |
| Age (mean ± SD) | 59 ± 11.6 | 59 ± 11.5 | 0.98 |
| (min–max; range) | (29–83; 54) | (26–87; 61) | |
| Stage | 0.67 | ||
| III (%) | 105 (85.4) | 345 (83.1) | |
| IV (%) | 18 (14.6) | 70 (16.9) | |
| Adiposity Estimates VAT (cm2) | 118.1 ± 58.9 | 130.8 ± 70 | 0.91 |
| VAT/SCAT | 0.49 ± 0.22 | 0.50 ± 0.22 | 0.55 |
| VAT/TAT | 0.32 ± 0.09 | 0.32 ± 0.09 | 0.55 |
| WBAT (kg) | 27.3 ± 9.2 | 30 ± 11.7 | 0.96 |
| Liver Steatosis (CTl-s) | 0.015 | ||
| Normal | 85 (81.9) | 65 (67) | |
| Steatosis | 19 (18.1) | 32 (33) | |
| Type of Surgery * | |||
| Upfront surgery | 63 (51.2) | 414 (99.8) | |
| Interval surgery | 46 (37.4) | 1 (0.2) | |
| Never surgery # | 14 (11.4) | N/A | |
| Residual Disease * | <0.0001 | ||
| Microscopic | 55 (44.7) | 86(20.7) | |
| Else residual disease | 68 (55.3) | 329 (79.3) | |
| Type of Response | 0.52 | ||
| Complete | 76 (61.8) | 286 (68.9) | |
| Partial/Stable | 35 (28.5) | 90 (21.7) | |
| Progressive | 12 (9.8) | 39 (9.4) | |
| Statin/Metformin Use | 31 (25.2) | N/A | |
| Mean/Median FU (months) | 58.9/44 | 44.5/38.3 | <0.0001 |
| (min-max; range) | (1–263; 262) | (1–183; 182) | |
| Deaths (%) | 98 (79.7) | 263 (63.4) | 0.0005 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuello, M.A.; Gómez, F.; Wichmann, I.; Suárez, F.; Kato, S.; Orlandini, E.; Brañes, J.; Ibañez, C. Body Composition and Metabolic Dysfunction Really Matter for the Achievement of Better Outcomes in High-Grade Serous Ovarian Cancer. Cancers 2023, 15, 1156. https://doi.org/10.3390/cancers15041156
Cuello MA, Gómez F, Wichmann I, Suárez F, Kato S, Orlandini E, Brañes J, Ibañez C. Body Composition and Metabolic Dysfunction Really Matter for the Achievement of Better Outcomes in High-Grade Serous Ovarian Cancer. Cancers. 2023; 15(4):1156. https://doi.org/10.3390/cancers15041156
Chicago/Turabian StyleCuello, Mauricio A., Fernán Gómez, Ignacio Wichmann, Felipe Suárez, Sumie Kato, Elisa Orlandini, Jorge Brañes, and Carolina Ibañez. 2023. "Body Composition and Metabolic Dysfunction Really Matter for the Achievement of Better Outcomes in High-Grade Serous Ovarian Cancer" Cancers 15, no. 4: 1156. https://doi.org/10.3390/cancers15041156
APA StyleCuello, M. A., Gómez, F., Wichmann, I., Suárez, F., Kato, S., Orlandini, E., Brañes, J., & Ibañez, C. (2023). Body Composition and Metabolic Dysfunction Really Matter for the Achievement of Better Outcomes in High-Grade Serous Ovarian Cancer. Cancers, 15(4), 1156. https://doi.org/10.3390/cancers15041156