Development and Internal Validation of a Risk Prediction Model to Identify Myeloma Based on Routine Blood Tests: A Case-Control Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Study Population
2.2. Blood Tests
2.3. Sample Size and Events per Parameter
2.4. Candidate Predictors
2.5. Model Development
2.6. Missing Data
2.7. Internal Validation
2.8. Diagnostic Accuracy Statistics
2.9. Sensitivity Analysis and Additional Models
- All ages, MGUS as controls, calcium excluded as a predictor.
- Over 60 s only, MGUS as controls, all predictors included.
- Over 60 s only, MGUS as controls, and calcium excluded as a predictor.
2.10. Software
3. Results
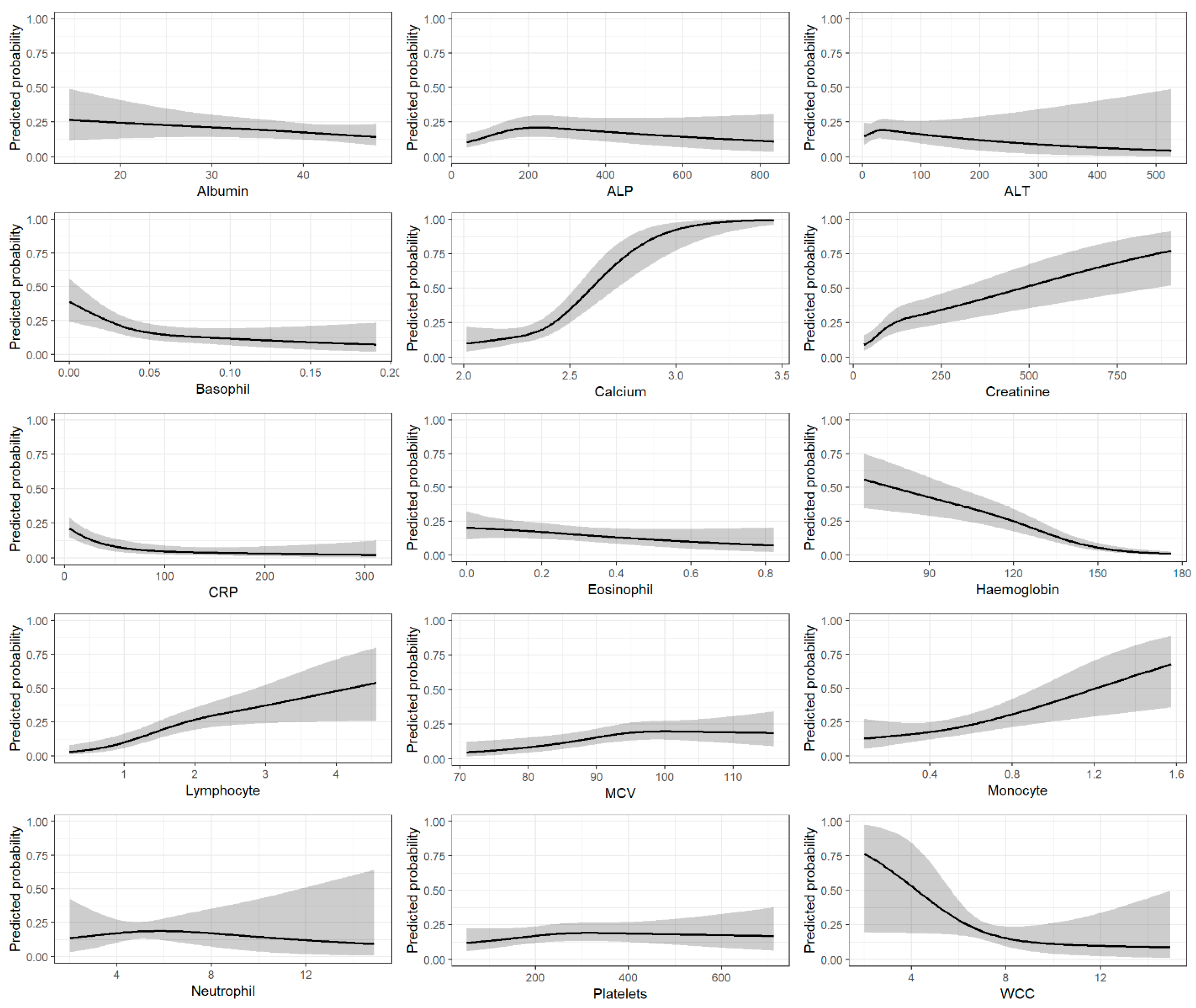
3.1. Predictor Variables
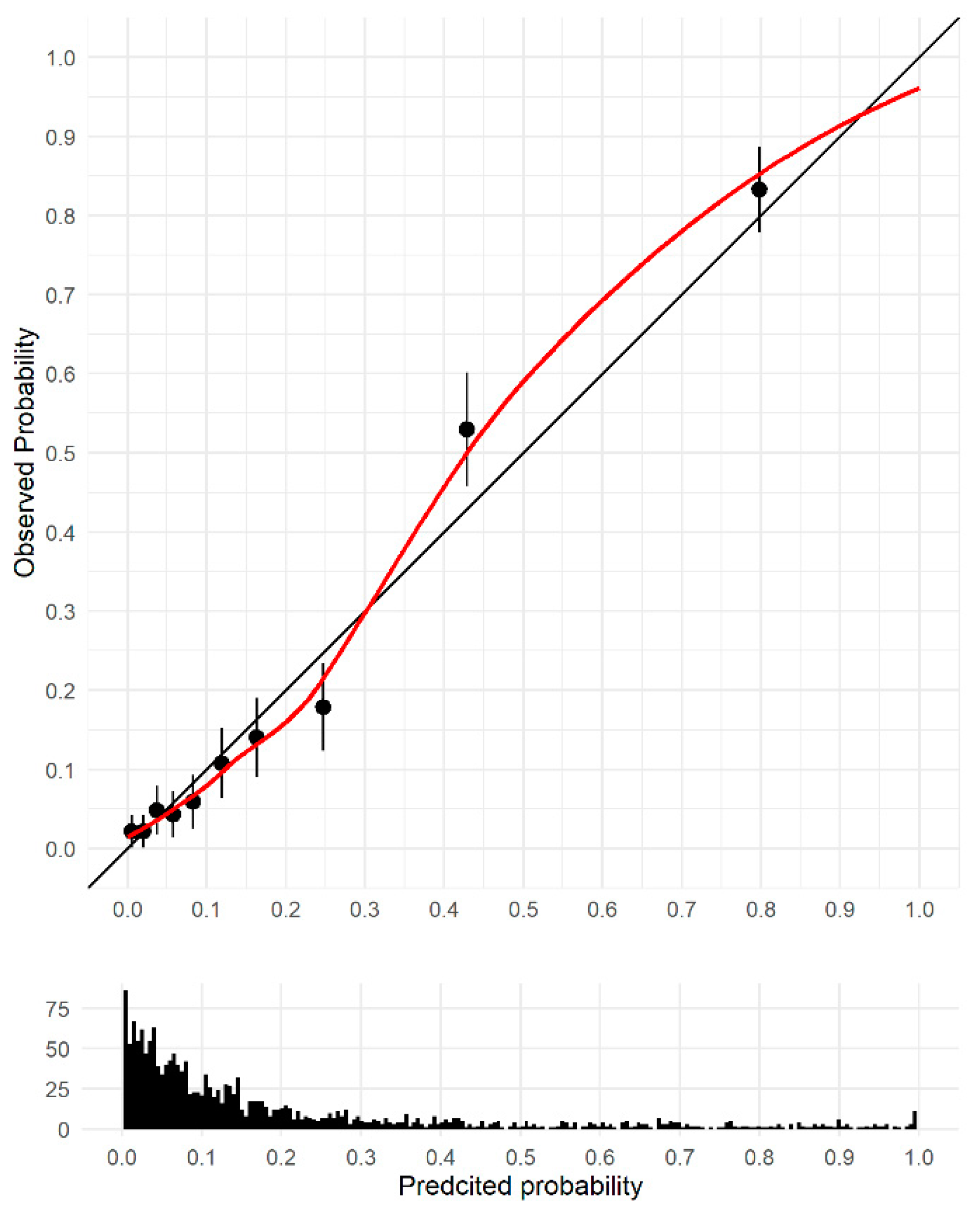
3.2. Model Discrimination and Validation
3.3. Sensitivity Analysis
3.4. Diagnostic Accuracy
4. Discussion
4.1. Key Findings
4.2. Comparison with Other Studies
4.3. Clinical Implications
4.4. Strengths and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cancer Research UK. Myeloma Statistics. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/myeloma (accessed on 28 July 2022).
- Lyratzopoulos, G.; Wardle, J.; Rubin, G. Rethinking diagnostic delay in cancer: How difficult is the diagnosis? BMJ 2014, 349, g7400. [Google Scholar] [CrossRef]
- Shephard, E.A.; Neal, R.D.; Rose, P.; Walter, F.M.; Litt, E.J.; Hamilton, W.T. Quantifying the risk of multiple myeloma from symptoms reported in primary care patients: A large case-control study using electronic records. Br. J. Gen. Pract. 2015, 65, e106–e113. [Google Scholar] [CrossRef]
- Koo, M.M.; Hamilton, W.; Walter, F.M.; Rubin, G.P.; Lyratzopoulos, G. Symptom Signatures and Diagnostic Timeliness in Cancer Patients: A Review of Current Evidence. Neoplasia 2018, 20, 165–174. [Google Scholar] [CrossRef]
- Howell, D.; Smith, A.; Appleton, S.; Bagguley, T.; Macleod, U.; Cook, G.; Patmore, R.; Roman, E. Multiple myeloma: Routes to diagnosis, clinical characteristics and survival—Findings from a UK population-based study. Br. J. Haematol. 2017, 177, 67–71. [Google Scholar] [CrossRef]
- Elliss-Brookes, L.; McPhail, S.; Ives, A.; Greenslade, M.; Shelton, J.; Hiom, S.; Richards, M. Routes to diagnosis for cancer—Determining the patient journey using multiple routine data sets. Br. J. Cancer 2012, 107, 1220–1226. [Google Scholar] [CrossRef]
- Lyratzopoulos, G.; Neal, R.D.; Barbiere, J.M.; Rubin, G.P.; Abel, G.A. Variation in number of general practitioner consultations before hospital referral for cancer: Findings from the 2010 National Cancer Patient Experience Survey in England. Lancet Oncol. 2012, 13, 353–365. [Google Scholar] [CrossRef]
- Atkin, C.; Iqbal, G.; Planche, T.; Pratt, G.; Yong, K.; Wood, J.; Raynes, K.; Low, E.; Higgins, H.; Neal, R.D.; et al. Diagnostic pathways in multiple myeloma and their relationship to end organ damage: An analysis from the Tackling Early Morbidity and Mortality in Myeloma (TEAMM) trial. Br. J. Haematol. 2021, 192, 997–1005. [Google Scholar] [CrossRef]
- National Disease Registration Service NHS Digital. CancerData. Survival. Available online: https://www.cancerdata.nhs.uk/survival/home (accessed on 16 September 2022).
- Smith, L.; Carmichael, J.; Cook, G.; Shinkins, B.; Neal, R.D. Diagnosing myeloma in general practice: How might earlier diagnosis be achieved? Br. J. Gen. Pract. 2022, 72, 462–463. [Google Scholar] [CrossRef]
- Watson, J.; Mounce, L.; Bailey, S.E.; Cooper, S.L.; Hamilton, W. Blood markers for cancer. BMJ 2019, 367, l5774. [Google Scholar] [CrossRef]
- Seesaghur, A.; Petruski-Ivleva, N.; Banks, V.L.; Wang, J.R.; Abbasi, A.; Neasham, D.; Ramasamy, K. Clinical features and diagnosis of multiple myeloma: A population-based cohort study in primary care. BMJ Open 2021, 11, e052759. [Google Scholar] [CrossRef]
- Cranfield, B.M.; Koo, M.M.; Abel, G.A.; Swann, R.; McPhail, S.; Rubin, G.P.; Lyratzopoulos, G. Primary care blood tests before cancer diagnosis: National Cancer Diagnosis Audit data. Br. J. Gen. Pract. 2022, 73, e95–e103. [Google Scholar] [CrossRef] [PubMed]
- Edgren, G.; Bagnardi, V.; Bellocco, R.; Hjalgrim, H.; Rostgaard, K.; Melbye, M.; Reilly, M.; Adami, H.O.; Hall, P.; Nyren, O. Pattern of declining hemoglobin concentration before cancer diagnosis. Int. J. Cancer 2010, 127, 1429–1436. [Google Scholar] [CrossRef] [PubMed]
- Koshiaris, C.; Van den Bruel, A.; Oke, J.L.; Nicholson, B.D.; Shephard, E.; Braddick, M.; Hamilton, W. Early detection of multiple myeloma in primary care using blood tests: A case-control study in primary care. Br. J. Gen. Pract. 2018, 68, E586–E593. [Google Scholar] [CrossRef] [PubMed]
- Koshiaris, C.; Van den Bruel, A.; Nicholson, B.D.; Lay-Flurrie, S.; Hobbs, F.R.; Oke, J.L. Clinical prediction tools to identify patients at highest risk of myeloma in primary care: A retrospective open cohort study. Br. J. Gen. Pract. 2021, 71, e347–e355. [Google Scholar] [CrossRef]
- Wadhera, R.K.; Rajkumar, S.V. Prevalence of monoclonal gammopathy of undetermined significance: A systematic review. Mayo Clin. Proc. 2010, 85, 933–942. [Google Scholar] [CrossRef]
- Riley, R.D.; Ensor, J.; Snell, K.I.E.; Harrell, F.E., Jr.; Martin, G.P.; Reitsma, J.B.; Moons, K.G.M.; Collins, G.; van Smeden, M. Calculating the sample size required for developing a clinical prediction model. BMJ 2020, 368, m441. [Google Scholar] [CrossRef]
- Harrell, F.E. rms: Regression Modeling Strategies. 2022. Available online: https://cran.r-project.org/web/packages/rms/index.html (accessed on 25 January 2023).
- White, I.R.; Royston, P.; Wood, A.M. Multiple imputation using chained equations: Issues and guidance for practice. Stat. Med. 2011, 30, 377–399. [Google Scholar] [CrossRef]
- National Disease Registration Service NHS Digital. CancerData. Cancer Incidence 2019. Available online: https://www.cancerdata.nhs.uk/incidence_and_mortality (accessed on 13 September 2022).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.A.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Van Buuren, S.; Groothuis-Oudshoorn, K. mice: Multivariate Imputation by Chained Equations in R. J. Stat. Softw. 2011, 45, 1–67. [Google Scholar] [CrossRef]
- Tierney, N.; Cook, D.; McBain, N.; Fay, C.; O’Hara-Wild, M.; Hester, J.; Smith, L.; Heiss, A. nanair: Data Structures, Summaries, and Visualisations for Missing Data. Available online: https://cran.r-project.org/web/packages/naniar/naniar.pdf (accessed on 12 September 2022).
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Muller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Heymans, M.W. psfmi: Prediction Model Selection and Performance Evaluation in Multiple Imputed Datasets. Available online: https://mwheymans.github.io/psfmi/ (accessed on 12 September 2022).
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): The TRIPOD statement. Ann. Intern. Med. 2015, 162, 55–63. [Google Scholar] [CrossRef]
- Watson, J.; Salisbury, C.; Banks, J.; Whiting, P.; Hamilton, W. Predictive value of inflammatory markers for cancer diagnosis in primary care: A prospective cohort study using electronic health records. Br. J. Cancer 2019, 120, 1045–1051. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, S.V.; Dimopoulos, M.A.; Palumbo, A.; Blade, J.; Merlini, G.; Mateos, M.V.; Kumar, S.; Hillengass, J.; Kastritis, E.; Richardson, P.; et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014, 15, E538–E548. [Google Scholar] [CrossRef]
- Hamilton, F.; Carroll, R.; Hamilton, W.; Salisbury, C. The risk of cancer in primary care patients with hypercalcaemia: A cohort study using electronic records. Br. J. Cancer 2014, 111, 1410–1412. [Google Scholar] [CrossRef] [PubMed]
- McKeeman, G.C.; Hall, S.L.; Freedman, D.B. Reflex and reflective testing practice in Clinical Biochemistry in the United Kingdom—A national survey. Ann. Clin. Biochem. 2020, 57, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Rognvaldsson, S.; Love, T.J.; Thorsteinsdottir, S.; Reed, E.R.; Oskarsson, J.; Petursdottir, I.; Sigurethardottir, G.A.; Vietharsson, B.; Onundarson, P.T.; Agnarsson, B.A.; et al. Iceland screens, treats, or prevents multiple myeloma (iStopMM): A population-based screening study for monoclonal gammopathy of undetermined significance and randomized controlled trial of follow-up strategies. Blood Cancer J. 2021, 11, 94. [Google Scholar] [CrossRef]
- Royston, P.; Altman, D.G.; Sauerbrei, W. Dichotomizing continuous predictors in multiple regression: A bad idea. Stat. Med. 2006, 25, 127–141. [Google Scholar] [CrossRef]
- Moons, K.G.; Altman, D.G.; Reitsma, J.B.; Ioannidis, J.P.; Macaskill, P.; Steyerberg, E.W.; Vickers, A.J.; Ransohoff, D.F.; Collins, G.S. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): Explanation and elaboration. Ann. Intern. Med. 2015, 162, W1–W73. [Google Scholar] [CrossRef]
- Watson, J.; Bailey, S.; Hamilton, F.; Hamilton, W.; Mounce, L. Lessons from biases in electronic health record data: The importance of clinical vigilance with negative test results. BMJ 2018, 361, k1479. [Google Scholar] [CrossRef]
- Bailey, S.E.; Ukoumunne, O.C.; Shephard, E.A.; Hamilton, W. Clinical relevance of thrombocytosis in primary care: A prospective cohort study of cancer incidence using English electronic medical records and cancer registry data. Br. J. Gen. Pract. 2017, 67, e405–e413. [Google Scholar] [CrossRef]
- Ankus, E.; Price, S.J.; Ukoumunne, O.C.; Hamilton, W.; Bailey, S.E.R. Cancer incidence in patients with a high normal platelet count: A cohort study using primary care data. Fam. Pract. 2018, 35, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Bull, L.M.; Lunt, M.; Martin, G.P.; Hyrich, K.; Sergeant, J.C. Harnessing repeated measurements of predictor variables for clinical risk prediction: A review of existing methods. Diagn. Progn. Res. 2020, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, B.A.; Navar, A.M.; Pencina, M.J.; Ioannidis, J.P. Opportunities and challenges in developing risk prediction models with electronic health records data: A systematic review. J. Am. Med. Inform. Assoc. 2017, 24, 198–208. [Google Scholar] [CrossRef] [PubMed]

| Variable | Cases N = 367 | Controls N = 1488 | ||
|---|---|---|---|---|
| Age, median (IQR) | 71 | (63, 79) | 70 | (60, 78) |
| Age group, n(%) | ||||
| <50 years | 18 | 4.9% | 111 | 7.5% |
| 50–59 | 48 | 13.1% | 237 | 15.9% |
| 60–69 | 98 | 26.7% | 389 | 26.1% |
| 70–79 | 125 | 34.1% | 452 | 30.4% |
| 80–89 | 71 | 19.3% | 275 | 18.5% |
| 90+ | 7 | 1.9% | 24 | 1.6% |
| Sex, n(%) | ||||
| Male | 214 | 58.3% | 875 | 58.8% |
| Female | 153 | 41.7% | 613 | 41.2% |
| Ethnicity, n(%) | ||||
| White British | 309 | 84.2% | 1229 | 82.6% |
| Black | 17 | 4.6% | 15 | 1.1% |
| South Asian | 10 | 2.7% | 68 | 4.6% |
| Other | 16 | 4.4% | 30 | 2.2% |
| Missing | 15 | 4.1% | 146 | 9.8% |
| Total number of comorbidities, n(%) | ||||
| 0 | 242 | 65.9% | 1018 | 68.4% |
| 1 | 88 | 24.0% | 326 | 21.9% |
| 2 | 29 | 7.9% | 111 | 7.5% |
| 3+ | 8 | 2.2% | 33 | 2.1% |
| Blood tests, median (IQR) | ||||
| Albumin | 36.0 | (31.0, 40.0) | 39.0 | (34.0, 42.0) |
| ALP | 139.0 | (89.0, 201.0) | 138.0 | (83.0, 213.0) |
| ALT | 19.0 | (14.0, 30.0) | 21.0 | (15.0, 31.0) |
| Basophils | 0.03 | (0.02, 0.04) | 0.04 | (0.03, 0.06) |
| Calcium | 2.41 | (2.31, 2.54) | 2.33 | (2.27, 2.40) |
| Creatinine | 89.50 | (72.0, 143.2) | 76.00 | (63.0, 95.25) |
| CRP | 5.00 | (5.0, 19.62) | 15.10 | (5.00, 54.50) |
| Eosinophils | 0.12 | (0.06, 0.19) | 0.13 | (0.06, 0.23) |
| Haemoglobin | 112.0 | (97.0, 127.2) | 132.0 | (115.0, 144.0) |
| Lymphocytes | 1.49 | (1.07, 2.03) | 1.46 | (1.03, 1.99) |
| MCV | 96 | (92, 101) | 94 | (90, 98) |
| Monocytes | 0.39 | (0.29, 0.50) | 0.45 | (0.34, 0.57) |
| Neutrophils | 3.64 | (2.75, 5.28) | 4.95 | (3.69, 6.94) |
| Platelets | 223.0 | (167.0, 288.5) | 248.0 | (199.0, 309.0) |
| WCC | 6.14 | (4.73, 7.94) | 7.58 | (5.95, 9.73) |
| Model | C-Statistic | C-Slope |
|---|---|---|
| Full model (all 15 blood tests included) | 0.85 (0.83, 0.87) | 0.87 (0.75, 0.90) |
| Calcium removed as predictor | 0.83 (0.81, 0.85) | 0.86 (0.73, 1.01) |
| Over 60s only included | 0.86 (0.84, 0.89) | 0.86 (0.73, 1.01) |
| MGUS cohort included as controls | 0.83 (0.81, 0.86) | 0.88 (0.76, 1.00) |
| All ages, MGUS as controls, calcium removed | 0.81 (0.79, 0.84) | 0.88 (0.75, 0.99) |
| Over 60s only, MGUS as controls, all predictors | 0.85 (0.82, 0.87) | 0.87 (0.75, 1.00) |
| Over 60s only, MGUS as controls, calcium removed | 0.83 (0.80, 0.85) | 0.87 (0.73, 1.00) |
| Prevalence 15 per 100,000 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | Threshold | Sensitivity (95% CI) | Specificity (95% CI) | PPV | NPV | Total to Reflex Test | Cancers Diagnosed | Cancers Missed | Number Needed to Reflex Test to Detect 1 Case |
| Full model | 0.1 | 90.2 (86.9, 92.9) | 60.2 (57.6, 62.8) | 0.0003 | 1.0000 | 39,808 | 14 | 1 | 2942 |
| 0.2 | 77.1 (72.8, 81.2) | 82.3 (80.3, 84.3) | 0.0007 | 1.0000 | 17,709 | 12 | 3 | 1531 | |
| 0.4 | 58.3 (53.1, 63.5) | 94.8 (93.6, 95.9) | 0.0017 | 0.9999 | 5208 | 9 | 6 | 596 | |
| Calcium removed | 0.1 | 88.8 (85.8, 92.1) | 55.6 (53.0, 58.0) | 0.0003 | 1.0000 | 44,407 | 13 | 2 | 3334 |
| 0.2 | 77.4 (73.3, 81.5) | 78.5 (76.3, 80.5) | 0.0006 | 1.0000 | 21,508 | 12 | 3 | 1853 | |
| 0.4 | 55.0 (50.1, 60.2) | 94.2 (92.8, 95.2) | 0.0014 | 0.9999 | 5807 | 8 | 7 | 704 | |
| Over 60s only | 0.1 | 91.4 (88.0, 94.4) | 61.6 (58.9, 64.2) | 0.0004 | 1.0000 | 38,408 | 14 | 1 | 2801 |
| 0.2 | 81.7 (77.1, 86.1) | 80.6 (78.5, 82.9) | 0.0006 | 1.0000 | 19,409 | 12 | 3 | 1584 | |
| 0.4 | 65.1 (59.5, 70.4) | 94.3 (92.9, 95.6) | 0.0017 | 0.9999 | 5709 | 10 | 5 | 585 | |
| MGUS as controls | 0.1 | 84.2 (80.1, 87.7) | 63.9 (61.7, 66.0) | 0.0003 | 1.0000 | 36,107 | 13 | 2 | 2859 |
| 0.2 | 73.6 (69.2, 77.9) | 86.0 (84.3, 87.5) | 0.0008 | 1.0000 | 14,009 | 11 | 4 | 1269 | |
| 0.4 | 46.6 (41.7, 51.8) | 96.0 (95.1, 96.9) | 0.0017 | 0.9999 | 4006 | 7 | 8 | 573 | |
| MGUS as controls and calcium removed | 0.1 | 85.6 (82.0, 89.1) | 59.4 (57.1, 61.6) | 0.0003 | 1.0000 | 40,607 | 13 | 2 | 3163 |
| 0.2 | 71.7 (67.0, 76.3) | 82.8 (81.1, 84.5) | 0.0006 | 0.9999 | 17,208 | 11 | 4 | 1600 | |
| 0.4 | 44.1 (39.5, 49.1) | 95.7 (94.7, 96.6) | 0.0015 | 0.9999 | 4306 | 7 | 8 | 651 | |
| Over 60s only, MGUS as controls, all predictors | 0.1 | 87.0 (83.1, 90.7) | 64.0 (61.5, 66.4) | 0.0004 | 1.0000 | 36,008 | 13 | 2 | 2759 |
| 0.2 | 75.4 (70.4, 80.0) | 84.6 (82.8, 86.3) | 0.0007 | 1.0000 | 15,409 | 11 | 4 | 1362 | |
| 0.4 | 48.8 (43.2, 54.5) | 94.9 (93.7, 96.0) | 0.0014 | 0.9999 | 5107 | 7 | 8 | 698 | |
| Over 60s only, MGUS as controls, calcium removed | 0.1 | 87.0 (83.4, 90.7) | 60.7 (58.1, 63.1) | 0.0003 | 1.0000 | 39,307 | 13 | 2 | 3012 |
| 0.2 | 74.8 (70.1, 79.7) | 81.5 (79.4, 83.4) | 0.0006 | 1.0000 | 18,508 | 11 | 4 | 1650 | |
| 0.4 | 48.5 (43.2, 54.2) | 94.6 (93.5, 95.7) | 0.0013 | 0.9999 | 5406 | 7 | 8 | 743 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, L.; Carmichael, J.; Cook, G.; Shinkins, B.; Neal, R.D. Development and Internal Validation of a Risk Prediction Model to Identify Myeloma Based on Routine Blood Tests: A Case-Control Study. Cancers 2023, 15, 975. https://doi.org/10.3390/cancers15030975
Smith L, Carmichael J, Cook G, Shinkins B, Neal RD. Development and Internal Validation of a Risk Prediction Model to Identify Myeloma Based on Routine Blood Tests: A Case-Control Study. Cancers. 2023; 15(3):975. https://doi.org/10.3390/cancers15030975
Chicago/Turabian StyleSmith, Lesley, Jonathan Carmichael, Gordon Cook, Bethany Shinkins, and Richard D. Neal. 2023. "Development and Internal Validation of a Risk Prediction Model to Identify Myeloma Based on Routine Blood Tests: A Case-Control Study" Cancers 15, no. 3: 975. https://doi.org/10.3390/cancers15030975
APA StyleSmith, L., Carmichael, J., Cook, G., Shinkins, B., & Neal, R. D. (2023). Development and Internal Validation of a Risk Prediction Model to Identify Myeloma Based on Routine Blood Tests: A Case-Control Study. Cancers, 15(3), 975. https://doi.org/10.3390/cancers15030975





