Exosomal LncRNAs in Gastrointestinal Cancer: Biological Functions and Emerging Clinical Applications
Abstract
Simple Summary
Abstract
1. Introduction
2. Exosomes and lncRNAs
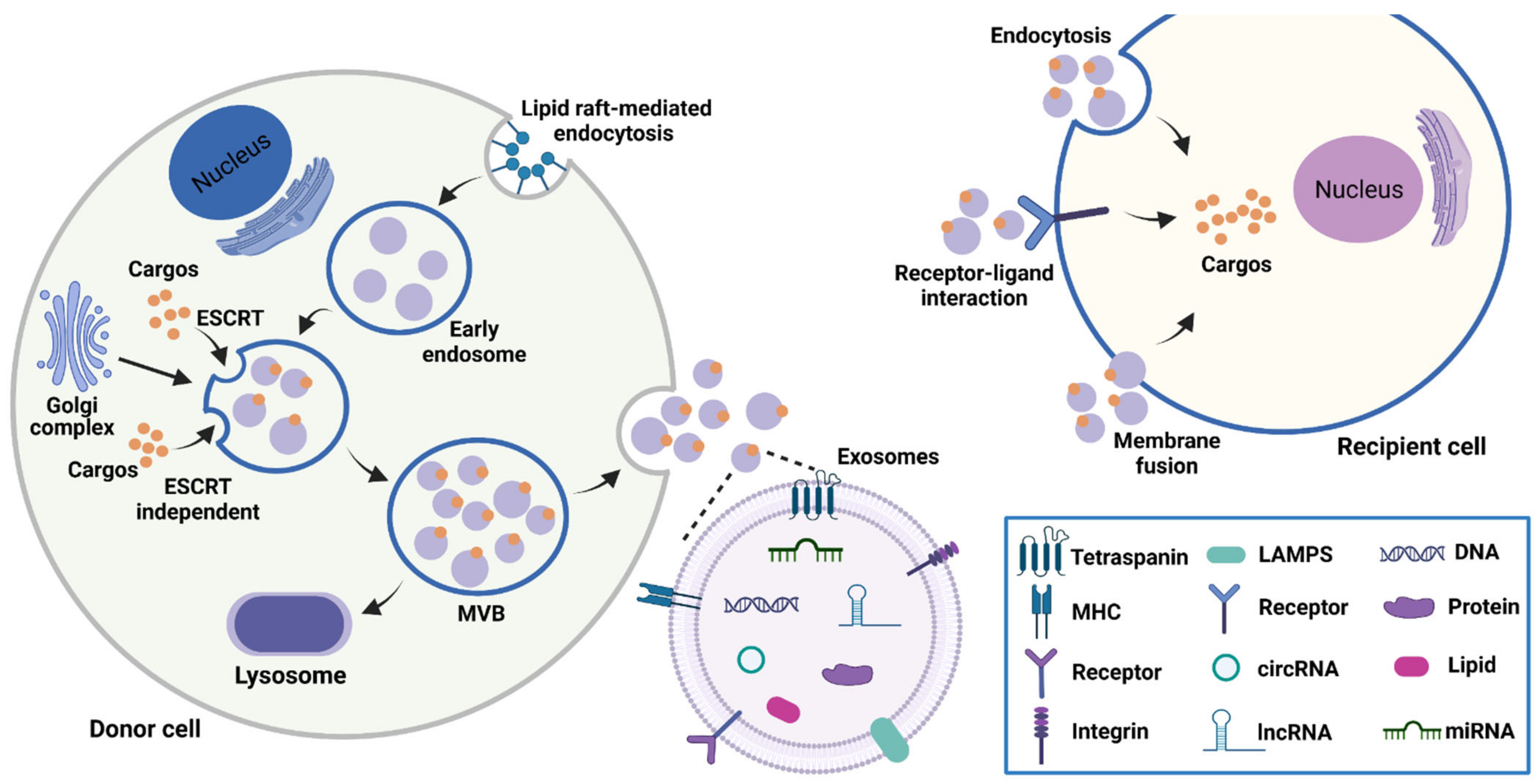
2.1. Biogenesis, Characteristics, and Functions of Exosomes
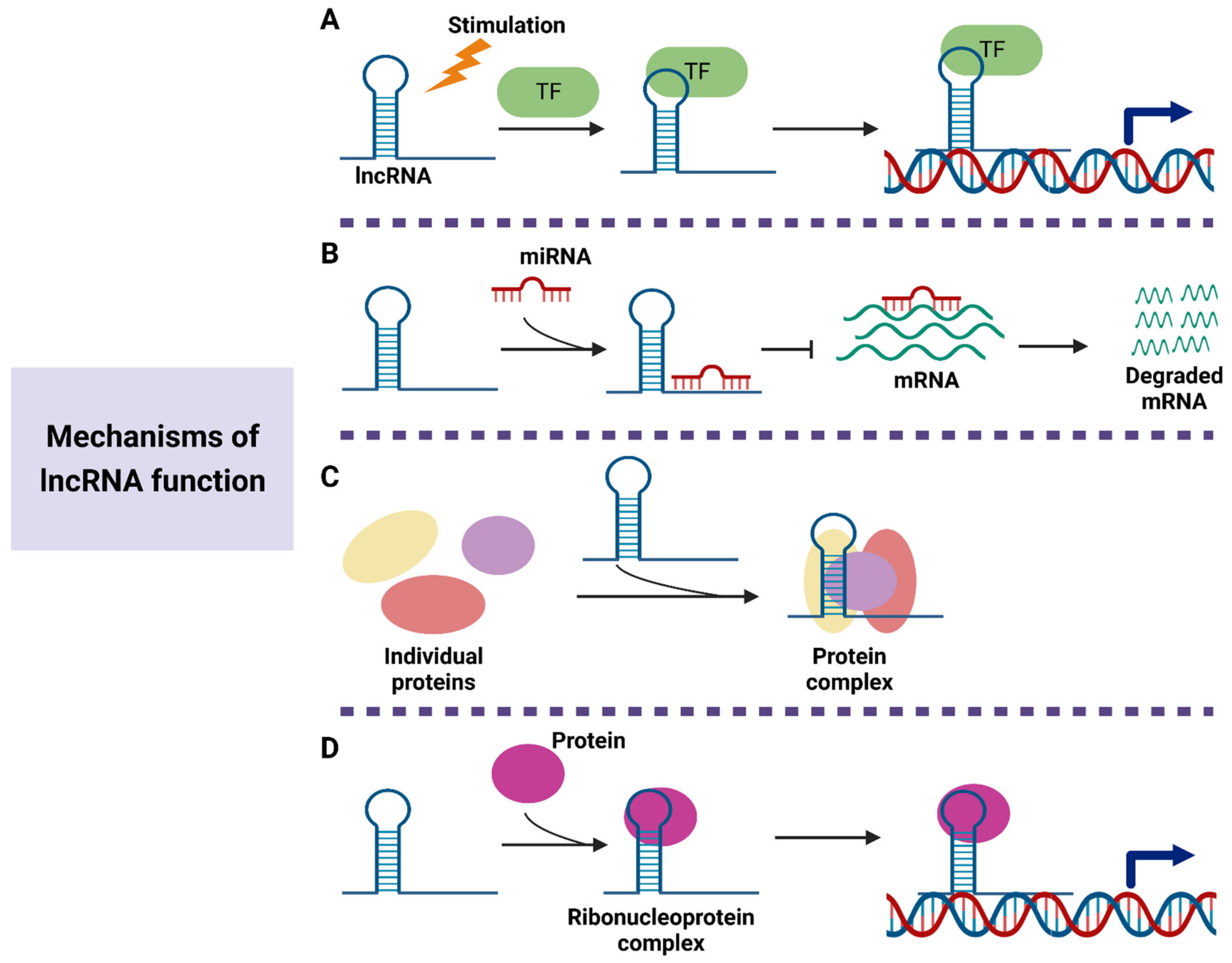
2.2. Properties and Functions of lncRNAs
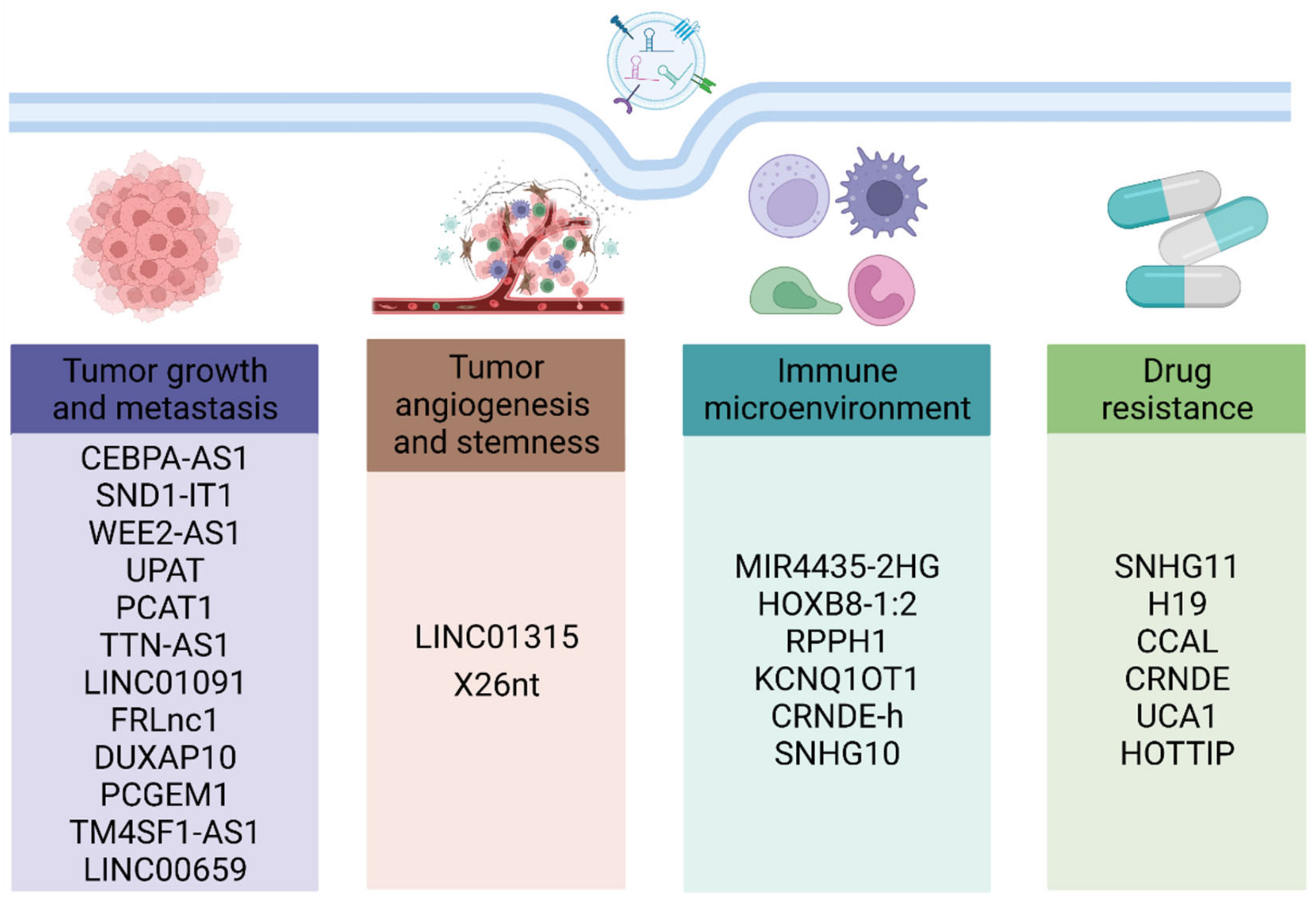
3. Biological Functions of Exosomal lncRNAs in Gastrointestinal Cancer
3.1. Roles of Exosomal lncRNAs in Tumor Proliferation, Metastasis, Angiogenesis, and Stemness
3.2. Roles of Exosomal lncRNAs in Tumor Immune Microenvironment
3.3. Roles of Exosomal lncRNAs in Drug Resistance
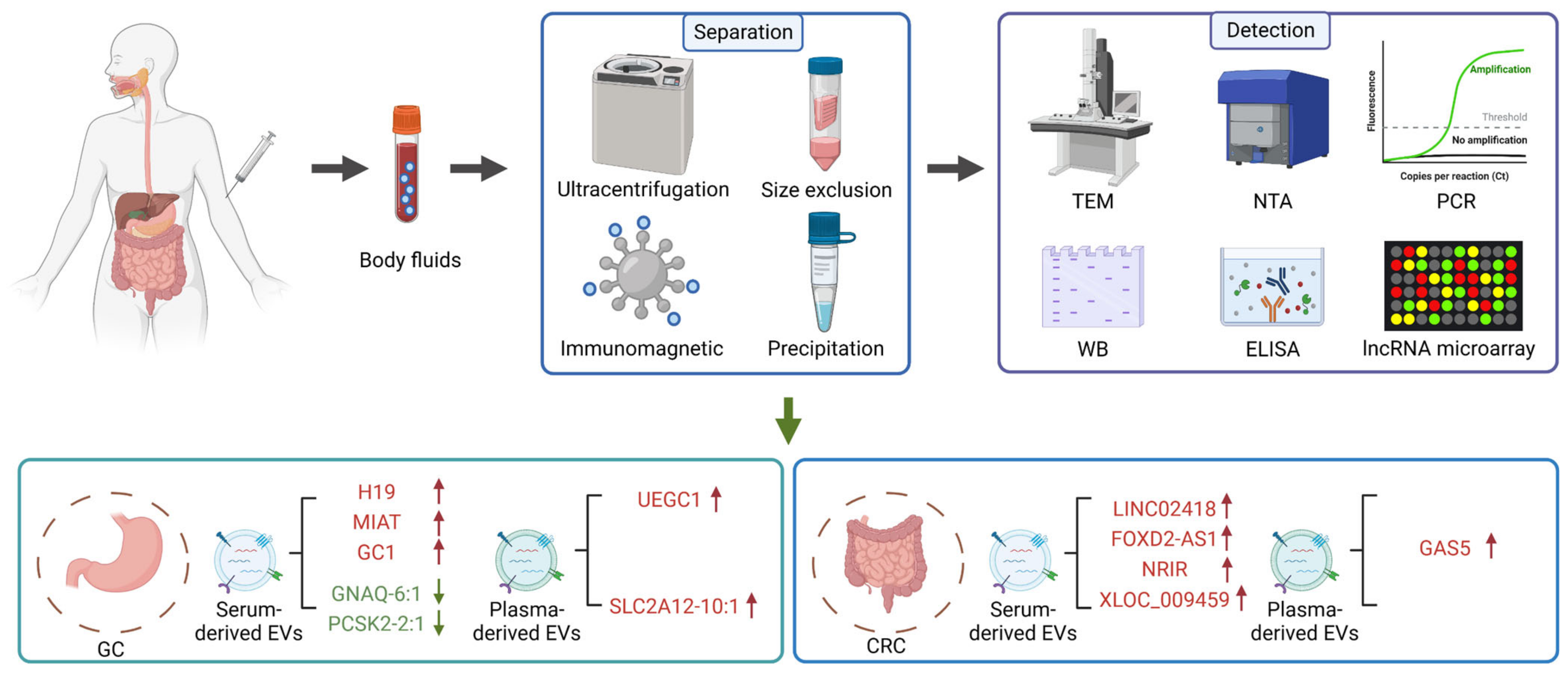
4. Exosomal lncRNAs as Potential Biomarkers of Gastrointestinal Cancer
5. Exosomal lncRNAs as Therapeutic Targets of Gastrointestinal Cancer
6. Challenges and Perspectives
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Arnold, M.; Abnet, C.C.; Neale, R.E.; Vignat, J.; Giovannucci, E.L.; McGlynn, K.A.; Bray, F. Global burden of 5 major types of gastrointestinal cancer. Gastroenterology 2020, 159, 335–349.e315. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, S.; Pal, S.; Chandan, G.; Saini, V.; Chakrabarti, S.; Saini, N.K.; Mittal, A.; Thakur, V.K.; Saini, A.K.; Saini, R.V. Understanding the cross-talk between human microbiota and gastrointestinal cancer for developing potential diagnostic and prognostic biomarkers. Semin. Cancer Biol. 2022, 86, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, D.; Zhang, C.; Liu, H.; Hao, M.; Kan, S.; Liu, D.; Liu, W. The applications of gold nanoparticles in the diagnosis and treatment of gastrointestinal cancer. Front. Oncol. 2021, 11, 5855. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Khan, A.Q.; Inchakalody, V.P.; Mestiri, S.; Yoosuf, Z.; Bedhiafi, T.; El-Ella, D.M.A.; Taib, N.; Hydrose, S.; Akbar, S.; et al. Dynamic liquid biopsy components as predictive and prognostic biomarkers in colorectal cancer. J. Exp. Clin. Cancer Res. 2022, 41, 99. [Google Scholar] [CrossRef] [PubMed]
- Yano, T.; Wang, K.K. Photodynamic therapy for gastrointestinal cancer. Photochem. Photobiol. 2020, 96, 517–523. [Google Scholar] [CrossRef]
- Yang, Q.; Xu, J.; Gu, J.; Shi, H.; Zhang, J.; Zhang, J.; Chen, Z.S.; Fang, X.; Zhu, T.; Zhang, X. Extracellular vesicles in cancer drug resistance: Roles, mechanisms, and implications. Adv. Sci. 2022, 9, 2201609. [Google Scholar] [CrossRef]
- Liu, X.; Li, Y.; Jiang, X.; Deng, Y.; Ma, C.; Yu, Q.; Gao, D. Long non-coding RNA: Multiple effects on the differentiation, maturity and cell function of dendritic cells. Clin. Immunol. 2022, 245, 109167. [Google Scholar] [CrossRef]
- Ruffo, P.; De Amicis, F.; Giardina, E.; Conforti, F.L. Long-noncoding RNAs as epigenetic regulators in neurodegenerative diseases. Neural Regen. Res. 2023, 18, 1243–1248. [Google Scholar] [CrossRef]
- Liu, J.; Ji, Q.; Cheng, F.; Chen, D.; Geng, T.; Huang, Y.; Zhang, J.; He, Y.; Song, T. The lncRNAs involved in regulating the RIG-I signaling pathway. Front. Cell. Infect. Microbiol. 2022, 12, 1664. [Google Scholar] [CrossRef]
- Wei, L.; Sun, J.; Zhang, N.; Zheng, Y.; Wang, X.; Lv, L.; Liu, J.; Xu, Y.; Shen, Y.; Yang, M. Noncoding RNAs in gastric cancer: Implications for drug resistance. Mol. Cancer 2020, 19, 62. [Google Scholar] [CrossRef]
- Chen, S.; Shen, X. Long noncoding RNAs: Functions and mechanisms in colon cancer. Mol. Cancer 2020, 19, 167. [Google Scholar] [CrossRef]
- Raziq, K.; Cai, M.; Dong, K.; Wang, P.; Afrifa, J.; Fu, S. Competitive endogenous network of lncRNA, miRNA, and mRNA in the chemoresistance of gastrointestinal tract adenocarcinomas. Biomed. Pharmacother. 2020, 130, 110570. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, Y.; Ma, L.; Chen, Y.; Liu, J.; Guo, Y.; Yu, T.; Zhang, L.; Zhu, L.; Shu, Y. Role of exosomal non-coding RNAs from tumor cells and tumor-associated macrophages in the tumor microenvironment. Mol. Ther. 2022, 30, 3133–3154. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Xiong, H.; Huang, Z.; Yang, Z.; Lin, Q.; Yang, B.; Fang, X.; Liu, B.; Chen, H.; Kong, J. Recent progress in detection and profiling of cancer cell-derived exosomes. Small 2021, 17, 2007971. [Google Scholar] [CrossRef]
- Tenchov, R.; Sasso, J.M.; Wang, X.; Liaw, W.S.; Chen, C.A.; Zhou, Q.A. Exosomes-nature’s lipid nanoparticles, a rising star in drug delivery and diagnostics. ACS Nano 2022, 16, 17802–17846. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Sun, F.; Sun, Y.; Wu, F.; Xu, W.; Qian, H. Mesenchymal stem cell-derived extracellular vesicles: A potential therapy for diabetes mellitus and diabetic complications. Pharmaceutics 2022, 14, 2208. [Google Scholar] [CrossRef]
- Liu, J.; Ren, L.; Li, S.; Li, W.; Zheng, X.; Yang, Y.; Fu, W.; Yi, J.; Wang, J.; Du, G. The biology, function, and applications of exosomes in cancer. Acta Pharm. Sin. B 2021, 11, 2783–2797. [Google Scholar] [CrossRef]
- Wei, H.; Chen, Q.; Lin, L.; Sha, C.; Li, T.; Liu, Y.; Yin, X.; Xu, Y.; Chen, L.; Gao, W.; et al. Regulation of exosome production and cargo sorting. Int. J. Biol. Sci. 2021, 17, 163–177. [Google Scholar] [CrossRef]
- Zhao, J.; An, Q.; Zhu, X.; Yang, B.; Gao, X.; Niu, Y.; Zhang, L.; Xu, K.; Ma, D. Research status and future prospects of extracellular vesicles in primary Sjögren’s syndrome. Stem Cell Res. Ther. 2022, 13, 230. [Google Scholar] [CrossRef] [PubMed]
- Noonin, C.; Thongboonkerd, V. Exosome-inflammasome crosstalk and their roles in inflammatory responses. Theranostics 2021, 11, 4436–4451. [Google Scholar] [CrossRef] [PubMed]
- Alptekin, A.; Parvin, M.; Chowdhury, H.I.; Rashid, M.H.; Arbab, A.S. Engineered exosomes for studies in tumor immunology. Immunol. Rev. 2022, 312, 76–102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, H.; Gu, J.; Zhang, J.; Shi, H.; Qian, H.; Wang, D.; Xu, W.; Pan, J.; Santos, H.A. Engineered extracellular vesicles for cancer therapy. Adv. Mater. 2021, 33, 2005709. [Google Scholar] [CrossRef] [PubMed]
- Tallon, C.; Hollinger, K.R.; Pal, A.; Bell, B.J.; Rais, R.; Tsukamoto, T.; Witwer, K.W.; Haughey, N.J.; Slusher, B.S. Nipping disease in the bud: nSMase2 inhibitors as therapeutics in extracellular vesicle-mediated diseases. Drug Discov. Today 2021, 26, 1656–1668. [Google Scholar] [CrossRef]
- Wu, P.; Zhang, B.; Ocansey, D.K.W.; Xu, W.; Qian, H. Extracellular vesicles: A bright star of nanomedicine. Biomaterials 2021, 269, 120467. [Google Scholar] [CrossRef]
- Wei, F.; Li, Y. The emerging roles of exosome-derived noncoding RNAs in the tumor immune microenvironment and their future applications. Biomed. Pharmacother. 2022, 156, 113863. [Google Scholar] [CrossRef]
- Isaac, R.; Reis, F.C.G.; Ying, W.; Olefsky, J.M. Exosomes as mediators of intercellular crosstalk in metabolism. Cell Metab. 2021, 33, 1744–1762. [Google Scholar] [CrossRef]
- Sun, Y.; Shi, H.; Yin, S.; Ji, C.; Zhang, X.; Zhang, B.; Wu, P.; Shi, Y.; Mao, F.; Yan, Y.; et al. Human mesenchymal stem cell derived exosomes alleviate type 2 diabetes mellitus by reversing peripheral insulin resistance and relieving β-cell destruction. ACS Nano 2018, 12, 7613–7628. [Google Scholar] [CrossRef]
- Ji, C.; Zhang, J.; Zhu, Y.; Shi, H.; Yin, S.; Sun, F.; Wang, Q.; Zhang, L.; Yan, Y.; Zhang, X.; et al. Exosomes derived from hucMSC attenuate renal fibrosis through CK1δ/β-TRCP-mediated YAP degradation. Cell Death Dis. 2020, 11, 327. [Google Scholar] [CrossRef]
- Sung, S.E.; Seo, M.S.; Kim, Y.I.; Kang, K.K.; Choi, J.H.; Lee, S.; Sung, M.; Yim, S.G.; Lim, J.H.; Seok, H.G.; et al. Human epidural AD-MSC exosomes improve function recovery after spinal cord injury in rats. Biomedicines 2022, 10, 678. [Google Scholar] [CrossRef]
- Luo, C.; Xin, H.; Zhou, Z.; Hu, Z.; Sun, R.; Yao, N.; Sun, Q.; Borjigin, U.; Wu, X.; Fan, J.; et al. Tumor-derived exosomes induce immunosuppressive macrophages to foster intrahepatic cholangiocarcinoma progression. Hepatology 2022, 76, 982–999. [Google Scholar] [CrossRef]
- Herman, A.B.; Tsitsipatis, D.; Gorospe, M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol. Cell 2022, 82, 2252–2266. [Google Scholar] [CrossRef]
- Liu, Y.; Ding, W.; Yu, W.; Zhang, Y.; Ao, X.; Wang, J. Long non-coding RNAs: Biogenesis, functions, and clinical significance in gastric cancer. Mol. Ther.-Oncolytics 2021, 23, 458–476. [Google Scholar] [CrossRef]
- Reggiardo, R.E.; Maroli, S.V.; Kim, D.H. LncRNA biomarkers of inflammation and cancer. Adv. Exp. Med. Biol. 2022, 1363, 121–145. [Google Scholar] [CrossRef]
- Marcia, M. The multiple molecular dimensions of long noncoding RNAs that regulate gene expression and tumorigenesis. Curr. Opin. Oncol. 2022, 34, 141–147. [Google Scholar] [CrossRef]
- Bermúdez, M.; Aguilar-Medina, M.; Lizárraga-Verdugo, E.; Avendaño-Félix, M.; Silva-Benítez, E.; López-Camarillo, C.; Ramos-Payán, R. LncRNAs as regulators of autophagy and drug resistance in colorectal cancer. Front. Oncol. 2019, 9, 1008. [Google Scholar] [CrossRef]
- Huang, W.; Li, H.; Yu, Q.; Xiao, W.; Wang, D.O. LncRNA-mediated DNA methylation: An emerging mechanism in cancer and beyond. J. Exp. Clin. Cancer Res. 2022, 41, 100. [Google Scholar] [CrossRef]
- Alkhathami, A.G.; Hadi, A.; Alfaifi, M.; Alshahrani, M.Y.; Verma, A.K.; Beg, M.M.A. Serum-based lncRNA ANRIL, TUG1, UCA1, and HIT expressions in breast cancer patients. Dis. Markers 2022, 2022, 9997212. [Google Scholar] [CrossRef]
- Adnane, S.; Marino, A.; Leucci, E. LncRNAs in human cancers: Signal from noise. Trends Cell Biol. 2022, 32, 565–573. [Google Scholar] [CrossRef]
- Shuman, S. Transcriptional interference at tandem lncRNA and protein-coding genes: An emerging theme in regulation of cellular nutrient homeostasis. Nucleic Acids Res. 2020, 48, 8243–8254. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Chen, J.; Yang, L.; Liu, J.; Zhang, X.; Zhang, Y.; Tu, Q.; Yin, D.; Lin, D.; Wong, P.P.; et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat. Cell Biol. 2019, 21, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, J.; Wasson, M.D.; Brown, J.M.; Fernando, W.; Marcato, P. LncRNA-miRNA axes in breast cancer: Novel points of interaction for strategic attack. Cancer Lett. 2021, 509, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Luo, M.; Cui, X.; O’Connell, D.; Yang, Y. Long noncoding RNA NEAT1 promotes ferroptosis by modulating the miR-362-3p/MIOX axis as a ceRNA. Cell Death Differ. 2022, 29, 1850–1863. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Zhuang, S.; Chen, X.; Du, J.; Zhong, L.; Ding, J.; Wang, L.; Yi, J.; Hu, G.; Tang, G.; et al. lncRNA ITGB8-AS1 functions as a ceRNA to promote colorectal cancer growth and migration through integrin-mediated focal adhesion signaling. Mol. Ther. 2022, 30, 688–702. [Google Scholar] [CrossRef]
- Han, Y.; Zhao, G.; Shi, X.; Wang, Y.; Wen, X.; Zhang, L.; Guo, X. The emerging role of long non-coding RNAs in esophageal cancer: Functions in tumorigenesis and clinical implications. Front. Pharmacol. 2022, 13, 885075. [Google Scholar] [CrossRef]
- He, D.; Xin, T.; Pang, B.; Sun, J.; Liu, Z.H.; Qin, Z.; Ji, X.S.; Yang, F.; Wei, Y.B.; Wang, Z.X.; et al. A novel lncRNA MDHDH suppresses glioblastoma multiforme by acting as a scaffold for MDH2 and PSMA1 to regulate NAD+ metabolism and autophagy. J. Exp. Clin. Cancer Res. 2022, 41, 349. [Google Scholar] [CrossRef]
- Shi, L.; Yang, Y.; Li, M.; Li, C.; Zhou, Z.; Tang, G.; Wu, L.; Yao, Y.; Shen, X.; Hou, Z.; et al. LncRNA IFITM4P promotes immune escape by up-regulating PD-L1 via dual mechanism in oral carcinogenesis. Mol. Ther. 2022, 30, 1564–1577. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, S.; He, Y.; Li, X.; Zhu, Y.; Lin, X.; Chen, L.; Zhao, Y.; Niu, L.; Zhang, S.; et al. LncRNA-mediated adipogenesis in different adipocytes. Int. J. Mol. Sci. 2022, 23, 7488. [Google Scholar] [CrossRef]
- Xiu, B.; Chi, Y.; Liu, L.; Chi, W.; Zhang, Q.; Chen, J.; Guo, R.; Si, J.; Li, L.; Xue, J.; et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol. Cancer 2019, 18, 187. [Google Scholar] [CrossRef]
- Lakshmi, S.; Hughes, T.A.; Priya, S. Exosomes and exosomal RNAs in breast cancer: A status update. Eur. J. Cancer 2021, 144, 252–268. [Google Scholar] [CrossRef]
- Wu, J.; Huang, H.; Huang, W.; Wang, L.; Xia, X.; Fang, X. Analysis of exosomal lncRNA, miRNA and mRNA expression profiles and ceRNA network construction in endometriosis. Epigenomics 2020, 12, 1193–1213. [Google Scholar] [CrossRef]
- Chang, W.; Wang, J. Exosomes and their noncoding RNA cargo are emerging as new modulators for diabetes mellitus. Cells 2019, 8, 853. [Google Scholar] [CrossRef]
- Fan, Q.; Yang, L.; Zhang, X.; Peng, X.; Wei, S.; Su, D.; Zhai, Z.; Hua, X.; Li, H. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 2018, 414, 107–115. [Google Scholar] [CrossRef]
- Li, L.; Bi, Y.; Diao, S.; Li, X.; Yuan, T.; Xu, T.; Huang, C.; Li, J. Exosomal lncRNAs and hepatocellular carcinoma: From basic research to clinical practice. Biochem. Pharmacol. 2022, 200, 115032. [Google Scholar] [CrossRef]
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim. Biophys. Acta 2014, 1841, 108–120. [Google Scholar] [CrossRef]
- Liu, Q.W.; He, Y.; Xu, W.W. Molecular functions and therapeutic applications of exosomal noncoding RNAs in cancer. Exp. Mol. Med. 2022, 54, 216–225. [Google Scholar] [CrossRef]
- Dang, X.; Zeng, X. Targeted therapeutic delivery using engineered exosomes and its applications in cardiovascular diseases. Gene 2016, 575 Pt 2, 377–384. [Google Scholar] [CrossRef]
- Kok, V.C.; Yu, C.C. Cancer-derived exosomes: Their role in cancer biology and biomarker development. Int. J. Nanomed. 2020, 15, 8019–8036. [Google Scholar] [CrossRef]
- Honarmand Tamizkar, K.; Gorji, P.; Gholipour, M.; Hussen, B.M.; Mazdeh, M.; Eslami, S.; Taheri, M.; Ghafouri-Fard, S. Parkinson’s disease is associated with dysregulation of circulatory levels of lncRNAs. Front. Immunol. 2021, 12, 4706. [Google Scholar] [CrossRef]
- Thakur, A.; Parra, D.C.; Motallebnejad, P.; Brocchi, M.; Chen, H.J. Exosomes: Small vesicles with big roles in cancer, vaccine development, and therapeutics. Bioact. Mater. 2022, 10, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Piao, H.Y.; Guo, S.; Wang, Y.; Zhang, J. Exosomal long non-coding RNA CEBPA-AS1 inhibits tumor apoptosis and functions as a non-invasive biomarker for diagnosis of gastric cancer. OncoTargets Ther. 2020, 13, 1365–1374. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Zhang, J.; Cao, T.; Chen, B.; Tian, Y.; Shi, Y. Exosome-mediated lncRNA SND1-IT1 from gastric cancer cells enhances malignant transformation of gastric mucosa cells via up-regulating SNAIL1. J. Transl. Med. 2022, 20, 284. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Zhang, D.; Wang, T.; Ji, J.; Jin, C.; Peng, C.; Tan, Y.; Zhou, J.; Wang, L.; Feng, Y.; et al. CAF-derived exosomal WEE2-AS1 facilitates colorectal cancer progression via promoting degradation of MOB1A to inhibit the Hippo pathway. Cell Death Dis. 2022, 13, 796. [Google Scholar] [CrossRef] [PubMed]
- Taniue, K.; Kurimoto, A.; Sugimasa, H.; Nasu, E.; Takeda, Y.; Iwasaki, K.; Nagashima, T.; Okada-Hatakeyama, M.; Oyama, M.; Kozuka-Hata, H.; et al. Long noncoding RNA UPAT promotes colon tumorigenesis by inhibiting degradation of UHRF1. Proc. Natl. Acad. Sci. USA 2016, 113, 1273–1278. [Google Scholar] [CrossRef]
- Fang, X.; Xu, Y.; Li, K.; Liu, P.; Zhang, H.; Jiang, Y.; Tang, J.; Li, Y. Exosomal lncRNA PCAT1 promotes tumor circulating cell-mediated colorectal cancer liver metastasis by regulating the activity of the miR-329-3p/Netrin-1-CD146 complex. J. Immunol. Res. 2022, 2022, 9916228. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, R.; Zhao, H.; Li, F.; Li, Y.; Zhu, M. TTN-AS1 delivered by gastric cancer cell-derived exosome induces gastric cancer progression through in vivo and in vitro studies. Cell Biol. Toxicol. 2022, 38, 1–15. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, C.; Cao, S.; Zhao, H.; Jiang, R.; Li, Y. Tumor-derived exosomes orchestrate the microRNA-128-3p/ELF4/CDX2 axis to facilitate the growth and metastasis of gastric cancer via delivery of LINC01091. Cell Biol. Toxicol. 2022, 38, 1–18. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, L.; Ye, X.; Wu, Z.; Zhang, Z.; Sun, B.; Fu, H.; Fu, C.; Liang, X.; Jiang, H. Expression and mechanism of exosome-mediated A FOXM1 related long noncoding RNA in gastric cancer. J. Nanobiotechnology 2021, 19, 133. [Google Scholar] [CrossRef]
- Lian, Y.; Xu, Y.; Xiao, C.; Xia, R.; Gong, H.; Yang, P.; Chen, T.; Wu, D.; Cai, Z.; Zhang, J.; et al. The pseudogene derived from long non-coding RNA DUXAP10 promotes colorectal cancer cell growth through epigenetically silencing of p21 and PTEN. Sci. Rep. 2017, 7, 7312. [Google Scholar] [CrossRef]
- Piao, H.Y.; Guo, S.; Wang, Y.; Zhang, J. Exosome-transmitted lncRNA PCGEM1 promotes invasive and metastasis in gastric cancer by maintaining the stability of SNAI1. Clin. Transl. Oncol. 2021, 23, 246–256. [Google Scholar] [CrossRef]
- He, C.; Qi, W.; Wang, Z. Effect and mechanism of downregulating the long-chain noncoding RNA TM4SF1-AS1 on the proliferation, apoptosis and invasion of gastric cancer cells. World J. Surg. Oncol. 2021, 19, 226. [Google Scholar] [CrossRef]
- Zhou, L.; Li, J.; Tang, Y.; Yang, M. Exosomal LncRNA LINC00659 transferred from cancer-associated fibroblasts promotes colorectal cancer cell progression via miR-342-3p/ANXA2 axis. J. Transl. Med. 2021, 19, 8. [Google Scholar] [CrossRef]
- Li, Y.; Wu, M.; Xu, S.; Huang, H.; Yan, L.; Gu, Y. Colorectal cancer stem cell-derived exosomal long intergenic noncoding RNA 01315 (LINC01315) promotes proliferation, migration, and stemness of colorectal cancer cells. Bioengineered 2022, 13, 10827–10842. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, S.; Du, K.; Zheng, N.; Liu, Y.; Chen, H.; Xie, G.; Ma, Y.; Zhou, Y.; Zheng, Y.; et al. Gastric cancer-secreted exosomal X26nt increases angiogenesis and vascular permeability by targeting VE-cadherin. Cancer Sci. 2021, 112, 1839–1852. [Google Scholar] [CrossRef]
- Li, C.; Chen, Z.; Gao, J.; Tang, T.; Zhou, L.; Zhang, G.; Zhang, D.; Shen, C.; Guo, L.; Fu, T. MIR4435-2HG in exosomes promotes gastric carcinogenesis by inducing M2 polarization in macrophages. Front. Oncol. 2022, 12, 1017745. [Google Scholar] [CrossRef]
- Li, X.; Lan, Q.; Lai, W.; Wu, H.; Xu, H.; Fang, K.; Chu, Z.; Zeng, Y. Exosome-derived lnc-HOXB8-1:2 induces tumor-associated macrophage infiltration to promote neuroendocrine differentiated colorectal cancer progression by sponging hsa-miR-6825-5p. BMC Cancer 2022, 22, 928. [Google Scholar] [CrossRef]
- Liang, Z.X.; Liu, H.S.; Wang, F.W.; Xiong, L.; Zhou, C.; Hu, T.; He, X.W.; Wu, X.J.; Xie, D.; Wu, X.R.; et al. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 2019, 10, 829. [Google Scholar] [CrossRef]
- Xian, D.; Niu, L.; Zeng, J.; Wang, L. LncRNA KCNQ1OT1 secreted by tumor cell-derived exosomes mediates immune escape in colorectal cancer by regulating PD-L1 ubiquitination via miR-30a-5p/USP22. Front. Cell Dev. Biol. 2021, 9, 653808. [Google Scholar] [CrossRef]
- Sun, J.; Jia, H.; Bao, X.; Wu, Y.; Zhu, T.; Li, R.; Zhao, H. Tumor exosome promotes Th17 cell differentiation by transmitting the lncRNA CRNDE-h in colorectal cancer. Cell Death Dis. 2021, 12, 123. [Google Scholar] [CrossRef]
- Huang, Y.; Luo, Y.; Ou, W.; Wang, Y.; Dong, D.; Peng, X.; Luo, Y. Exosomal lncRNA SNHG10 derived from colorectal cancer cells suppresses natural killer cell cytotoxicity by upregulating INHBC. Cancer Cell Int. 2021, 21, 528. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zhang, H.; Tian, Y.; Li, Y.; Li, J.; Zhong, X.; Yuan, X. LncRNA SNHG11 enhances bevacizumab resistance in colorectal cancer by mediating miR-1207-5p/ABCC1 axis. Anti-Cancer Drugs 2022, 33, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Ding, L.; Zhang, D.; Shi, G.; Xu, Q.; Shen, S.; Wang, Y.; Wang, T.; Hou, Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics 2018, 8, 3932–3948. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Ruan, H.; Zhang, X.; Xu, X.; Zhu, Y.; Peng, H.; Zhang, X.; Kong, F.; Guan, M. Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. Int. J. Cancer 2020, 146, 1700–1716. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.; Zhou, L.Q.; Liu, C.; Zeng, F.; Yuan, Y.W.; Zhou, Q.; Li, S.H.; Wu, Y.; Wang, J.L.; Wu, D.Z.; et al. Transfer of lncRNA CRNDE in TAM-derived exosomes is linked with cisplatin resistance in gastric cancer. EMBO Rep. 2021, 22, e52124. [Google Scholar] [CrossRef]
- Yang, Y.N.; Zhang, R.; Du, J.W.; Yuan, H.H.; Li, Y.J.; Wei, X.L.; Du, X.X.; Jiang, S.L.; Han, Y. Predictive role of UCA1-containing exosomes in cetuximab-resistant colorectal cancer. Cancer Cell Int. 2018, 18, 164. [Google Scholar] [CrossRef]
- Wang, J.; Lv, B.; Su, Y.; Wang, X.; Bu, J.; Yao, L. Exosome-mediated transfer of lncRNA HOTTIP promotes cisplatin resistance in gastric cancer cells by regulating HMGA1/miR-218 axis. OncoTargets Ther. 2019, 12, 11325–11338. [Google Scholar] [CrossRef]
- Chen, X.; Liu, Y.; Zhang, Q.; Liu, B.; Cheng, Y.; Zhang, Y.; Sun, Y.; Liu, J.; Gen, H. Exosomal long non-coding RNA HOTTIP increases resistance of colorectal cancer cells to mitomycin via impairing miR-214-mediated degradation of KPNA3. Front. Cell Dev. Biol. 2020, 8, 582723. [Google Scholar] [CrossRef]
- Zhi, J.; Jia, X.J.; Yan, J.; Wang, H.C.; Feng, B.; Xing, H.Y.; Jia, Y.T. BRAF(V600E) mutant colorectal cancer cells mediate local immunosuppressive microenvironment through exosomal long noncoding RNAs. World J. Gastrointest. Oncol. 2021, 13, 2129–2148. [Google Scholar] [CrossRef]
- Poznanski, S.M.; Singh, K.; Ritchie, T.M.; Aguiar, J.A.; Fan, I.Y.; Portillo, A.L.; Rojas, E.A.; Vahedi, F.; El-Sayes, A.; Xing, S.; et al. Metabolic flexibility determines human NK cell functional fate in the tumor microenvironment. Cell Metab. 2021, 33, 1205–1220.e5. [Google Scholar] [CrossRef]
- Kobayashi, H.; Enomoto, A.; Woods, S.L.; Burt, A.D.; Takahashi, M.; Worthley, D.L. Cancer-associated fibroblasts in gastrointestinal cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 282–295. [Google Scholar] [CrossRef]
- Yu, D.; Li, Y.; Wang, M.; Gu, J.; Xu, W.; Cai, H.; Fang, X.; Zhang, X. Exosomes as a new frontier of cancer liquid biopsy. Mol. Cancer 2022, 21, 56. [Google Scholar] [CrossRef]
- Zhou, B.; Xu, K.; Zheng, X.; Chen, T.; Wang, J.; Song, Y.; Shao, Y.; Zheng, S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct. Target. Ther. 2020, 5, 144. [Google Scholar] [CrossRef]
- Dong, L.; Lin, W.; Qi, P.; Xu, M.D.; Wu, X.; Ni, S.; Huang, D.; Weng, W.W.; Tan, C.; Sheng, W.; et al. Circulating long RNAs in serum extracellular vesicles: Their characterization and potential application as biomarkers for diagnosis of colorectal cancer. Cancer Epidemiol. Biomark. Prev. 2016, 25, 1158–1166. [Google Scholar] [CrossRef]
- Zhou, H.; Shen, W.; Zou, H.; Lv, Q.; Shao, P. Circulating exosomal long non-coding RNA H19 as a potential novel diagnostic and prognostic biomarker for gastric cancer. J. Int. Med. Res. 2020, 48, 300060520934297. [Google Scholar] [CrossRef]
- Wei, S.; Dai, S.; Zhang, C.; Zhao, R.; Zhao, Z.; Song, Y.; Shan, B.; Zhao, L. LncRNA NR038975, a serum-based biomarker, promotes gastric tumorigenesis by interacting with NF90/NF45 complex. Front. Oncol. 2021, 11, 721604. [Google Scholar] [CrossRef]
- Xu, H.; Zhou, J.; Tang, J.; Min, X.; Yi, T.; Zhao, J.; Ren, Y. Identification of serum exosomal lncRNA MIAT as a novel diagnostic and prognostic biomarker for gastric cancer. J. Clin. Lab. Anal. 2020, 34, e23323. [Google Scholar] [CrossRef]
- Li, S.; Zhang, M.; Zhang, H.; Hu, K.; Cai, C.; Wang, J.; Shi, L.; Ma, P.; Xu, Y.; Zheng, P. Exosomal long noncoding RNA lnc-GNAQ-6:1 may serve as a diagnostic marker for gastric cancer. Clin. Chim. Acta 2020, 501, 252–257. [Google Scholar] [CrossRef]
- Lin, L.Y.; Yang, L.; Zeng, Q.; Wang, L.; Chen, M.L.; Zhao, Z.H.; Ye, G.D.; Luo, Q.C.; Lv, P.Y.; Guo, Q.W.; et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol. Cancer 2018, 17, 84. [Google Scholar] [CrossRef]
- Guo, X.; Lv, X.; Ru, Y.; Zhou, F.; Wang, N.; Xi, H.; Zhang, K.; Li, J.; Chang, R.; Xie, T.; et al. Circulating exosomal gastric cancer-associated long noncoding RNA1 as a biomarker for early detection and monitoring progression of gastric cancer: A multiphase study. JAMA Surg. 2020, 155, 572–579. [Google Scholar] [CrossRef]
- Pan, L.; Liang, W.; Fu, M.; Huang, Z.H.; Li, X.; Zhang, W.; Zhang, P.; Qian, H.; Jiang, P.C.; Xu, W.R.; et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J. Cancer Res. Clin. Oncol. 2017, 143, 991–1004. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Zhang, H.; Gao, H.; Sun, J.; Li, J.; Zhang, X.; Gao, L.; Ma, P.; Li, S. Plasma exosomal long noncoding RNA lnc-SLC2A12-10:1 as a novel diagnostic biomarker for gastric cancer. OncoTargets Ther. 2020, 13, 4009–4018. [Google Scholar] [CrossRef]
- Cai, C.; Zhang, H.; Zhu, Y.; Zheng, P.; Xu, Y.; Sun, J.; Zhang, M.; Lan, T.; Gu, B.; Li, S.; et al. Serum exosomal long noncoding RNA pcsk2-2:1 as a potential novel diagnostic biomarker for gastric cancer. OncoTargets Ther. 2019, 12, 10035–10041. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Meng, T.; Yang, X.H.; Sayim, P.; Lei, C.; Jin, B.; Ge, L.; Wang, H.J. Prognostic and predictive value of long non-coding RNA GAS5 and mircoRNA-221 in colorectal cancer and their effects on colorectal cancer cell proliferation, migration and invasion. Cancer Biomark. 2018, 22, 283–299. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Duan, W.; Yan, S.; Xie, Y.; Wang, C. Circulating long non-coding RNA colon cancer-associated transcript 2 protected by exosome as a potential biomarker for colorectal cancer. Biomed. Pharmacother. 2019, 113, 108758. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Du, T.; Du, L.; Li, P.; Li, J.; Duan, W.; Wang, Y.; Wang, C. Long noncoding RNA LINC02418 regulates MELK expression by acting as a ceRNA and may serve as a diagnostic marker for colorectal cancer. Cell Death Dis. 2019, 10, 568. [Google Scholar] [CrossRef]
- Yu, M.; Song, X.G.; Zhao, Y.J.; Dong, X.H.; Niu, L.M.; Zhang, Z.J.; Shang, X.L.; Tang, Y.Y.; Song, X.R.; Xie, L. Circulating serum exosomal long non-coding RNAs FOXD2-AS1, NRIR, and XLOC_009459 as diagnostic biomarkers for colorectal cancer. Front. Oncol. 2021, 11, 618967. [Google Scholar] [CrossRef]
- Oehme, F.; Krahl, S.; Gyorffy, B.; Muessle, B.; Rao, V.; Greif, H.; Ziegler, N.; Lin, K.; Thepkaysone, M.L.; Polster, H.; et al. Low level of exosomal long non-coding RNA HOTTIP is a prognostic biomarker in colorectal cancer. RNA Biol. 2019, 16, 1339–1345. [Google Scholar] [CrossRef]
- Gao, T.; Liu, X.; He, B.; Nie, Z.; Zhu, C.; Zhang, P.; Wang, S. Exosomal lncRNA 91H is associated with poor development in colorectal cancer by modifying HNRNPK expression. Cancer Cell Int. 2018, 18, 11. [Google Scholar] [CrossRef]
- Lai, S.W.; Chen, M.Y.; Bamodu, O.A.; Hsieh, M.S.; Huang, T.Y.; Yeh, C.T.; Lee, W.H.; Cherng, Y.G. Exosomal lncRNA PVT1/VEGFA axis promotes colon cancer metastasis and stemness by downregulation of tumor suppressor miR-152-3p. Oxidative Med. Cell. Longev. 2021, 2021, 9959807. [Google Scholar] [CrossRef]
- Sun, Y.; Li, B.; Cao, Q.; Liu, T.; Li, J. Targeting cancer stem cells with polymer nanoparticles for gastrointestinal cancer treatment. Stem Cell Res. Ther. 2022, 13, 489. [Google Scholar] [CrossRef]
- Li, C.; Li, W.; Zhang, Y.; Zhang, X.; Liu, T.; Zhang, Y.; Yang, Y.; Wang, L.; Pan, H.; Ji, J.; et al. Increased expression of antisense lncRNA SPINT1-AS1 predicts a poor prognosis in colorectal cancer and is negatively correlated with its sense transcript. OncoTargets Ther. 2018, 11, 3969–3978. [Google Scholar] [CrossRef]
- Hui, B.; Lu, C.; Wang, J.; Xu, Y.; Yang, Y.; Ji, H.; Li, X.; Xu, L.; Wang, J.; Tang, W.; et al. Engineered exosomes for co-delivery of PGM5-AS1 and oxaliplatin to reverse drug resistance in colon cancer. J. Cell. Physiol. 2022, 237, 911–933. [Google Scholar] [CrossRef]
- Li, N.; Li, J.; Mi, Q.; Xie, Y.; Li, P.; Wang, L.; Binang, H.; Wang, Q.; Wang, Y.; Chen, Y.; et al. Long non-coding RNA ADAMTS9-AS1 suppresses colorectal cancer by inhibiting the Wnt/β-catenin signalling pathway and is a potential diagnostic biomarker. J. Cell. Mol. Med. 2020, 24, 11318–11329. [Google Scholar] [CrossRef]
- Yin, H.; Hu, J.; Ye, Z.; Chen, S.; Chen, Y. Serum long non-coding RNA NNT-AS1 protected by exosome is a potential biomarker and functions as an oncogene via the miR-496/RAP2C axis in colorectal cancer. Mol. Med. Rep. 2021, 24, 585. [Google Scholar] [CrossRef]
- Liu, D.; Yin, H.; Wang, Y.; Cao, Y.; Yin, J.; Zhang, J.; Yin, H.; Zhao, X. Development of a highly sensitive digital PCR assay to quantify long non-coding RNA MYU in urine samples which exhibited great potential as an alternative diagnostic biomarker for prostate cancer. Transl. Androl. Urol. 2021, 10, 3815–3825. [Google Scholar] [CrossRef]
- Lin, Q.; Huang, Z.; Ye, X.; Yang, B.; Fang, X.; Liu, B.; Chen, H.; Kong, J. Lab in a tube: Isolation, extraction, and isothermal amplification detection of exosomal long noncoding RNA of gastric cancer. Talanta 2021, 225, 122090. [Google Scholar] [CrossRef]
| Cancer | lncRNA | Function | Mechanism | Reference |
|---|---|---|---|---|
| GC | CEBPA-AS1 | Promoting proliferation and inhibiting apoptosis | N/A | [62] |
| GC | SND1-IT1 | Inducing malignant transformation of GES-1 cells | MiRNA sponge for miR-1245b-5b to enhance USP3 expression | [63] |
| CRC | WEE2-AS1 | Promoting proliferation | Inducing MOB1A degradation to inhibit Hippo pathway | [64] |
| CRC | UPAT | Promoting survival and tumorigenicity of CRC cells | stabilizing UHRF1 expression | [65] |
| CRC | PCAT1 | Promoting colorectal cancer liver metastasis | MiRNA sponge for miR-329-3p to enhance Netrin-1 and CD146 expression | [66] |
| GC | TTN-AS1 | Promoting growth and metastasis | MiRNA sponge for miR-499a-5p to enhance ZEB1 and CDX2 expression | [67] |
| GC | LINC01091 | Promoting growth and metastasis | MiRNA sponge for miR-128-3p to enhance ELF4 and CDX2 expression | [68] |
| GC | FRLnc1 | Promoting growth and metastasis | N/A | [69] |
| CRC | DUXAP10 | Promoting growth and lymph node metastasis | Binding to LSD1 to inhibit the expression of p21 and PTEN | [70] |
| GC | PCGEM1 | Promoting EMT process | Maintaining the stability of SNAI1 | [71] |
| GC | TM4SF1-AS1 | Promoting the proliferation, invasion and EMT | Activating PI3K/AKT signalling pathway | [72] |
| CRC | LINC00659 | Promoting proliferation, migration, invasion and EMT progression | MiRNA sponge for miR-342-3p to enhance annexin A2 expression | [73] |
| CRC | LINC01315 | Promoting proliferation, migration, and stemness | N/A | [74] |
| GC | X26nt | Increasing angiogenesis and vascular permeability | Reducing VE-cadherin expression | [75] |
| GC | MIR4435-2HG | Inducing macrophage M2 polarization to promote tumor growth | Activating the Jagged1/Notch and JAK1/STAT3 pathways | [76] |
| CRC | HOXB8-1:2 | Inducing macrophage infiltration and M2 polarization to promote CRC progression | MiRNA sponge for miR-6825-5p to enhance CXCR3 expression | [77] |
| CRC | RPPH1 | Inducing macrophage M2 polarization to promote the metastasis and proliferation of CRC cells | Inhibiting TUBB3 ubiquitination | [78] |
| CRC | KCNQ1OT1 | Inhibiting CD8+ T-cell response to promote CRC cell immune escape | MiRNA sponge for miR-30a-5p to enhance USP22 expression | [79] |
| CRC | CRNDE-h | Activating Th17 cell differentiation to promote tumor growth | Inhibiting the Itch-mediated ubiquitination and degradation of RORγt | [80] |
| CRC | SNHG10 | Suppressing the function of NK cells to promote CRC cell immune escape | Upregulating INHBC expression | [81] |
| CRC | SNHG11 | Enhancing bevacizumab resistance in CRC cells | MiRNA sponge for miR-1207-5p to enhance ABCC1 expression | [82] |
| CRC | H19 | Promoting the stemness and chemoresistance of CRC cells | MiRNA sponge for miR-141 to activate the β-catenin pathway | [83] |
| CRC | CCAL | Promoting chemoresistance of CRC cells | Interacting with HuR to enhance β-catenin expression | [84] |
| GC | CRNDE | Inducing cisplatin resistance in GC cells | Promoting NEDD4-1-mediated PTEN ubiquitination | [85] |
| CRC | UCA1 | Promoting cetuximab resistance in CRC cells | N/A | [86] |
| GC | HOTTIP | Promoting cisplatin resistance in GC cells | MiRNA sponge for miR-218 to enhance HMGA1 expression | [87] |
| CRC | HOTTIP | Increasing resistance of CRC cells to mitomycin | MiRNA sponge for miR-214 to enhance KPNA3 expression | [88] |
| Cancer | lncRNA | Expression | Source | Case Number | AUC | Reference |
|---|---|---|---|---|---|---|
| GC | H19 | Upregulation | Serum-derived EVs | 81 | 0.849 | [95] |
| GC | MIAT | Upregulation | Serum-derived EVs | 109 | N/A | [97] |
| GC | GNAQ-6:1 | Downregulation | Serum-derived EVs | 43 | 0.732 | [98] |
| GC | UEGC1 | Upregulation | Plasma-derived EVs | 10 | 0.876 (distinguish GC patients from healthy individuals) 0.8406 (distinguish GC patients from chronic atrophic gastritis patients) | [99] |
| GC | GC1 | Upregulation | Serum-derived EVs | 826 | 0.9033 | [100] |
| GC | SLC2A12-10:1 | Upregulation | Plasma-derived EVs | 120 | 0.776 | [102] |
| GC | PCSK2-2:1 | Downregulation | Serum-derived EVs | 63 | 0.896 | [103] |
| CRC | GAS5 | Upregulation | Plasma-derived EVs | 158 | 0.964 | [104] |
| CRC | LINC02418 | Upregulation | Serum-derived EVs | 155 | 0.8978 | [106] |
| CRC | FOXD2-AS1 | Upregulation | Serum-derived EVs | 203 | 0.728 | [107] |
| CRC | NRIR | Upregulation | Serum-derived EVs | 203 | 0.660 | [107] |
| CRC | XLOC_009459 | Upregulation | Serum-derived EVs | 203 | 0.682 | [107] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Sun, F.; Jin, J.; Xu, W.; Qian, H. Exosomal LncRNAs in Gastrointestinal Cancer: Biological Functions and Emerging Clinical Applications. Cancers 2023, 15, 959. https://doi.org/10.3390/cancers15030959
Sun Y, Sun F, Jin J, Xu W, Qian H. Exosomal LncRNAs in Gastrointestinal Cancer: Biological Functions and Emerging Clinical Applications. Cancers. 2023; 15(3):959. https://doi.org/10.3390/cancers15030959
Chicago/Turabian StyleSun, Yuntong, Fengtian Sun, Jianhua Jin, Wenrong Xu, and Hui Qian. 2023. "Exosomal LncRNAs in Gastrointestinal Cancer: Biological Functions and Emerging Clinical Applications" Cancers 15, no. 3: 959. https://doi.org/10.3390/cancers15030959
APA StyleSun, Y., Sun, F., Jin, J., Xu, W., & Qian, H. (2023). Exosomal LncRNAs in Gastrointestinal Cancer: Biological Functions and Emerging Clinical Applications. Cancers, 15(3), 959. https://doi.org/10.3390/cancers15030959





