Endocrine Tumor Classification via Machine-Learning-Based Elastography: A Systematic Scoping Review
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
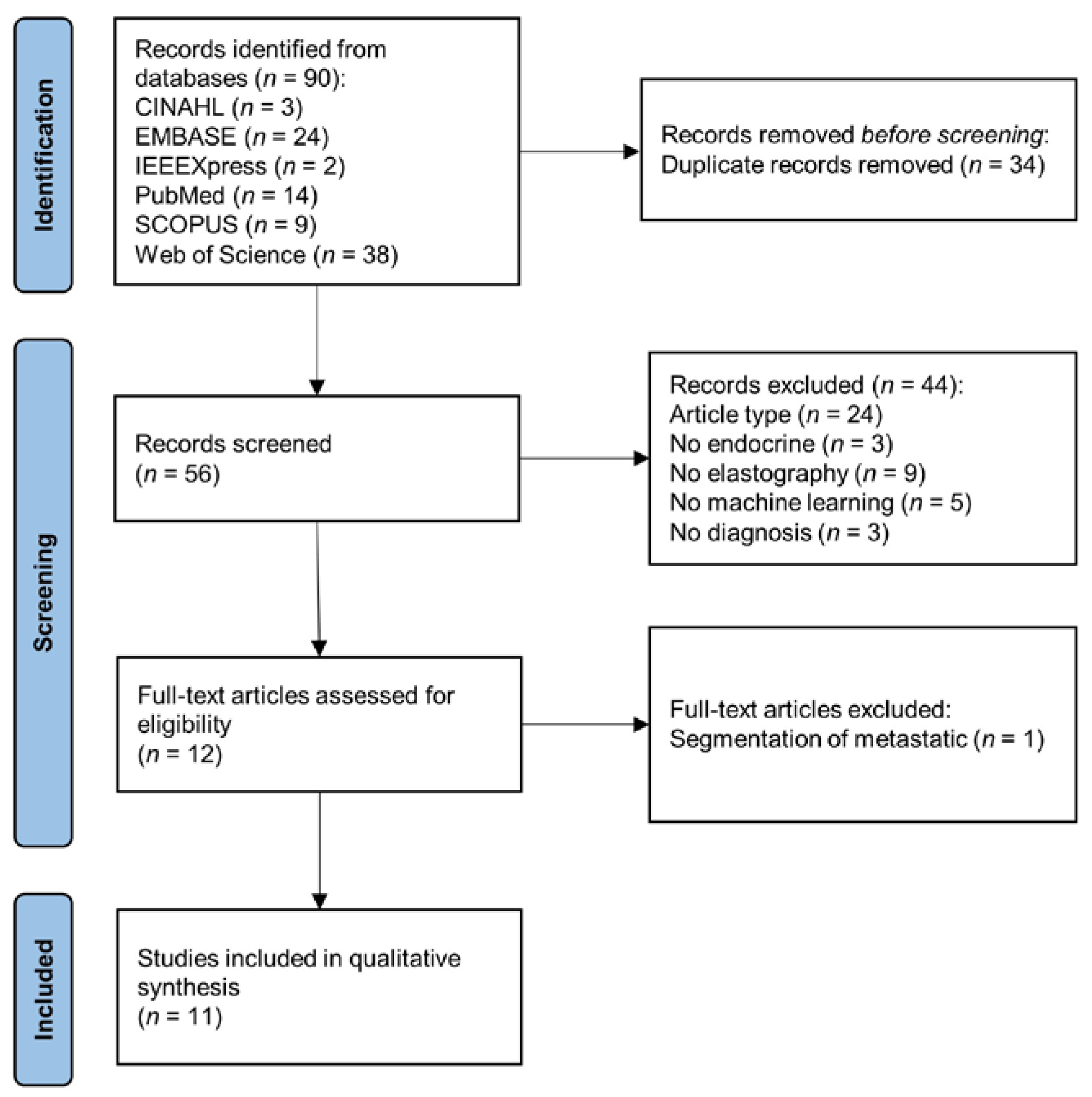
2.1. Search Strategy
2.2. Screening and Data Extraction
3. Results
3.1. Search Results
3.2. Basic Information and Dataset
4. Review Theme and Context
4.1. Data Processing and Segmentation
4.2. Feature Extraction and Data Fusion
4.3. Classification and Modeling
4.4. Classification Performance
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Laycock, J.; Meeran, K. Integrated Endocrinology; Laycock, J., Meeran, K., Eds.; John Wiley & Sons: West Sussex, UK, 2012. [Google Scholar]
- Jolly, E.; Fry, A.; Chaudhry, A. Training in Medicine; Jolly, E., Fry, A., Chaudhry, A., Eds.; Oxford University Press: Oxford, UK, 2016. [Google Scholar]
- Leoncini, E.; Carioli, G.; La Vecchia, C.; Boccia, S.; Rindi, G. Risk factors for neuroendocrine neoplasms: A systematic review and meta-analysis. Ann. Oncol. 2016, 27, 68–81. [Google Scholar] [CrossRef] [PubMed]
- Kitahara, C.M.; Sosa, J.A. The changing incidence of thyroid cancer. Nat. Rev. Endocrinol. 2016, 12, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Soerjomataram, I.; Ervik, M.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Kardosh, A.; Lichtensztajn, D.Y.; Gubens, M.A.; Kunz, P.L.; Fisher, G.A.; Clarke, C.A. Long-term survivors of pancreatic cancer: A California population-based study. Pancreas 2018, 47, 958. [Google Scholar] [CrossRef]
- Bikas, A.; Burman, K.D. Epidemiology of thyroid cancer. In The Thyroid and Its Diseases; Luster, M., Duntas, L.H., Wartofsky, L., Eds.; Springer: New York, NY, USA, 2019; pp. 541–547. [Google Scholar]
- Han, L.; Wu, Z.; Li, W.; Li, Y.; Ma, J.; Wu, X.; Wen, W.; Li, R.; Yao, Y.; Wang, Y. The real world and thinking of thyroid cancer in China. IJS Oncol. 2019, 4, e81. [Google Scholar] [CrossRef]
- Uppal, N.; Cunningham, C.; James, B. The Cost and Financial Burden of Thyroid Cancer on Patients in the US: A Review and Directions for Future Research. JAMA Otolaryngol.–Head Neck Surg. 2022, 148, 568–575. [Google Scholar] [CrossRef]
- Barrows, C.E.; Belle, J.M.; Fleishman, A.; Lubitz, C.C.; James, B.C. Financial burden of thyroid cancer in the United States: An estimate of economic and psychological hardship among thyroid cancer survivors. Surgery 2020, 167, 378–384. [Google Scholar] [CrossRef]
- Mongelli, M.N.; Giri, S.; Peipert, B.J.; Helenowski, I.B.; Yount, S.E.; Sturgeon, C. Financial burden and quality of life among thyroid cancer survivors. Surgery 2020, 167, 631–637. [Google Scholar] [CrossRef]
- Klein, A.P. Pancreatic cancer epidemiology: Understanding the role of lifestyle and inherited risk factors. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 493–502. [Google Scholar] [CrossRef]
- Strobel, O.; Neoptolemos, J.; Jäger, D.; Büchler, M.W. Optimizing the outcomes of pancreatic cancer surgery. Nat. Rev. Clin. Oncol. 2019, 16, 11–26. [Google Scholar] [CrossRef]
- Zhang, L.; Sanagapalli, S.; Stoita, A. Challenges in diagnosis of pancreatic cancer. World J. Gastroenterol. 2018, 24, 2047. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.T.; Lee, E.J.; Huang, M.G.; Park, Y.I.; Khullar, A.; Plodkowski, R.A. Diagnosis and treatment of patients with thyroid cancer. Am. Health Drug Benefits 2015, 8, 30. [Google Scholar] [PubMed]
- Thomasian, N.M.; Kamel, I.R.; Bai, H.X. Machine intelligence in non-invasive endocrine cancer diagnostics. Nat. Rev. Endocrinol. 2022, 18, 81–95. [Google Scholar] [CrossRef]
- Baig, F.; Liu, S.; Yip, S.; Law, H.; Ying, M. Update on ultrasound diagnosis for thyroid cancer. Hong Kong J. Radiol. 2018, 21, 82–93. [Google Scholar] [CrossRef]
- Abu-Ghanem, S.; Cohen, O.; Lazutkin, A.; Abu-Ghanem, Y.; Fliss, D.M.; Yehuda, M. Evaluation of clinical presentation and referral indications for ultrasound-guided fine-needle aspiration biopsy of the thyroid as possible predictors of thyroid cancer. Head Neck 2016, 38, E991–E995. [Google Scholar] [CrossRef]
- Bowman, A.W.; Bolan, C.W. MRI evaluation of pancreatic ductal adenocarcinoma: Diagnosis, mimics, and staging. Abdom. Radiol. 2019, 44, 936–949. [Google Scholar] [CrossRef]
- Siddiqi, A.J.; Miller, F. Chronic pancreatitis: Ultrasound, computed tomography, and magnetic resonance imaging features. Semin. Ultrasound CT MRI 2007, 28, 384–394. [Google Scholar] [CrossRef]
- Zamora, C.; Castillo, M. Sellar and parasellar imaging. Neurosurgery 2017, 80, 17–38. [Google Scholar] [CrossRef]
- Connor, S.; Penney, C. MRI in the differential diagnosis of a sellar mass. Clin. Radiol. 2003, 58, 20–31. [Google Scholar] [CrossRef]
- Beregi, J.P.; Greffier, J. Low and ultra-low dose radiation in CT: Opportunities and limitations. Diagn. Interv. Imaging 2019, 100, 63–64. [Google Scholar] [CrossRef]
- Pilmeyer, J.; Huijbers, W.; Lamerichs, R.; Jansen, J.F.; Breeuwer, M.; Zinger, S. Functional MRI in major depressive disorder: A review of findings, limitations, and future prospects. J. Neuroimaging 2022, 32, 582–595. [Google Scholar] [CrossRef] [PubMed]
- Wallyn, J.; Anton, N.; Akram, S.; Vandamme, T.F. Biomedical Imaging: Principles, Technologies, Clinical Aspects, Contrast Agents, Limitations and Future Trends in Nanomedicines. Pharm. Res. 2019, 36, 78. [Google Scholar] [CrossRef] [PubMed]
- Garra, B.S. Elastography: History, principles, and technique comparison. Abdom. Imaging 2015, 40, 680–697. [Google Scholar] [CrossRef] [PubMed]
- Sigrist, R.M.S.; Liau, J.; Kaffas, A.E.; Chammas, M.C.; Willmann, J.K. Ultrasound Elastography: Review of Techniques and Clinical Applications. Theranostics 2017, 7, 1303–1329. [Google Scholar] [CrossRef]
- Lee, T.T.-Y.; Cheung, J.C.-W.; Law, S.-Y.; To, M.K.T.; Cheung, J.P.Y.; Zheng, Y.-P. Analysis of sagittal profile of spine using 3D ultrasound imaging: A phantom study and preliminary subject test. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2020, 8, 232–244. [Google Scholar] [CrossRef]
- Cheung, C.-W.J.; Zhou, G.-Q.; Law, S.-Y.; Lai, K.-L.; Jiang, W.-W.; Zheng, Y.-P. Freehand three-dimensional ultrasound system for assessment of scoliosis. J. Orthop. Transl. 2015, 3, 123–133. [Google Scholar] [CrossRef]
- Mao, Y.-J.; Lim, H.-J.; Ni, M.; Yan, W.-H.; Wong, D.W.-C.; Cheung, J.C.-W. Breast tumor classification using ultrasound elastography with machine learning: A systematic scoping review. Cancers 2022, 14, 367. [Google Scholar] [CrossRef]
- Zheng, Y.-P.; Mak, T.-M.; Huang, Z.-M.; Cheung, C.-W.J.; Zhou, Y.-J.; He, J.-F. Liver fibrosis assessment using transient elastography guided with real-time B-mode ultrasound imaging. In Proceedings of the 6th World Congress of Biomechanics (WCB 2010), Singapore, 1–6 August 2010; pp. 1036–1039. [Google Scholar]
- Liao, J.; Yang, H.; Yu, J.; Liang, X.; Chen, Z. Progress in the application of ultrasound elastography for brain diseases. J. Ultrasound Med. 2020, 39, 2093–2104. [Google Scholar] [CrossRef]
- Ullah, Z.; Farooq, M.U.; Lee, S.-H.; An, D. A hybrid image enhancement based brain MRI images classification technique. Med. Hypotheses 2020, 143, 109922. [Google Scholar] [CrossRef]
- Ullah, Z.; Usman, M.; Jeon, M.; Gwak, J. Cascade multiscale residual attention cnns with adaptive roi for automatic brain tumor segmentation. Inf. Sci. 2022, 608, 1541–1556. [Google Scholar] [CrossRef]
- Yuen, Q.W.-H.; Zheng, Y.-P.; Huang, Y.-P.; He, J.-F.; Cheung, J.C.-W.; Ying, M. In-vitro strain and modulus measurements in porcine cervical lymph nodes. Open Biomed. Eng. J. 2011, 5, 39. [Google Scholar] [CrossRef] [PubMed]
- Goss, B.; McGee, K.P.; Ehman, E.; Manduca, A.; Ehman, R. Magnetic resonance elastography of the lung: Technical feasibility. Magn. Reson. Med. Off. J. Int. Soc. Magn. Reson. Med. 2006, 56, 1060–1066. [Google Scholar] [CrossRef] [PubMed]
- Yin, M.; Talwalkar, J.A.; Glaser, K.J.; Manduca, A.; Grimm, R.C.; Rossman, P.J.; Fidler, J.L.; Ehman, R.L. Assessment of hepatic fibrosis with magnetic resonance elastography. Clin. Gastroenterol. Hepatol. 2007, 5, 1207–1213.e1202. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, A.; Mahowald, J.; Manduca, A.; Rossman, P.; Hartmann, L.; Ehman, R. Magnetic resonance elastography of breast cancer. Radiology 2000, 214, 612–613. [Google Scholar]
- Yoo, Y.J.; Ha, E.J.; Cho, Y.J.; Kim, H.L.; Han, M.; Kang, S.Y. Computer-aided diagnosis of thyroid nodules via ultrasonography: Initial clinical experience. Korean J. Radiol. 2018, 19, 665–672. [Google Scholar] [CrossRef]
- Chambara, N.; Ying, M. The diagnostic efficiency of ultrasound computer–aided diagnosis in differentiating thyroid nodules: A systematic review and narrative synthesis. Cancers 2019, 11, 1759. [Google Scholar] [CrossRef]
- Blanco-Montenegro, I.; De Ritis, R.; Chiappini, M. Imaging and modelling the subsurface structure of volcanic calderas with high-resolution aeromagnetic data at Vulcano (Aeolian Islands, Italy). Bull. Volcanol. 2007, 69, 643–659. [Google Scholar] [CrossRef]
- Zhang, X.; Lee, V.C.; Rong, J.; Lee, J.C.; Liu, F. Deep convolutional neural networks in thyroid disease detection: A multi-classification comparison by ultrasonography and computed tomography. Comput. Methods Programs Biomed. 2022, 220, 106823. [Google Scholar] [CrossRef]
- Chan, H.P.; Hadjiiski, L.M.; Samala, R.K. Computer-aided diagnosis in the era of deep learning. Med. Phys. 2020, 47, e218–e227. [Google Scholar] [CrossRef]
- Elmohr, M.; Fuentes, D.; Habra, M.; Bhosale, P.; Qayyum, A.; Gates, E.; Morshid, A.; Hazle, J.; Elsayes, K. Machine learning-based texture analysis for differentiation of large adrenal cortical tumors on CT. Clin. Radiol. 2019, 74, 818, e811–818.e817. [Google Scholar] [CrossRef]
- Barat, M.; Chassagnon, G.; Dohan, A.; Gaujoux, S.; Coriat, R.; Hoeffel, C.; Cassinotto, C.; Soyer, P. Artificial intelligence: A critical review of current applications in pancreatic imaging. Jpn. J. Radiol. 2021, 39, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, L.; Qi, L.; Yi, X.; Li, M.; Zhou, M.; Chen, D.; Xiao, Q.; Wang, C.; Pang, Y.; et al. Machine Learning: Applications and Advanced Progresses of Radiomics in Endocrine Neoplasms. J. Oncol. 2021, 2021, 8615450. [Google Scholar] [CrossRef] [PubMed]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.; Horsley, T.; Weeks, L. PRISMA extension for scoping reviews (PRISMA-ScR): Checklist and explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Pei, C.; Xie, L.; Liu, Z.; He, N.; Lv, W. Convolutional Neural Network for predicting thyroid cancer based on ultrasound elastography image of perinodular region. Endocrinology 2022, 163, bqac135. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Dighe, M.; Alessio, A.M. Comparison of machine learned approaches for thyroid nodule characterization from shear wave elastography images. Med. Imaging 2018 Comput.-Aided Diagn. 2018, 10575, 437–443. [Google Scholar]
- Qin, P.; Wu, K.; Hu, Y.; Zeng, J.; Chai, X. Diagnosis of benign and malignant thyroid nodules using combined conventional ultrasound and ultrasound elasticity imaging. IEEE J. Biomed. Health Inform. 2019, 24, 1028–1036. [Google Scholar] [CrossRef]
- Săftoiu, A.; Vilmann, P.; Gorunescu, F.; Gheonea, D.I.; Gorunescu, M.; Ciurea, T.; Popescu, G.L.; Iordache, A.; Hassan, H.; Iordache, S. Neural network analysis of dynamic sequences of EUS elastography used for the differential diagnosis of chronic pancreatitis and pancreatic cancer. Gastrointest. Endosc. 2008, 68, 1086–1094. [Google Scholar] [CrossRef]
- Săftoiu, A.; Vilmann, P.; Gorunescu, F.; Janssen, J.; Hocke, M.; Larsen, M.; Iglesias–Garcia, J.; Arcidiacono, P.; Will, U.; Giovannini, M. Efficacy of an artificial neural network–based approach to endoscopic ultrasound elastography in diagnosis of focal pancreatic masses. Clin. Gastroenterol. Hepatol. 2012, 10, 84–90.e81. [Google Scholar] [CrossRef]
- Sun, H.; Yu, F.; Xu, H. Discriminating the nature of thyroid nodules using the hybrid method. Math. Probl. Eng. 2020, 2020, 6147037. [Google Scholar] [CrossRef]
- Udriștoiu, A.L.; Cazacu, I.M.; Gruionu, L.G.; Gruionu, G.; Iacob, A.V.; Burtea, D.E.; Ungureanu, B.S.; Costache, M.I.; Constantin, A.; Popescu, C.F. Real-time computer-aided diagnosis of focal pancreatic masses from endoscopic ultrasound imaging based on a hybrid convolutional and long short-term memory neural network model. PLoS ONE 2021, 16, e0251701. [Google Scholar] [CrossRef]
- Zhang, B.; Tian, J.; Pei, S.; Chen, Y.; He, X.; Dong, Y.; Zhang, L.; Mo, X.; Huang, W.; Cong, S. Machine learning–assisted system for thyroid nodule diagnosis. Thyroid 2019, 29, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.-N.; Liu, J.-Y.; Lin, Q.-Z.; He, Y.-S.; Luo, H.-H.; Peng, Y.-L.; Ma, B.-Y. Partially cystic thyroid cancer on conventional and elastographic ultrasound: A retrospective study and a machine learning–assisted system. Ann. Transl. Med. 2020, 8, 495. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.-K.; Ren, T.-T.; Yin, Y.-F.; Shi, H.; Wang, H.-X.; Zhou, B.-Y.; Wang, X.-R.; Li, X.; Zhang, Y.-F.; Liu, C. A comparative analysis of two machine learning-based diagnostic patterns with thyroid imaging reporting and data system for thyroid nodules: Diagnostic performance and unnecessary biopsy rate. Thyroid 2021, 31, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Liu, B.; Liu, Y.; Huang, Q.; Yan, W. Ultrasonic Intelligent Diagnosis of Papillary Thyroid Carcinoma Based on Machine Learning. J. Healthc. Eng. 2022, 2022, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Huang, H.; Fang, B. Application of weighted cross-entropy loss function in intrusion detection. J. Comput. Commun. 2021, 9, 1–21. [Google Scholar] [CrossRef]
- Peng, J.; Liu, Y.; Tang, S.; Hao, Y.; Chu, L.; Chen, G.; Wu, Z.; Chen, Z.; Yu, Z.; Du, Y. PP-LiteSeg: A Superior Real-Time Semantic Segmentation Model. arXiv 2022, arXiv:2204.02681. [Google Scholar]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Muhammad, U.; Wang, W.; Chattha, S.P.; Ali, S. Pre-trained VGGNet architecture for remote-sensing image scene classification. In Proceedings of the 2018 24th International Conference on Pattern Recognition (ICPR), Beijing, China, 20–24 August 2018; pp. 1622–1627. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE conference on computer vision and pattern recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- He, K.; Girshick, R.; Dollár, P. Rethinking imagenet pre-training. In Proceedings of the Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Korea, 27 October–2 November 2019; pp. 4918–4927. [Google Scholar]
- Branke, J.; Chick, S.E.; Schmidt, C. Selecting a selection procedure. Manag. Sci. 2007, 53, 1916–1932. [Google Scholar] [CrossRef]
- Bahn, M.M.; Brennan, M.D.; Bahn, R.S.; Dean, D.S.; Kugel, J.L.; Ehman, R.L. Development and application of magnetic resonance elastography of the normal and pathological thyroid gland in vivo. J. Magn. Reson. Imaging 2009, 30, 1151–1154. [Google Scholar] [CrossRef]
- Menzo, E.L.; Szomstein, S.; Rosenthal, R.J. Pancreas, Liver, and Adrenal Glands in Obesity. In The Globesity Challenge to General Surgery; Foletto, M., Rosenthal, R.J., Eds.; Springer: New York, NY, USA, 2014; pp. 155–170. [Google Scholar]
- Shi, Y.; Glaser, K.J.; Venkatesh, S.K.; Ben-Abraham, E.I.; Ehman, R.L. Feasibility of using 3D MR elastography to determine pancreatic stiffness in healthy volunteers. J. Magn. Reson. Imaging 2015, 41, 369–375. [Google Scholar] [CrossRef]
- Qian, X.; Ma, T.; Yu, M.; Chen, X.; Shung, K.K.; Zhou, Q. Multi-functional ultrasonic micro-elastography imaging system. Sci. Rep. 2017, 7, 1230. [Google Scholar] [CrossRef] [PubMed]
- Suh, C.H.; Yoon, H.M.; Jung, S.C.; Choi, Y.J. Accuracy and precision of ultrasound shear wave elasticity measurements according to target elasticity and acquisition depth: A phantom study. PLoS ONE 2019, 14, e0219621. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, M.; Atal, I.; Li, J.; Smith, P.; Ravaud, P.; Fergie, M.; Callaghan, M.; Selfe, J. Reporting quality of studies using machine learning models for medical diagnosis: A systematic review. BMJ Open 2020, 10, e034568. [Google Scholar] [CrossRef]
- Pastor-López, I.; Sanz, B.; Tellaeche, A.; Psaila, G.; de la Puerta, J.G.; Bringas, P.G. Quality assessment methodology based on machine learning with small datasets: Industrial castings defects. Neurocomputing 2021, 456, 622–628. [Google Scholar] [CrossRef]
- Lai, D.K.-H.; Zha, L.-W.; Leung, T.Y.-N.; Tam, A.Y.-C.; So, B.P.-H.; Lim, H.J.; Cheung, D.S.-K.; Wong, D.W.-C.; Cheung, J.C.-W. Dual ultra-wideband (UWB) radar-based sleep posture recognition system: Towards ubiquitous sleep monitoring. Eng. Regen. 2022, 4, 36–43. [Google Scholar] [CrossRef]
- Wang, K. An Overview of Deep Learning Based Small Sample Medical Imaging Classification. In Proceedings of the 2021 International Conference on Signal Processing and Machine Learning (CONF-SPML), Beijing, China, 18–20 August 2021; pp. 278–281. [Google Scholar]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? Adv. Neural Inf. Process. Syst. 2014, 27, 3320–3328. [Google Scholar]
- Vabalas, A.; Gowen, E.; Poliakoff, E.; Casson, A.J. Machine learning algorithm validation with a limited sample size. PLoS ONE 2019, 14, e0224365. [Google Scholar] [CrossRef]
- Probst, P.; Boulesteix, A.-L.; Bischl, B. Tunability: Importance of hyperparameters of machine learning algorithms. J. Mach. Learn. Res. 2019, 20, 1934–1965. [Google Scholar]
- Larracy, R.; Phinyomark, A.; Scheme, E. Machine learning model validation for early stage studies with small sample sizes. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Online, 1–5 November 2021; pp. 2314–2319. [Google Scholar]
- Liashchynskyi, P.; Liashchynskyi, P. Grid search, random search, genetic algorithm: A big comparison for NAS. arXiv 2019, arXiv:1905.12787. [Google Scholar]
- Bhatia, K.S.S.; Lam, A.C.L.; Pang, S.W.A.; Wang, D.; Ahuja, A.T. Feasibility study of texture analysis using ultrasound shear wave elastography to predict malignancy in thyroid nodules. Ultrasound Med. Biol. 2016, 42, 1671–1680. [Google Scholar] [CrossRef]
- Sagheer, S.V.M.; George, S.N. A review on medical image denoising algorithms. Biomed. Signal Process. Control 2020, 61, 102036. [Google Scholar] [CrossRef]
- Jeong, J.J.; Tariq, A.; Adejumo, T.; Trivedi, H.; Gichoya, J.W.; Banerjee, I. Systematic review of generative adversarial networks (gans) for medical image classification and segmentation. J. Digit. Imaging 2022, 35, 137–152. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Walia, E.; Babyn, P. Generative adversarial network in medical imaging: A review. Med. Image Anal. 2019, 58, 101552. [Google Scholar] [CrossRef] [PubMed]
- Tam, A.Y.-C.; So, B.P.-H.; Chan, T.T.-C.; Cheung, A.K.-Y.; Wong, D.W.-C.; Cheung, J.C.-W. A Blanket Accommodative Sleep Posture Classification System Using an Infrared Depth Camera: A Deep Learning Approach with Synthetic Augmentation of Blanket Conditions. Sensors 2021, 21, 5553. [Google Scholar] [CrossRef] [PubMed]
| Reference | Sample Size | Sex (M:F) | Mean Age (years) | Type (B:M) | Size (mm) | Reference Standard |
|---|---|---|---|---|---|---|
| Hu et al. [48] | 1582 patients 1747 nodules | 567:1015 | 46.40 (SD: 9.65) | Thyroid 701:1046 | 14.51 ± 3.51 | FNA |
| Pereira et al. [49] | 165 patients 964 images | - | - | Thyroid 752:212 | - | - |
| Qin et al. [50] | 233 patients 1156 nodules | - | - | Thyroid 539:617 | - | Verified by clinical pathology |
| Săftoiu et al. [51] | 68 patients | 47:21 | Normal: 49.4 (SD: 15.4) ChPan: 55.1 (SD: 17.0) PanCA: 62.3 (SD: 12.9) | Normal: 22 ChPan: 11 PanCA: 35 | - | CT and biopsy |
| Săftoiu et al. [52] | 258 patients 774 recordings | 172:76 | PanCA: 64 (SD: 15.40) ChPan: 56 (SD: 13.25) | Pancreatic 47:211 | PanCA: 31.97 (SD: 11.69, 6–85) ChPan: 28.36 (SD: 12.23, 9–60) | FNA biopsy, verified by clinical, biological exams, and repeated imaging tests |
| Sun et al. [53] | 245 patients 490 images | - | - | Thyroid 145:100 | - | Biopsy |
| Udriștoiu et al. [54] | 65 patients 1300 images | - | - | PDAC: 30 CPP: 20 PNET: 15 | - | FNA biopsy |
| Zhang et al. [55] | 2032 patients 2064 nodules | 695:1337 | 45.25 (SD: 13.49) | Thyroid 1314:750 | ≤25 | - |
| Zhao et al. [56] | 174 patients 177 nodules | 45:132 | B: 47.9 M: 41.9 | Thyroid 81:96 | B: 23.4 M: 20.0 | FNA biopsy |
| Zhao et al. [57] | 720 patients 743 nodules | 168:552 | 49.61 (15–89) | Thyroid 469:274 | ≥10 | Biopsy |
| Zhou et al. [58] | 70 patients 107 nodes | 10:60 | 30 | Thyroid 32:75 | ≤10 | FNA biopsy |
| Articles | Mode | Type | System | Processing and Segmentation |
|---|---|---|---|---|
| Hu et al. [48] | US | SWE + B-mode | ACUSON Sequoia Redwood US diagnostic system (Siemens, Munich, Germany) | Use PP-LiteSeg to segment SWE by B-mode |
| Pereira et al. [49] | US | SWE + B-mode | - | Segmented SWE region with stress corresponding to 0.7 max stress value |
| Qin et al. [50] | US | SWE + B-mode | Aixplorer ultrasonic machine | Pre-extracted ROI by color channel transformation and segmented by radiologists |
| Săftoiu et al. [51] | US | EUS, SE + B-mode | HITACHI 8500 (Hitachi Medical Systems, Zug, Switzerland) Pentax Linear Endoscope EG3830UT and EG3870 UTK (Pentax, Hamburg Germany) | Processed using ImageJ software to extract hue histogram matrix. Manual selection or tumor area |
| Săftoiu et al. [52] | US | EUS | - | Processed using a special software plugin based on ImageJ software to extract hue histogram matrix. Manual selection or tumor area. |
| Sun et al. [53] | US | SWE + B-mode | - | ROI manually segmented using ITK-SNAP Denoise with Median Filter and outlined by radiologists. |
| Udriștoiu et al. [54] | US | EUS, SE, Doppler | HITACHI Preirus EG3870UTK, Pentax Medical Corporation | Contrast enhancement, ROI manual segmentation |
| Zhang et al. [55] | US | SE + B-mode | HI Vision 900, HI Vision Ascendus, HI Vision Preirus color US units from Hitachi (Tokyo, Japan) | Conducted by experienced radiologist |
| Zhao et al. [56] | US | SE + B-mode | HITACHI Vision 900 system (Hitachi Medical System, Tokyo, Japan), - | - |
| B-mode | Philips iu222 (Philips, Bothell, WA, USA) | |||
| Zhao et al. [57] | US | SWE + B-mode | Aixplorer; Supersonic Imagine (Paris, France), SWE | ROI extracted by Q-Box quantification tool |
| Zhou et al. [58] | US | - | - | Contrast enhancement |
| Articles | Feature Extraction Strategy | Classifier/Model | Validation (trn:tst) |
|---|---|---|---|
| Hu et al. [48] | 7 ResNet18 models on different segmentation approaches | 71:29 | |
| Pereira et al. [49] | Predetermined SWE statistical features and SWE features extracted by circular Hough transform | Logistic regression, naïve Bayes, SVM, decision tree | 82:18 |
| Fully trained CNN (2-layer) model for B-mode and SWE Pretrained CNN18 for B-mode and SWE Combine classification by averaging class probabilities of trained B-mode and SWE models | |||
| Qin et al. [50] | Pretrained VGG16 with 3 fused methods (MT, FEx-reFus, and Fus-reFEx) and 3 classifier layers (FCL, SPP, and GAP) | 82:18 | |
| Săftoiu et al. [51] | MLP (3- and 4-layer) | 10-fold cxv | |
| Săftoiu et al. [52] | MLP (4-layer) | 10-fold cxv | |
| Sun et al. [53] | Deep feature extractor on SWE US Predetermined statistical and radiomics features on B-mode US | Logistic regression, naïve Bayes, and SVM on both SWE and B-mode features. Classifications of both models were combined and hybridized by uncertainty decision-theory-based voting system (pessimistic, optimistic, and compromise approaches). | 5-fold cxv |
| Udriștoiu et al. [54] | CNN on B-mode, contrast harmonic sequential images taken at 0, 10, 20, 30, 40 s, color Doppler, and elastography LSTM on contrast harmonic sequential images taken at 0, 10, 20, 30, 40 s CNN and LSTM merged by concatenation layer. | 80:20 | |
| Zhang et al. [55] | 11 predetermined B-mode features 1 predetermined elastography feature | Logistic regression, linear discriminant analysis, random forest, kernel SVM, adaptive boosting, KNN, neural network, naïve Bayes, CNN | 60:40, 10-fold cxv |
| Zhao et al. [56] | 20 predetermined radiomics features | Logistic regression, random forest, XGBoost, SVM, MLP, KNN | - |
| Zhao et al. [57] | Machine-learning-assisted approach (6 predetermined B-mode and 5 SWE features) Radiomics features | Decision tree, naïve Bayes, KNN, logistic regression, SVM, KNN-based bagging, random forest, XGBoost, MLP, gradient boosting tree | Training: 520 Testing: 223 External Testing: 106 |
| Zhou et al. [58] | Predetermined statistical features, Feature extraction by GLCOM-GLRLM, MSCOM | RBM + Bayesian | - |
| Articles | Model and Approach | Evaluation Metrics and Outcomes | |||||
|---|---|---|---|---|---|---|---|
| Acc (%) | Sn/Rc (%) | Sp (%) | PPV/Pc (%) | NPV (%) | AUC (%) | ||
| Hu et al. [44] | B-mode + SWE (1.0 mm offset) ResNet18 | 86.45 | 85.15 | 91.93 | 82.12 | 73.54 | 93 |
| Pereira et al. [45] | SWE Pretrained CNN18 | 83 | - | - | - | - | 80 |
| Qin et al. [46] | Pretrained VGG16 Ex-reFus with SPP | 94.7 | 92.77 | 97.96 | - | - | 98.77 |
| Săftoiu et al. [47] | MLP (3-layer) | 89.7 | 91.4 | 87.9 | 88.9 | 90.6 | 95 |
| Săftoiu et al. [48] | MLP (2-layer) | 84.27 | 87.59 | 82.94 | 96.25 | 57.22 | 94 |
| Sun et al. [49] | Hybridized model with voting system (compromise approach) | 86.5 | 82 | 89.7 | - | - | 92.1 |
| Udriștoiu et al. [50] | CNN-LSTM | 98.26 | 98.6 | 97.4 | 98.7 | 97.4 | 98 |
| Zhang et al. [51] | Random forest | 85.7 | 89.1 | 85.3 | - | - | 93.8 |
| Zhao et al. [52] 2020 | Random forest | 86.0 | 86.6 | 85.5 | - | - | 93.4 |
| Zhao et al. [53] 2021 | Machine-learning-assisted approach (B-mode + SWE) using KNN-based bagging model | 93.4 | 93.9 | 93.2 | 86.1 | 97.1 | 95.3 |
| Zhou et al. [54] | RBM + Bayesian (UE) | - | 90.21 | 78.45 | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, Y.-J.; Zha, L.-W.; Tam, A.Y.-C.; Lim, H.-J.; Cheung, A.K.-Y.; Zhang, Y.-Q.; Ni, M.; Cheung, J.C.-W.; Wong, D.W.-C. Endocrine Tumor Classification via Machine-Learning-Based Elastography: A Systematic Scoping Review. Cancers 2023, 15, 837. https://doi.org/10.3390/cancers15030837
Mao Y-J, Zha L-W, Tam AY-C, Lim H-J, Cheung AK-Y, Zhang Y-Q, Ni M, Cheung JC-W, Wong DW-C. Endocrine Tumor Classification via Machine-Learning-Based Elastography: A Systematic Scoping Review. Cancers. 2023; 15(3):837. https://doi.org/10.3390/cancers15030837
Chicago/Turabian StyleMao, Ye-Jiao, Li-Wen Zha, Andy Yiu-Chau Tam, Hyo-Jung Lim, Alyssa Ka-Yan Cheung, Ying-Qi Zhang, Ming Ni, James Chung-Wai Cheung, and Duo Wai-Chi Wong. 2023. "Endocrine Tumor Classification via Machine-Learning-Based Elastography: A Systematic Scoping Review" Cancers 15, no. 3: 837. https://doi.org/10.3390/cancers15030837
APA StyleMao, Y.-J., Zha, L.-W., Tam, A. Y.-C., Lim, H.-J., Cheung, A. K.-Y., Zhang, Y.-Q., Ni, M., Cheung, J. C.-W., & Wong, D. W.-C. (2023). Endocrine Tumor Classification via Machine-Learning-Based Elastography: A Systematic Scoping Review. Cancers, 15(3), 837. https://doi.org/10.3390/cancers15030837









