The NOTCH-RIPK4-IRF6-ELOVL4 Axis Suppresses Squamous Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Plasmid Constructs
2.3. Lentivirus Production and Transduction
2.4. In Utero Lentiviral Transduction
2.5. Deep Sequencing: Sample Preparation, Pre-Amplification, and Sequence Processing
2.6. Immunoprecipitation
2.7. Antibodies
2.8. Cell Culture
2.9. RNA Isolation, cDNA Synthesis, and Real-Time QPCR Analysis
2.10. RNA-seq and GSEA Analyses
2.11. Statistics and Reproducibility
3. Results
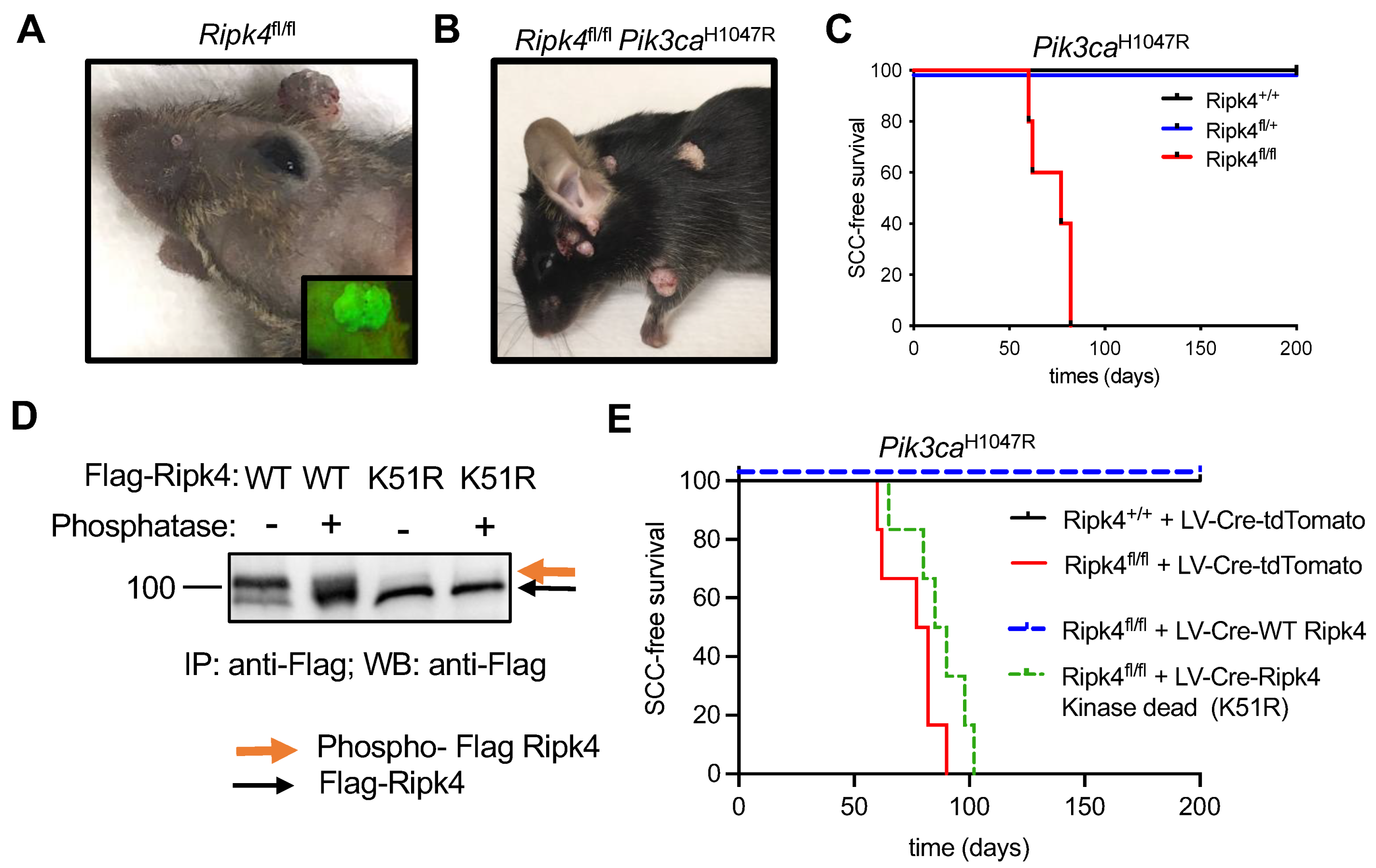
3.1. Ripk4 Tumor Suppressive Function Is Dependent on Its Kinase Activity
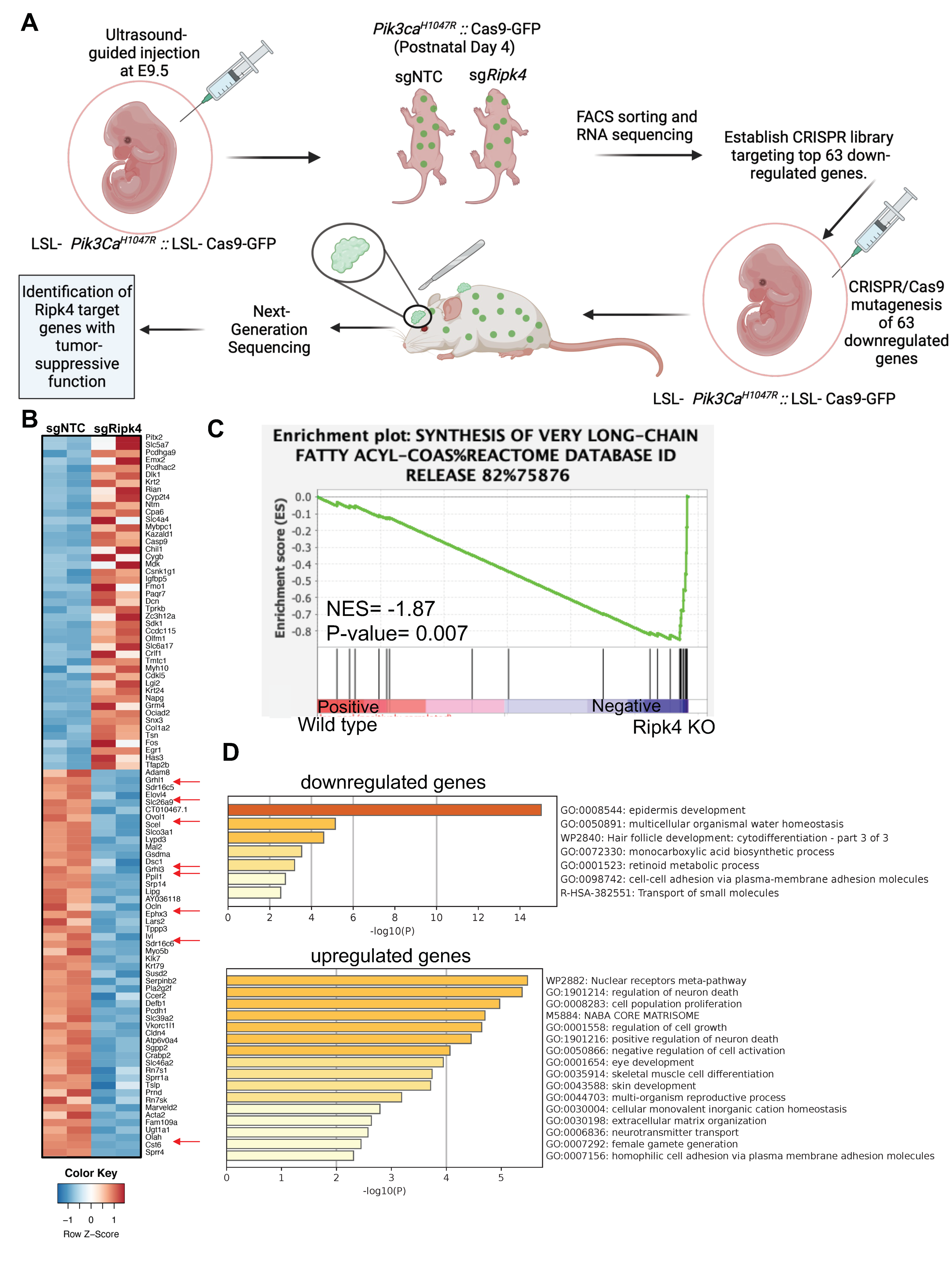
3.2. Ripk4 Regulates Expression of Differentiation Genes in Keratinocytes
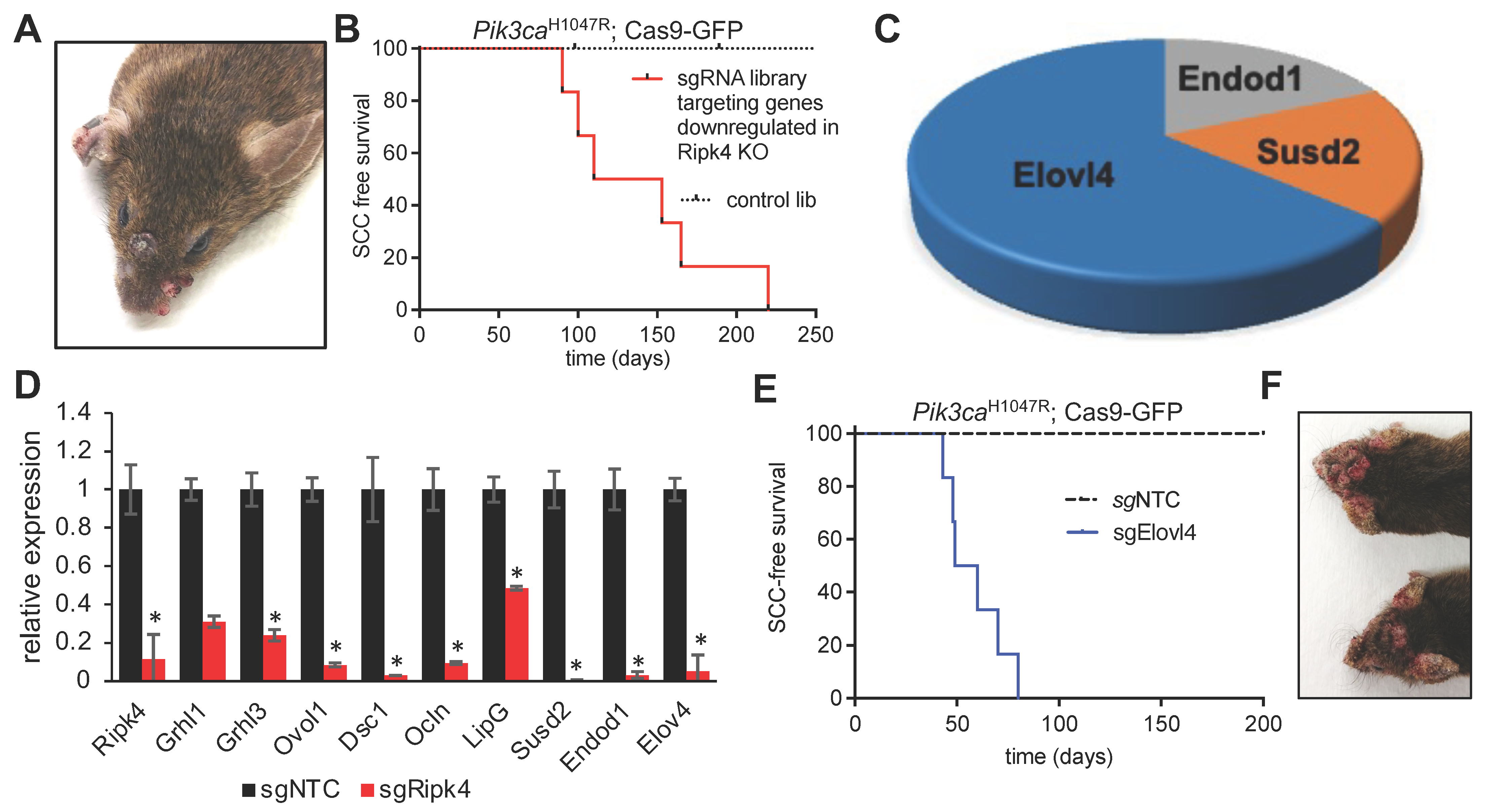
3.3. In vivo CRISPR Screen to Identify Mediators of Ripk4-Mediated Tumor Suppression
3.4. Elovl4 Functions As a Tumor Suppressor in Pik3caH1047R-Mutant Mice
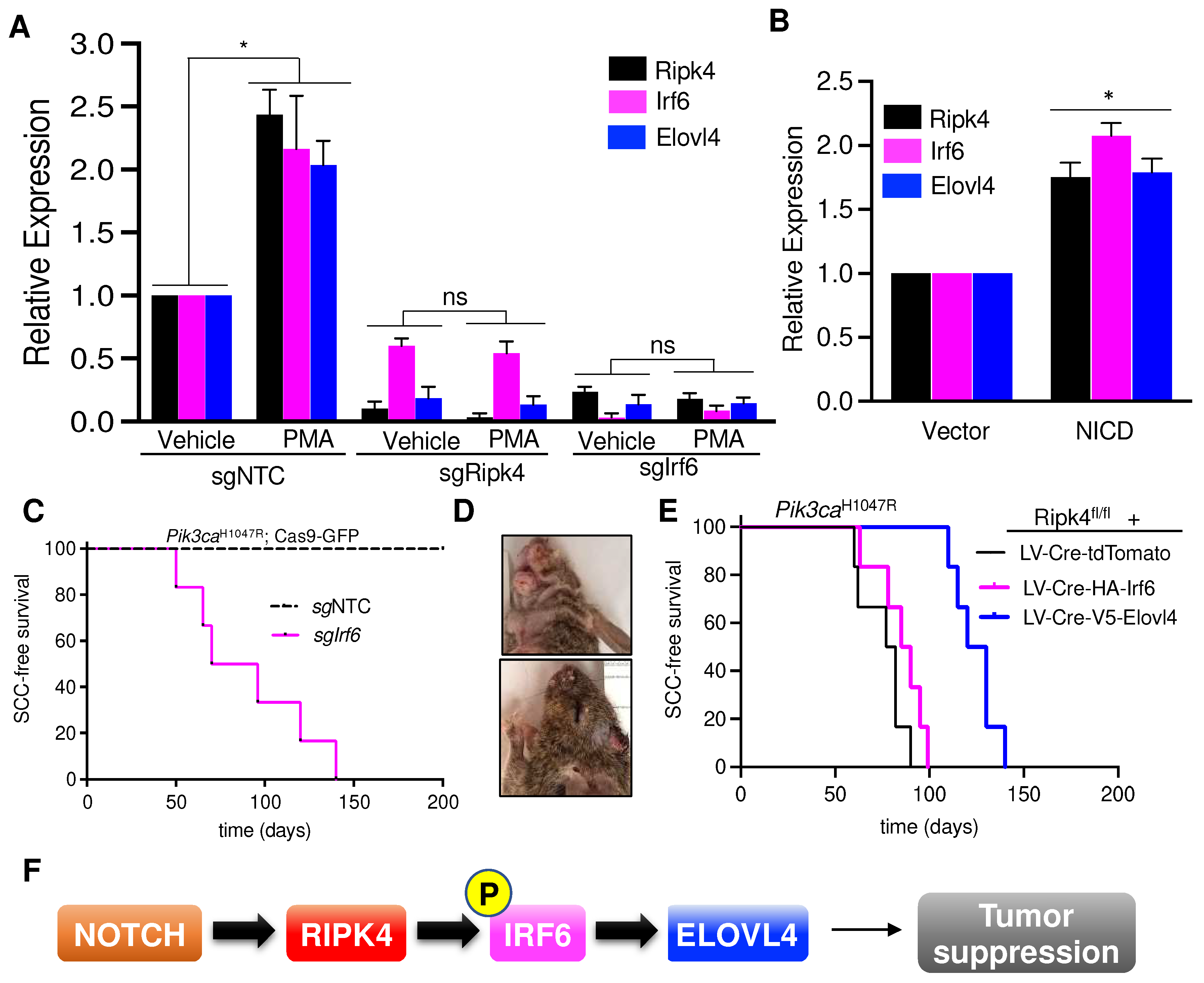
3.5. Elovl4 Is a Target Gene of NOTCH-Ripk4-Irf6 Tumor Suppressor Axis in Keratinocytes
3.6. Elovl4 Overexpression Rescues Tumor Suppression in Ripk4-KO Skin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, J.; Wei, Q.; He, Z. Insight Into the Function of RIPK4 in Keratinocyte Differentiation and Carcinogenesis. Front. Oncol. 2020, 10, 1562. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Haider, K.; Ponda, M.; Cariappa, A.; Rowitch, D.; Pillai, S. Protein kinase C-associated kinase (PKK), a novel membrane-associated, ankyrin repeat-containing protein kinase. J. Biol. Chem. 2001, 276, 21737–21744. [Google Scholar] [CrossRef] [PubMed]
- Tanghe, G.; Urwyler-Rosselet, C.; De Groote, P.; Dejardin, E.; De Bock, P.J.; Gevaert, K.; Vandenabeele, P.; Declercq, W. RIPK4 activity in keratinocytes is controlled by the SCF(beta-TrCP) ubiquitin ligase to maintain cortical actin organization. Cell. Mol. Life Sci. 2018, 75, 2827–2841. [Google Scholar] [CrossRef] [PubMed]
- Kwa, M.Q.; Huynh, J.; Aw, J.; Zhang, L.; Nguyen, T.; Reynolds, E.C.; Sweet, M.J.; Hamilton, J.A.; Scholz, G.M. Receptor-interacting protein kinase 4 and interferon regulatory factor 6 function as a signaling axis to regulate keratinocyte differentiation. J. Biol. Chem. 2014, 289, 31077–31087. [Google Scholar] [CrossRef] [PubMed]
- Meylan, E.; Martinon, F.; Thome, M.; Gschwendt, M.; Tschopp, J. RIP4 (DIK/PKK), a novel member of the RIP kinase family, activates NF-kappa B and is processed during apoptosis. EMBO Rep. 2002, 3, 1201–1208. [Google Scholar] [CrossRef]
- Huang, X.; McGann, J.C.; Liu, B.Y.; Hannoush, R.N.; Lill, J.R.; Pham, V.; Newton, K.; Kakunda, M.; Liu, J.; Yu, C.; et al. Phosphorylation of Dishevelled by protein kinase RIPK4 regulates Wnt signaling. Science 2013, 339, 1441–1445. [Google Scholar] [CrossRef]
- Moran, S.T.; Haider, K.; Ow, Y.; Milton, P.; Chen, L.; Pillai, S. Protein kinase C-associated kinase can activate NFkappaB in both a kinase-dependent and a kinase-independent manner. J. Biol. Chem. 2003, 278, 21526–21533. [Google Scholar] [CrossRef]
- De Groote, P.; Tran, H.T.; Fransen, M.; Tanghe, G.; Urwyler, C.; De Craene, B.; Leurs, K.; Gilbert, B.; Van Imschoot, G.; De Rycke, R.; et al. A novel RIPK4-IRF6 connection is required to prevent epithelial fusions characteristic for popliteal pterygium syndromes. Cell Death Differ. 2015, 22, 1012–1024. [Google Scholar] [CrossRef]
- Oberbeck, N.; Pham, V.C.; Webster, J.D.; Reja, R.; Huang, C.S.; Zhang, Y.; Roose-Girma, M.; Warming, S.; Li, Q.; Birnberg, A.; et al. The RIPK4-IRF6 signalling axis safeguards epidermal differentiation and barrier function. Nature 2019, 574, 249–253. [Google Scholar] [CrossRef]
- Mitchell, K.; O’Sullivan, J.; Missero, C.; Blair, E.; Richardson, R.; Anderson, B.; Antonini, D.; Murray, J.C.; Shanske, A.L.; Schutte, B.C.; et al. Exome sequence identifies RIPK4 as the Bartsocas-Papas syndrome locus. Am. J. Hum. Genet. 2012, 90, 69–75. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.; Zhang, H.; Xiao, M.; Li, J.; Yang, C.; Lin, X.; Wu, Z.; Hu, L.; Kong, X. Novel mutations in the IRF6 gene for Van der Woude syndrome. Hum. Genet. 2003, 113, 382–386. [Google Scholar] [CrossRef]
- Ingraham, C.R.; Kinoshita, A.; Kondo, S.; Yang, B.; Sajan, S.; Trout, K.J.; Malik, M.I.; Dunnwald, M.; Goudy, S.L.; Lovett, M.; et al. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat. Genet. 2006, 38, 1335–1340. [Google Scholar] [CrossRef]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.; Sougnez, C.; McKenna, A.; et al. The Mutational Landscape of Head and Neck Squamous Cell Carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef]
- Agrawal, N.; Frederick, M.J.; Pickering, C.R.; Bettegowda, C.; Chang, K.; Li, R.J.; Fakhry, C.; Xie, T.X.; Zhang, J.; Wang, J.; et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011, 333, 1154–1157. [Google Scholar] [CrossRef]
- Cancer Genome Atlas, N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef]
- Botti, E.; Spallone, G.; Moretti, F.; Marinari, B.; Pinetti, V.; Galanti, S.; De Meo, P.D.; De Nicola, F.; Ganci, F.; Castrignano, T.; et al. Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. Proc. Natl. Acad. Sci. USA 2011, 108, 13710–13715. [Google Scholar] [CrossRef]
- Pickering, C.R.; Zhou, J.H.; Lee, J.J.; Drummond, J.A.; Peng, S.A.; Saade, R.E.; Tsai, K.Y.; Curry, J.L.; Tetzlaff, M.T.; Lai, S.Y.; et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin. Cancer Res. 2014, 20, 6582–6592. [Google Scholar] [CrossRef]
- Li, Y.Y.; Hanna, G.J.; Laga, A.C.; Haddad, R.I.; Lorch, J.H.; Hammerman, P.S. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin. Cancer Res. 2015, 21, 1447–1456. [Google Scholar] [CrossRef]
- Loganathan, S.K.; Schleicher, K.; Malik, A.; Quevedo, R.; Langille, E.; Teng, K.; Oh, R.H.; Rathod, B.; Tsai, R.; Samavarchi-Tehrani, P.; et al. Rare driver mutations in head and neck squamous cell carcinomas converge on NOTCH signaling. Science 2020, 367, 1264–1269. [Google Scholar] [CrossRef]
- Chen, L.; Hayden, M.S.; Gilmore, E.S.; Alexander-Savino, C.; Oleksyn, D.; Gillespie, K.; Zhao, J.; Poligone, B. PKK deletion in basal keratinocytes promotes tumorigenesis after chemical carcinogenesis. Carcinogenesis 2018, 39, 418–428. [Google Scholar] [CrossRef]
- Lee, P.; Jiang, S.; Li, Y.; Yue, J.; Gou, X.; Chen, S.Y.; Zhao, Y.; Schober, M.; Tan, M.; Wu, X. Phosphorylation of Pkp1 by RIPK4 regulates epidermal differentiation and skin tumorigenesis. EMBO J. 2017, 36, 1963–1980. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.R.; Xu, K.; Liu, J.C.; Agamez, N.M.; Loch, A.J.; Wong, R.G.; Wang, W.; Wright, K.L.; Lane, T.F.; Zacksenhaus, E.; et al. Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer Res. 2011, 71, 2706–2717. [Google Scholar] [CrossRef] [PubMed]
- Loganathan, S.K.; Malik, A.; Langille, E.; Luxenburg, C.; Schramek, D. In Vivo CRISPR/Cas9 Screening to Simultaneously Evaluate Gene Function in Mouse Skin and Oral Cavity. J. Vis. Exp. 2020, 165, e61693. [Google Scholar] [CrossRef] [PubMed]
- Beronja, S.; Fuchs, E. RNAi-mediated gene function analysis in skin. Methods Mol. Biol. 2013, 961, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Beronja, S.; Janki, P.; Heller, E.; Lien, W.H.; Keyes, B.E.; Oshimori, N.; Fuchs, E. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature 2013, 501, 185–190. [Google Scholar] [CrossRef]
- Beronja, S.; Livshits, G.; Williams, S.; Fuchs, E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat. Med. 2010, 16, 821–827. [Google Scholar] [CrossRef]
- Schramek, D.; Sendoel, A.; Segal, J.P.; Beronja, S.; Heller, E.; Oristian, D.; Reva, B.; Fuchs, E. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science 2014, 343, 309–313. [Google Scholar] [CrossRef]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 2014, 11, 783–784. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Genovese, G.; Kahler, A.K.; Handsaker, R.E.; Lindberg, J.; Rose, S.A.; Bakhoum, S.F.; Chambert, K.; Mick, E.; Neale, B.M.; Fromer, M.; et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014, 371, 2477–2487. [Google Scholar] [CrossRef]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef]
- Genovese, G.; Jaiswal, S.; Ebert, B.L.; McCarroll, S.A. Clonal hematopoiesis and blood-cancer risk. N. Engl. J. Med. 2015, 372, 1071–1072. [Google Scholar] [CrossRef]
- McKerrell, T.; Park, N.; Moreno, T.; Grove, C.S.; Ponstingl, H.; Stephens, J.; Understanding Society Scientific, G.; Crawley, C.; Craig, J.; Scott, M.A.; et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015, 10, 1239–1245. [Google Scholar] [CrossRef]
- Martincorena, I.; Campbell, P.J. Somatic mutation in cancer and normal cells. Science 2015, 349, 1483–1489. [Google Scholar] [CrossRef]
- Martincorena, I.; Roshan, A.; Gerstung, M.; Ellis, P.; Van Loo, P.; McLaren, S.; Wedge, D.C.; Fullam, A.; Alexandrov, L.B.; Tubio, J.M.; et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015, 348, 880–886. [Google Scholar] [CrossRef]
- Martincorena, I.; Raine, K.M.; Gerstung, M.; Dawson, K.J.; Haase, K.; Van Loo, P.; Davies, H.; Stratton, M.R.; Campbell, P.J. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell 2018, 173, 1823. [Google Scholar] [CrossRef]
- Jonason, A.S.; Kunala, S.; Price, G.J.; Restifo, R.J.; Spinelli, H.M.; Persing, J.A.; Leffell, D.J.; Tarone, R.E.; Brash, D.E. Frequent clones of p53-mutated keratinocytes in normal human skin. Proc. Natl. Acad. Sci. USA 1996, 93, 14025–14029. [Google Scholar] [CrossRef]
- Meier-Stiegen, F.; Schwanbeck, R.; Bernoth, K.; Martini, S.; Hieronymus, T.; Ruau, D.; Zenke, M.; Just, U. Activated Notch1 target genes during embryonic cell differentiation depend on the cellular context and include lineage determinants and inhibitors. PLoS ONE 2010, 5, e11481. [Google Scholar] [CrossRef]
- Nowell, C.S.; Radtke, F. Notch as a tumour suppressor. Nat. Rev. Cancer 2017, 17, 145–159. [Google Scholar] [CrossRef]
- Li, H.; Luo, D.; Huttad, L.; Zhang, M.; Wang, Y.; Feng, J.; Ding, Y.; Han, B. RIPK4 Suppresses the Invasion and Metastasis of Hepatocellular Carcinoma by Inhibiting the Phosphorylation of STAT3. Front. Mol. Biosci. 2021, 8, 654766. [Google Scholar] [CrossRef] [PubMed]
- Yeboah, G.K.; Lobanova, E.S.; Brush, R.S.; Agbaga, M.P. Very long chain fatty acid-containing lipids: A decade of novel insights from the study of ELOVL4. J. Lipid Res. 2021, 62, 100030. [Google Scholar] [CrossRef] [PubMed]
- Blanpain, C.; Lowry, W.E.; Pasolli, H.A.; Fuchs, E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 2006, 20, 3022–3035. [Google Scholar] [CrossRef] [PubMed]
- Nowell, C.; Radtke, F. Cutaneous Notch signaling in health and disease. Cold Spring Harb. Perspect. Med. 2013, 3, a017772. [Google Scholar] [CrossRef]
- Fortugno, P.; Monetta, R.; Belli, M.; Botti, E.; Angelucci, F.; Palmerini, M.G.; Nottola, S.A.; De Luca, C.; Ceccarini, M.; Salvatore, M.; et al. RIPK4 regulates cell-cell adhesion in epidermal development and homeostasis. Hum. Mol. Genet. 2022, 31, 2535–2547. [Google Scholar] [CrossRef]
- Urwyler-Rosselet, C.; Tanghe, G.; Leurs, K.; Gilbert, B.; De Rycke, R.; De Bruyne, M.; Lippens, S.; Bartunkova, S.; De Groote, P.; Niessen, C.; et al. Keratinocyte-Specific Ablation of RIPK4 Allows Epidermal Cornification but Impairs Skin Barrier Formation. J. Investig. Dermatol. 2018, 138, 1268–1278. [Google Scholar] [CrossRef]
- Kondo, S.; Schutte, B.C.; Richardson, R.J.; Bjork, B.C.; Knight, A.S.; Watanabe, Y.; Howard, E.; de Lima, R.L.; Daack-Hirsch, S.; Sander, A.; et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat. Genet. 2002, 32, 285–289. [Google Scholar] [CrossRef]
- Hopiavuori, B.R.; Deak, F.; Wilkerson, J.L.; Brush, R.S.; Rocha-Hopiavuori, N.A.; Hopiavuori, A.R.; Ozan, K.G.; Sullivan, M.T.; Wren, J.D.; Georgescu, C.; et al. Homozygous Expression of Mutant ELOVL4 Leads to Seizures and Death in a Novel Animal Model of Very Long-Chain Fatty Acid Deficiency. Mol. Neurobiol. 2018, 55, 1795–1813. [Google Scholar] [CrossRef]
- Vasireddy, V.; Uchida, Y.; Salem, N., Jr.; Kim, S.Y.; Mandal, M.N.; Reddy, G.B.; Bodepudi, R.; Alderson, N.L.; Brown, J.C.; Hama, H.; et al. Loss of functional ELOVL4 depletes very long-chain fatty acids (> or =C28) and the unique omega-O-acylceramides in skin leading to neonatal death. Hum. Mol. Genet. 2007, 16, 471–482. [Google Scholar] [CrossRef]
- Marinari, B.; Moretti, F.; Botti, E.; Giustizieri, M.L.; Descargues, P.; Giunta, A.; Stolfi, C.; Ballaro, C.; Papoutsaki, M.; Alema, S.; et al. The tumor suppressor activity of IKKalpha in stratified epithelia is exerted in part via the TGF-beta antiproliferative pathway. Proc. Natl. Acad. Sci. USA 2008, 105, 17091–17096. [Google Scholar] [CrossRef]
- Poligone, B.; Gilmore, E.S.; Alexander, C.V.; Oleksyn, D.; Gillespie, K.; Zhao, J.; Ibrahim, S.F.; Pentland, A.P.; Brown, M.D.; Chen, L. PKK suppresses tumor growth and is decreased in squamous cell carcinoma of the skin. J. Investig. Dermatol. 2015, 135, 869–876. [Google Scholar] [CrossRef]
- Zheng, J.M.; Gan, M.F.; Yu, H.Y.; Ye, L.X.; Yu, Q.X.; Xia, Y.H.; Zhou, H.X.; Bao, J.Q.; Guo, Y.Q. KDF1, a Novel Tumor Suppressor in Clear Cell Renal Cell Carcinoma. Front. Oncol. 2021, 11, 686678. [Google Scholar] [CrossRef]
- Li, Y.; Tang, L.; Yue, J.; Gou, X.; Lin, A.; Weatherbee, S.D.; Wu, X. Regulation of epidermal differentiation through KDF1-mediated deubiquitination of IKKalpha. EMBO Rep. 2020, 21, e48566. [Google Scholar] [CrossRef]
- Fomenkov, A.; Zangen, R.; Huang, Y.P.; Osada, M.; Guo, Z.; Fomenkov, T.; Trink, B.; Sidransky, D.; Ratovitski, E.A. RACK1 and stratifin target DeltaNp63alpha for a proteasome degradation in head and neck squamous cell carcinoma cells upon DNA damage. Cell Cycle 2004, 3, 1285–1295. [Google Scholar] [CrossRef]
| Gene Name | Forward Primers (5′-3′) | Reverse Primers (5′-3′) |
|---|---|---|
| Ppib | caacgataagaagaagggacctaaa | cgtcctacagattcatctccaattt |
| Ripk4 | tagacctgaagccagcgaac | tgctgaggtcatgagagtgg |
| Irf6 | tccccttcctgaacatcaac | gttctgttttgggccacact |
| Elovl4 | actatgggctgactgcgttc | gggcagtcggtgtagagaga |
| Grhl3 | tgtggaatgtcaacgaggaa | gcgaggagaagtctgtgctc |
| Ovol1 | gcgagatctacgtgccagtc | caccgatgcctctggttc |
| Grhl1 | ataacgccatttccttcacg | ttgaatgttgagtggcaagc |
| Dsc1 | gccccatattttgaaaccaa | ttcctggctcatccttgtct |
| Ocln | cgtctagataaagagctggatga | ctgcagatcccttaacttgctt |
| LipG | ggcgaattcgtgtcaaatct | tgggtcttgagtgcaaaatg |
| Susd2 | tgctgaatcagaaagtgctca | gccactgacaggaacatgc |
| Endod1 | ctggagccgcagattgat | attcaaggcttgcttgcttc |
| Hprt1 | gatcagtcaacgggggacataaa | cttgcgctcatcttaggctttgt |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Y.; Gauthier, M.-A.; Malik, A.; Fotiadou, I.; Ostrovski, M.; Dervovic, D.; Ghadban, L.; Tsai, R.; Gish, G.; Loganathan, S.K.; et al. The NOTCH-RIPK4-IRF6-ELOVL4 Axis Suppresses Squamous Cell Carcinoma. Cancers 2023, 15, 737. https://doi.org/10.3390/cancers15030737
Yan Y, Gauthier M-A, Malik A, Fotiadou I, Ostrovski M, Dervovic D, Ghadban L, Tsai R, Gish G, Loganathan SK, et al. The NOTCH-RIPK4-IRF6-ELOVL4 Axis Suppresses Squamous Cell Carcinoma. Cancers. 2023; 15(3):737. https://doi.org/10.3390/cancers15030737
Chicago/Turabian StyleYan, Yue, Marc-Andre Gauthier, Ahmad Malik, Iosifina Fotiadou, Michael Ostrovski, Dzana Dervovic, Logine Ghadban, Ricky Tsai, Gerald Gish, Sampath Kumar Loganathan, and et al. 2023. "The NOTCH-RIPK4-IRF6-ELOVL4 Axis Suppresses Squamous Cell Carcinoma" Cancers 15, no. 3: 737. https://doi.org/10.3390/cancers15030737
APA StyleYan, Y., Gauthier, M.-A., Malik, A., Fotiadou, I., Ostrovski, M., Dervovic, D., Ghadban, L., Tsai, R., Gish, G., Loganathan, S. K., & Schramek, D. (2023). The NOTCH-RIPK4-IRF6-ELOVL4 Axis Suppresses Squamous Cell Carcinoma. Cancers, 15(3), 737. https://doi.org/10.3390/cancers15030737





