Generation of Stable Epithelial–Mesenchymal Hybrid Cancer Cells with Tumorigenic Potential
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
2.2. Antibodies and Reagents
2.3. Anoikis Assay
2.4. Invasion Assay
2.5. RNA Sequencing
2.6. q-PCR
2.7. Protein Extraction and Quantification
2.8. SDS Polyacrylamide Gel Electrophoresis and Western Blot Analysis
2.9. Tumorigenic Potential In Vivo Assay
2.10. Statistical Analysis
3. Results
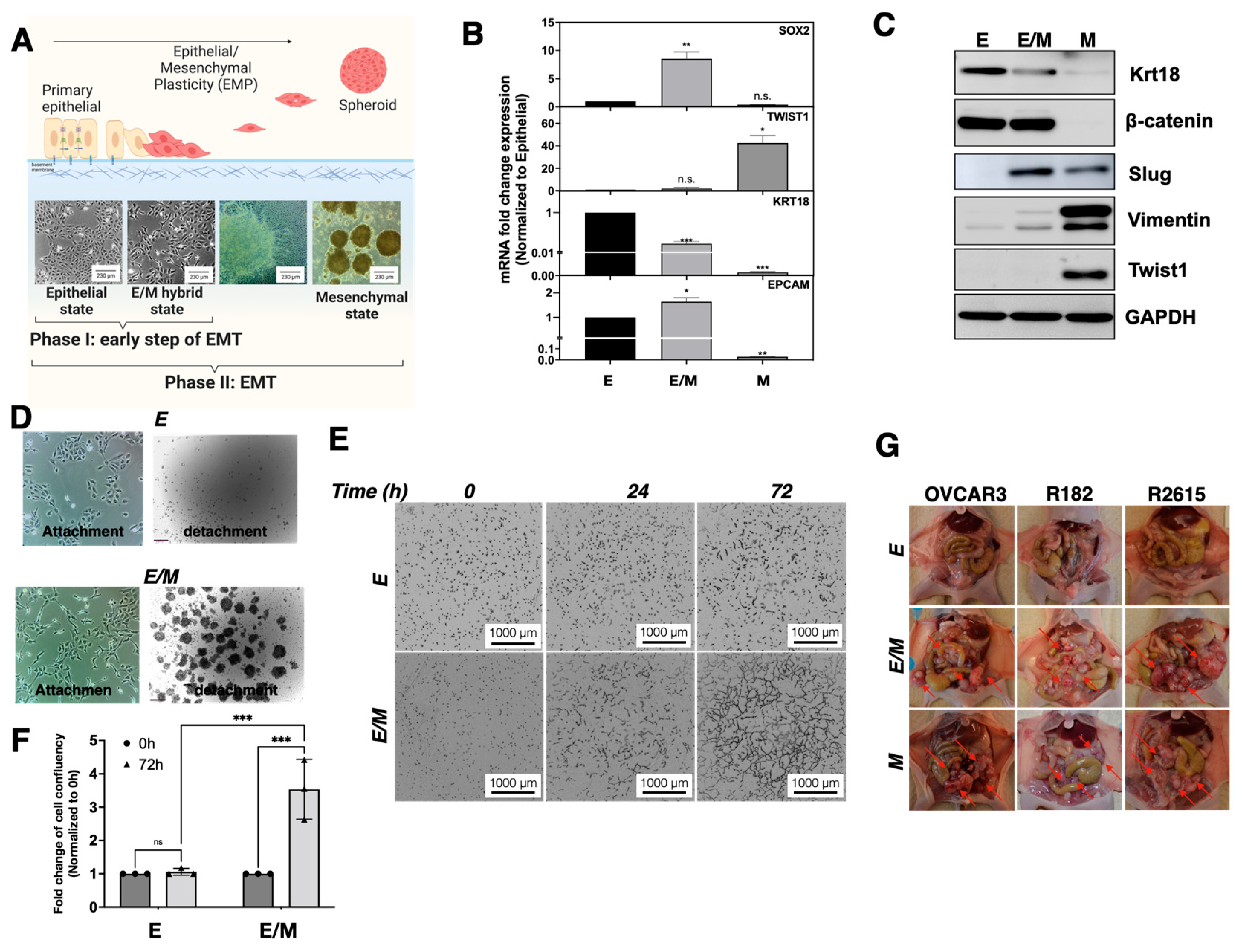
3.1. Characterization of Epithelial Ovarian Cancer Cell Lines Based on Their Epithelial and Mesenchymal Markers
3.2. Identification of a Tumorigenic E/M Hybrid State in Ovarian Cancer
3.3. Characterization of E/M Hybrid State
3.4. Characterization of Late EMT Differentiation Stage
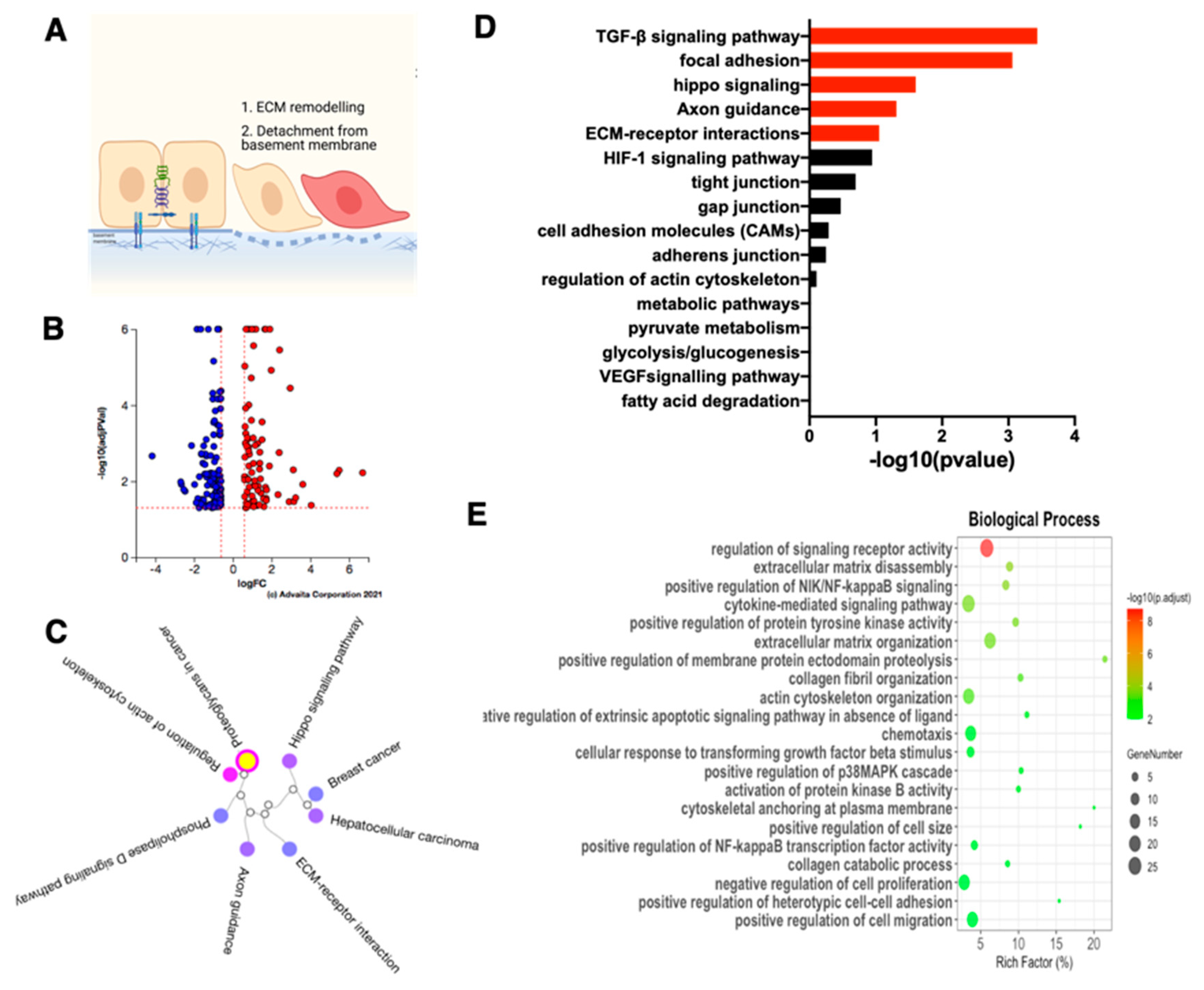
3.5. ECM Disassembly and Organization Are the First Step in Early Phase of EMT
3.6. Rearrangement of Focal Adhesion Is the Next Step in E/M Hybrid State
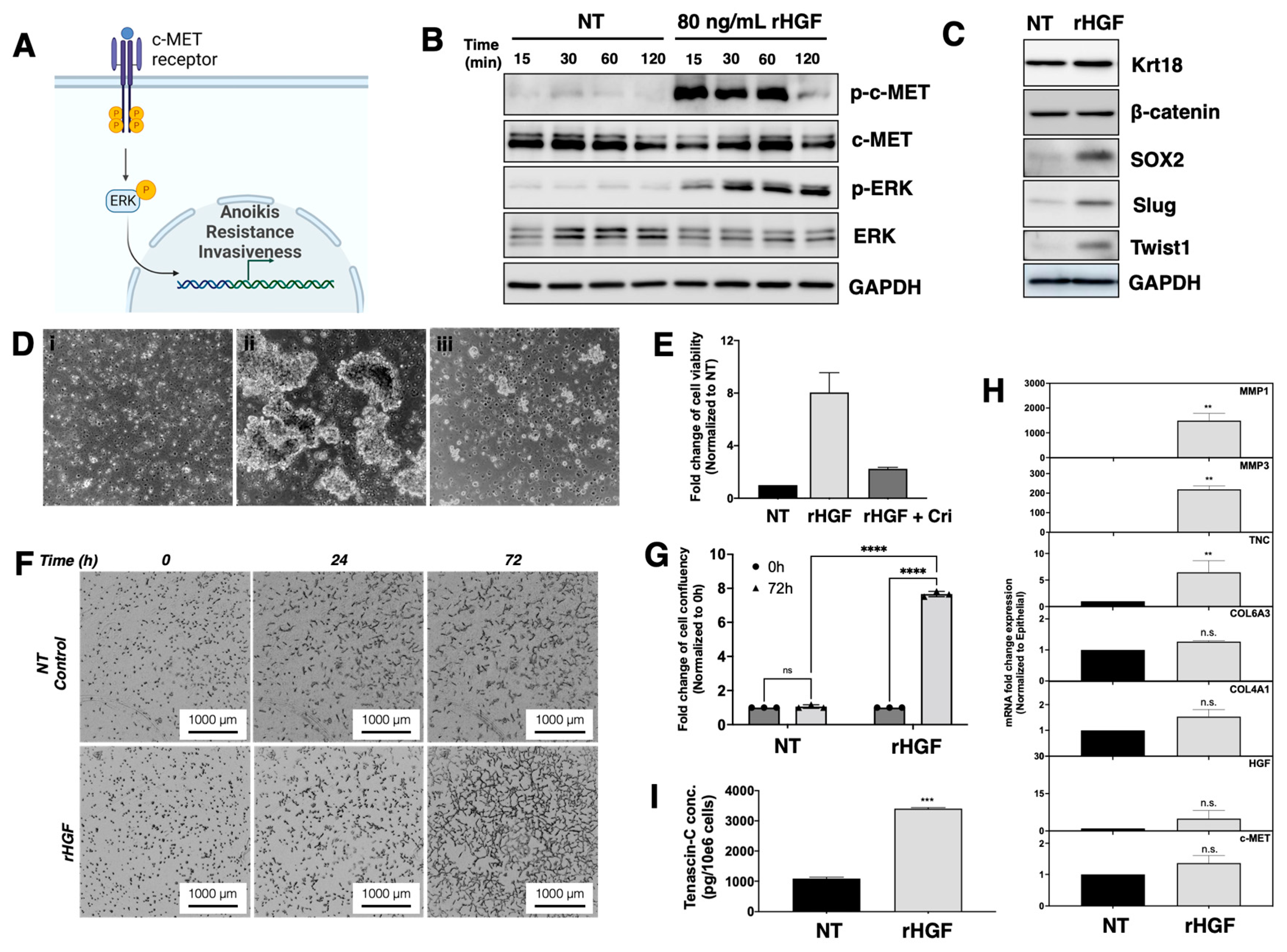
3.7. HGF Induce Epithelial Cells to Enter the E/M Hybrid State
3.8. HGF Induces Anoikis Resistance in EOCCs
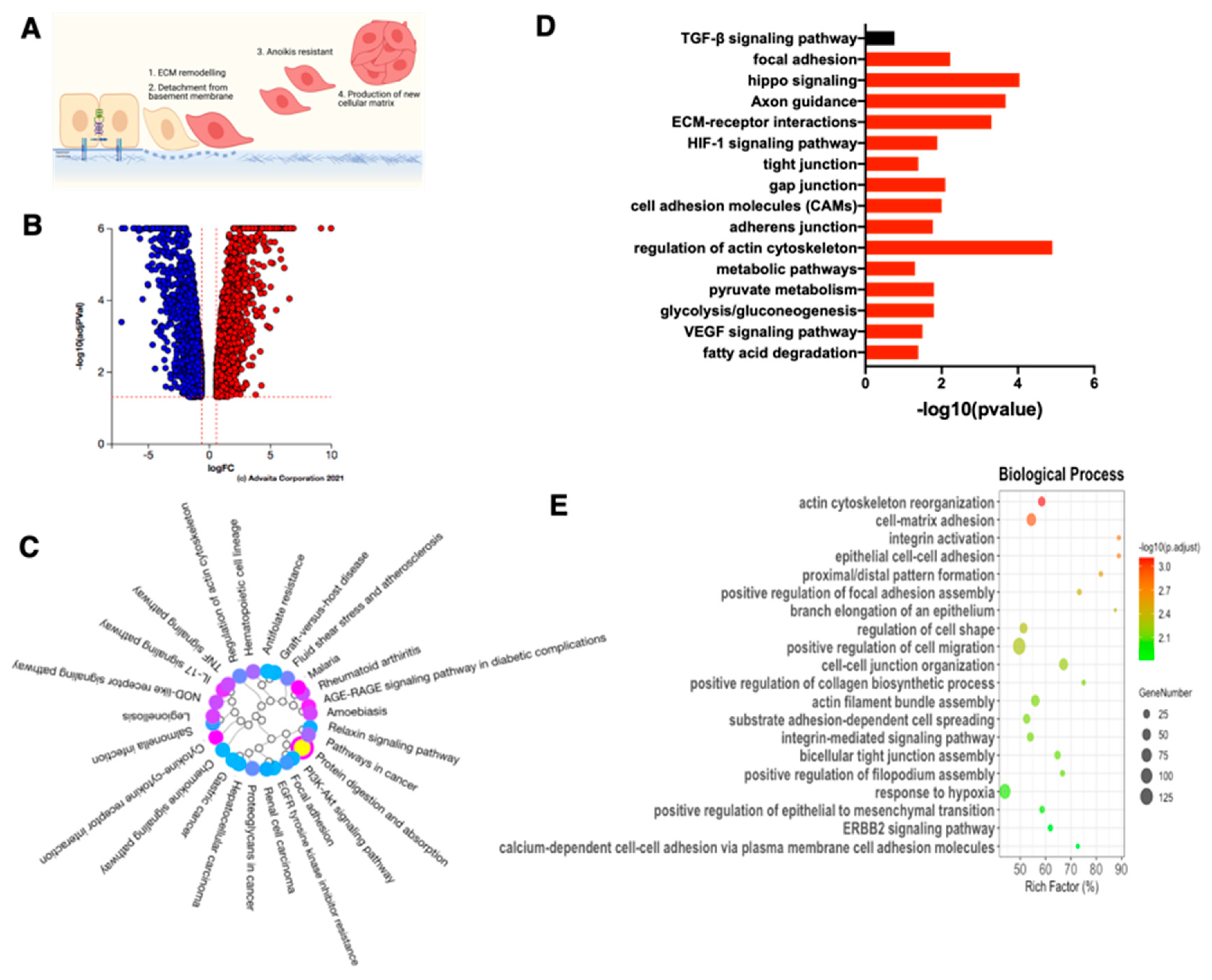
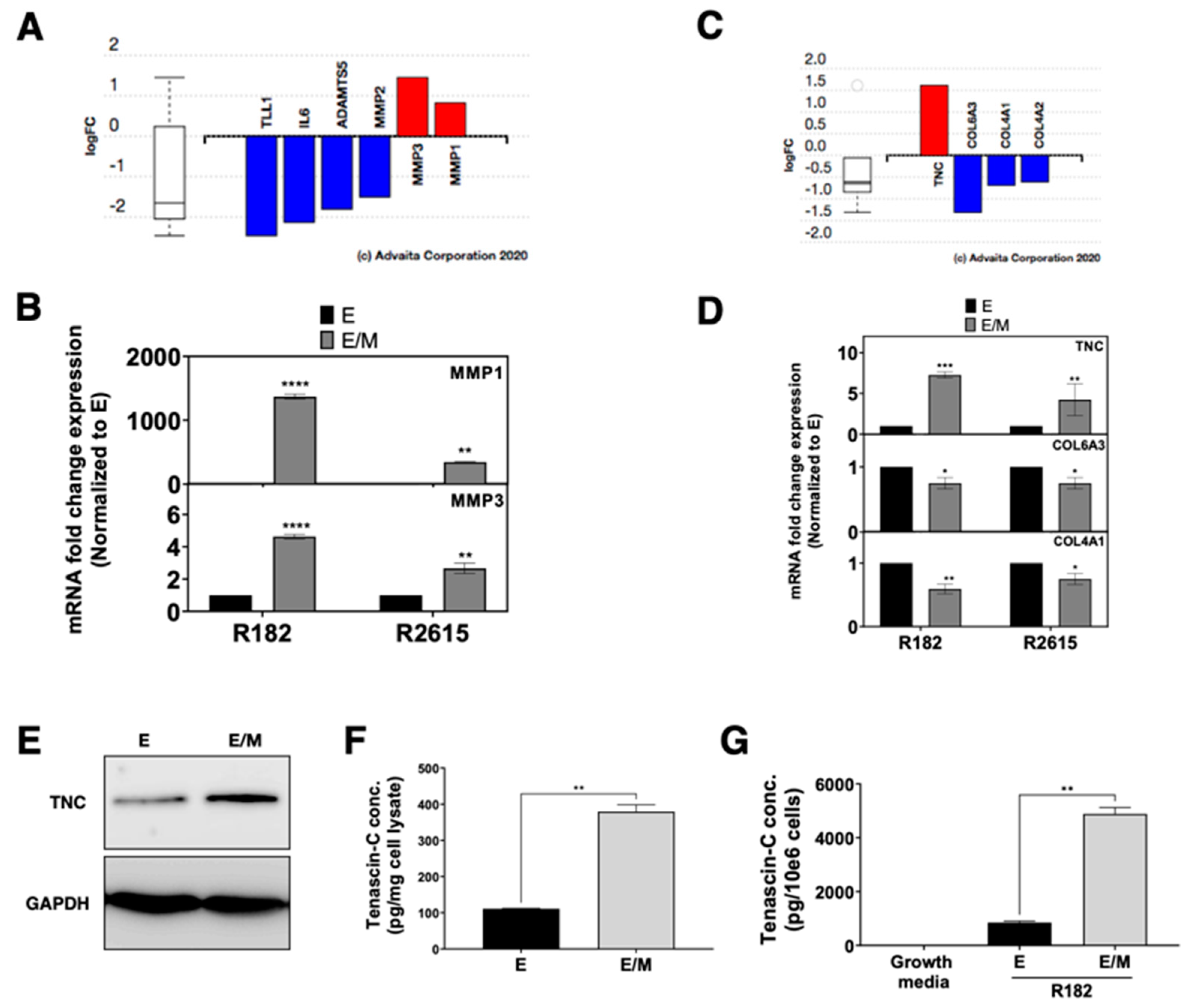
3.9. Role of HGF on ECM Assembly and Focal Adhesion
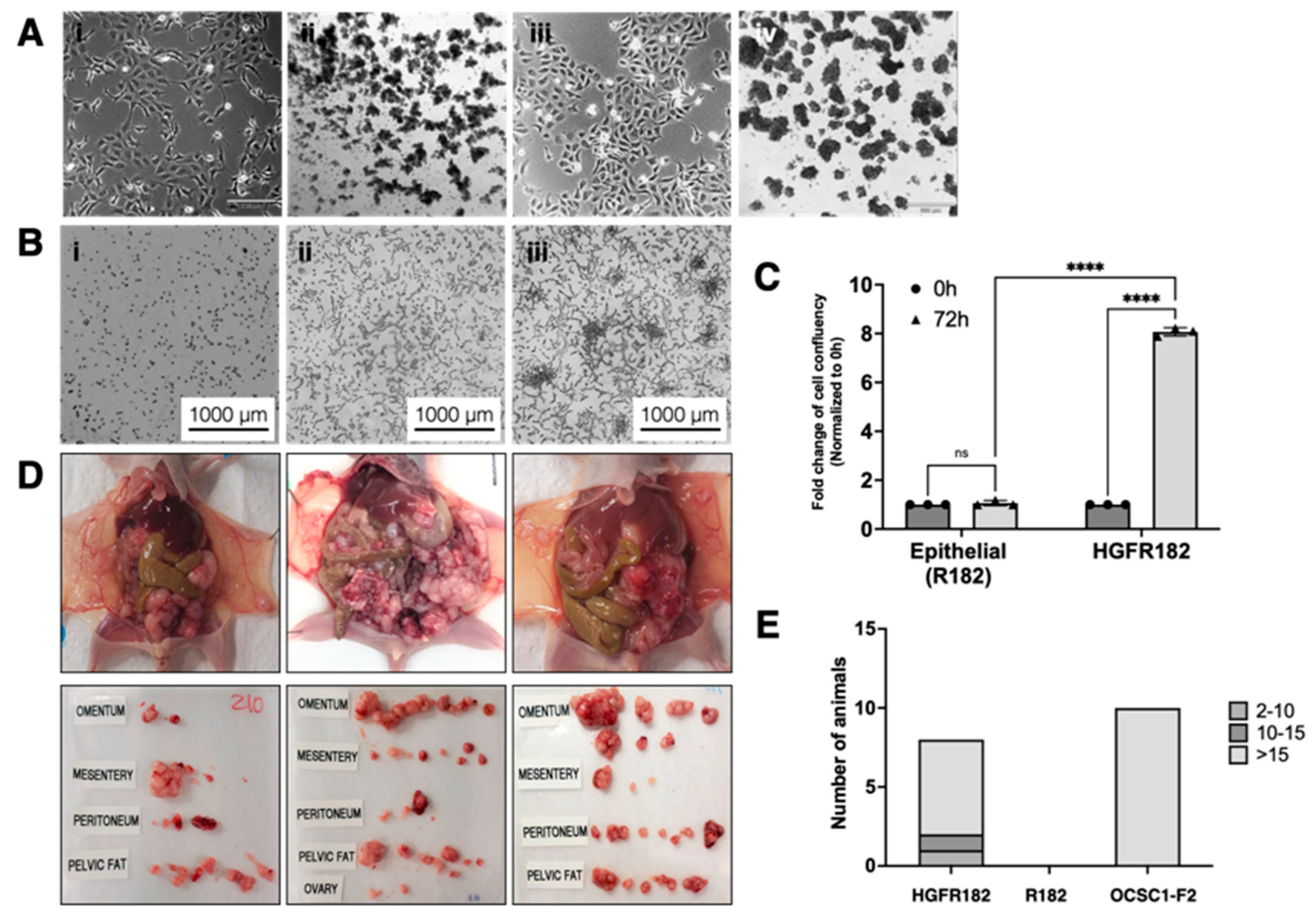
3.10. HGF Promotes Matrix Degradation and Invasion Capacity in EOCCs
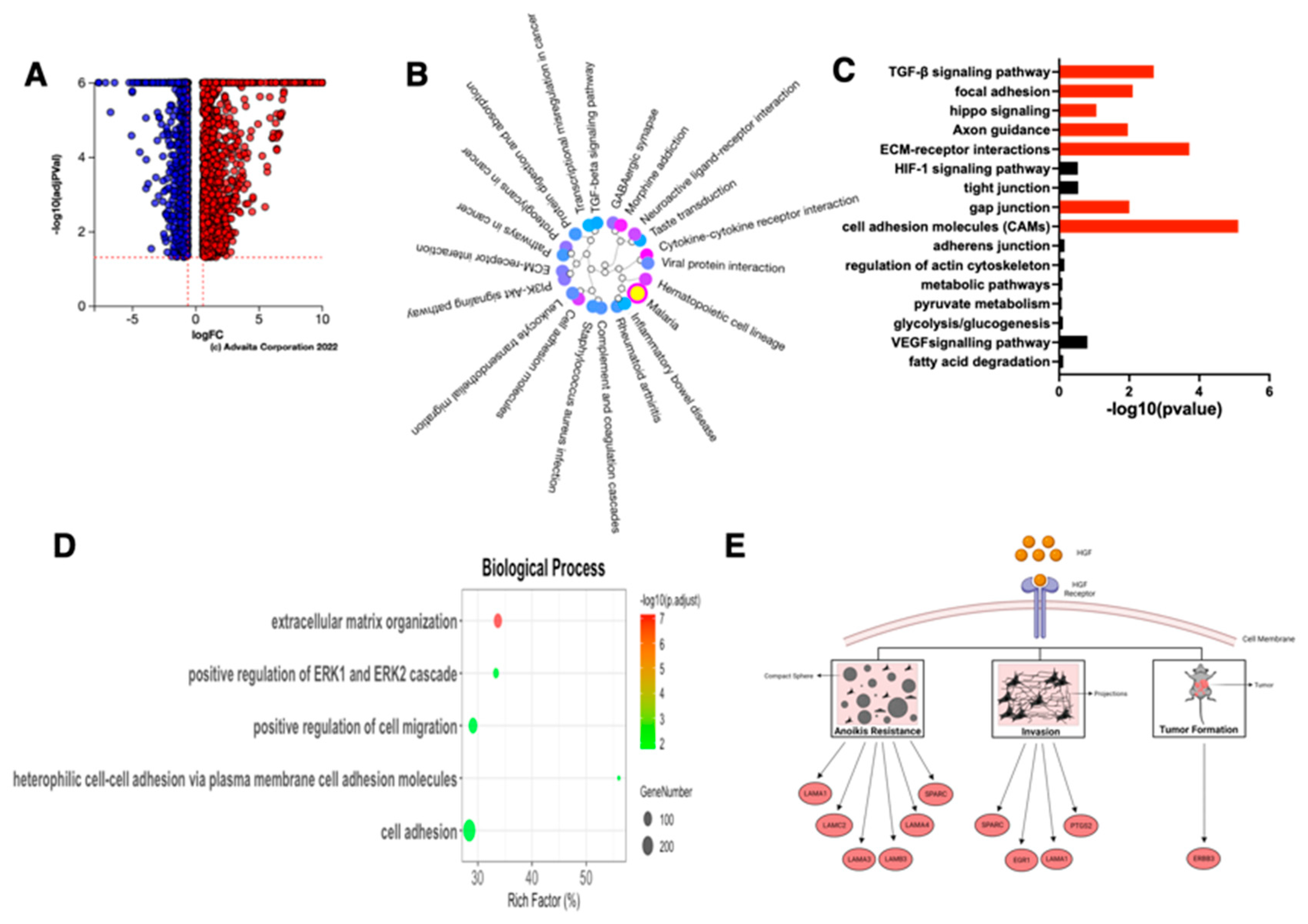
3.11. HGF Induce Reprograming in the Epithelial Ovarian Cancer Cells
3.12. HGF-Induced E/M Hybrid State Cells Acquire Tumorigenic Potential
3.13. Characteristics of the HGF-Induced E/M Hybrid State EOCCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional review board statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thiery, J.P. Epithelial—Mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol. 2003, 15, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Banyard, J.; Bielenberg, D.R. The role of EMT and MET in cancer dissemination. Connect. Tissue Res. 2015, 56, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.Y.-J.; Guilford, P.; Thiery, J.P. Early events in cell adhesion and polarity during epithelial-mesenchymal transition. J. Cell Sci. 2012, 125, 4417–4422. [Google Scholar] [CrossRef] [PubMed]
- Wells, A.; Chao, Y.L.; Grahovac, J.; Wu, Q.; Lauffenburger, D.A. Epithelial and mesenchymal phenotypic switchings modulate cell motility in metastasis. Front. Biosci. 2011, 16, 815–837. [Google Scholar] [CrossRef]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, C.; Alvero, A.B.; Yun, B.S.; Mor, G. Redefining the origin and evolution of ovarian cancer: A hormonal connection. Endocrine-Related Cancer 2016, 23, R411–R422. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.M.; Cardenas, C.; Tedja, R. The Role of Intra-Tumoral Heterogeneity and Its Clinical Relevance in Epithelial Ovarian Cancer Recurrence and Metastasis. Cancers 2019, 11, 1083. [Google Scholar] [CrossRef]
- Ahmed, N.; Thompson, E.W.; Quinn, M.A. Epithelial—mesenchymal interconversions in normal ovarian surface epithelium and ovarian carcinomas: An exception to the norm. J. Cell Physiol. 2007, 213, 581–588. [Google Scholar] [CrossRef]
- Li, J.; Alvero, A.B.; Nuti, S.; Tedja, R.; Roberts, C.M.; Pitruzzello, M.; Li, Y.; Xiao, Q.; Zhang, S.; Gan, Y.; et al. CBX7 binds the E-box to inhibit TWIST-1 function and inhibit tumorigenicity and metastatic potential. Oncogene 2020, 39, 3965–3979. [Google Scholar] [CrossRef]
- Yang-Hartwich, Y.; Tedja, R.; Roberts, C.M.; Goodner-Bingham, J.; Cardenas, C.; Gurea, M.; Sumi, N.J.; Alvero, A.B.; Glackin, C.A.; Mor, G. p53—Pirh2 Complex Promotes Twist1 Degradation and Inhibits EMT. Mol. Cancer Res. 2019, 17, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, C.; Tedja, R.; Roberts, C.M.; Goodner-Bingham, J.; Cardenas, C.; Gurea, M.; Sumi, N.J.; Alvero, A.B.; Glackin, C.A.; Mor, G. Adipocyte microenvironment promotes Bclxl expression and confers chemoresistance in ovarian cancer cells. Apoptosis 2006, 22, 558–569. [Google Scholar] [CrossRef]
- Kelly, M.G.; Alvero, A.B.; Chen, R.; Silasi, D.-A.; Abrahams, V.M.; Chan, S.; Visintin, I.; Rutherford, T.; Mor, G. TLR-4 Signaling Promotes Tumor Growth and Paclitaxel Chemoresistance in Ovarian Cancer. Cancer Res. 2006, 66, 3859–3868. [Google Scholar] [CrossRef] [PubMed]
- Alvero, A.B.; Chen, R.; Fu, H.-H.; Montagna, M.; Schwartz, P.E.; Rutherford, T.; Silasi, D.-A.; Steffensen, K.D.; Waldstrom, M.; Visintin, I.; et al. Molecular phenotyping of human ovarian cancer stem cells unravels the mechanisms for repair and chemoresistance. Cell Cycle 2009, 8, 158–166. [Google Scholar] [CrossRef]
- Alvero, A.B.; Montagna, M.K.; Craveiro, V.; Liu, L.; Mor, G. Distinct subpopulations of epithelial ovarian cancer cells can differentially induce macrophages and T regulatory cells toward a pro-tumor phenotype. Am. J. Reprod. Immunol. 2012, 67, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Tedja, R.; Fox, A.; Alvero, A.B. Detection of Cell Death Mechanisms: Methods and Protocols; Alvero, A.B., Moreds, G.G., Eds.; Springer: Louisville, KY, USA, 2021; pp. 69–76. [Google Scholar]
- Tedja, R.; Roberts, C.M.; Alvero, A.B.; Cardenas, C.; Yang-Hartwich, Y.; Spadinger, S.; Pitruzzello, M.; Yin, G.; Glackin, C.A.; Mor, G. Protein kinase Cα-mediated phosphorylation of Twist1 at Ser-144 prevents Twist1 ubiquitination and stabilizes it. J. Biol. Chem. 2019, 294, 5082–5093. [Google Scholar] [CrossRef] [PubMed]
- Alvero, A.B.; Heaton, A.; Lima, E.; Pitruzzello, M.; Sumi, N.; Yang-Hartwich, Y.; Cardenas, C.; Steinmacher, S.; Silasi, D.-A.; Brown, D.; et al. TRX-E-002-1 Induces c-Jun—Dependent Apoptosis in Ovarian Cancer Stem Cells and Prevents Recurrence In Vivo. Mol. Cancer Ther. 2016, 15, 1279–1290. [Google Scholar] [CrossRef]
- Yin, G.; Alvero, A.B.; Craveiro, V.; Holmberg, J.C.; Fu, H.-H.; Montagna, M.K.; Yang, Y.; Chefetz-Menaker, I.; Nuti, S.; Rossi, M.; et al. Constitutive proteasomal degradation of TWIST-1 in epithelial–ovarian cancer stem cells impacts differentiation and metastatic potential. Oncogene 2013, 32, 39–49. [Google Scholar] [CrossRef]
- Yang-Hartwich, Y.; Romanoff, E.; Bingham, J.; Alvero, A.B.; Mor, G. Detection of p53 protein transcriptional activity by chromatin immunoprecipitation. Methods Mol. Biol. 2015, 1219, 87–93. [Google Scholar] [CrossRef]
- Yang-Hartwich, Y.; Bingham, J.; Garofalo, F.; Alvero, A.B.; Mor, G. Detection of p53 protein aggregation in cancer cell lines and tumor samples. Methods Mol. Biol. 2015, 1219, 75–86. [Google Scholar] [CrossRef]
- Tan, T.Z.; Miow, Q.H.; Miki, Y.; Noda, T.; Mori, S.; Huang, R.Y.-J.; Thiery, J.P. Epithelial-mesenchymal transition spectrum quantification and its efficacy in deciphering survival and drug responses of cancer patients. EMBO Mol. Med. 2014, 6, 1279–1293. [Google Scholar] [CrossRef]
- Vesuna, F.; van Diest, P.; Chen, J.H.; Raman, V. Twist is a transcriptional repressor of E-cadherin gene expression in breast cancer. Biochem. Biophys. Res. Commun. 2008, 367, 235–241. [Google Scholar] [CrossRef]
- Conacci-Sorrell, M.; Simcha, I.; Ben-Yedidia, T.; Blechman, J.; Savagner, P.; Ben-Ze’Ev, A. Autoregulation of E-cadherin expression by cadherin—cadherin interactions: The roles of beta-catenin signaling, Slug, and MAPK. J. Cell Biol. 2003, 163, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.Y.-J.; Wong, M.K.; Tan, T.Z.; Kuay, K.T.; Ng, A.H.C.; Chung, V.Y.; Chu, Y.-S.; Matsumura, N.; Lai, H.-C.; Lee, Y.F.; et al. An EMT spectrum defines an anoikis-resistant and spheroidogenic intermediate mesenchymal state that is sensitive to e-cadherin restoration by a src-kinase inhibitor, saracatinib (AZD0530). Cell Death Dis. 2013, 4, e915. [Google Scholar] [CrossRef]
- Kapoor, C.; Vaidya, S.; Wadhwan, V.; Hitesh; Kaur, G.; Pathak, A. Seesaw of matrix metalloproteinases (MMPs). J. Cancer Res. Ther. 2016, 12, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Mehner, C.; Miller, E.; Nassar, A.; Bamlet, W.R.; Radisky, E.S.; Radisky, D.C. Tumor cell expression of MMP3 as a prognostic factor for poor survival in pancreatic, pulmonary, and mammary carcinoma. Genes Cancer 2015, 6, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, C.M.; Dolgonos, L.; Zemans, R.L.; Young, S.K.; Robertson, J.; Briones, N.; Suzuki, T.; Campbell, M.N.; Gauldie, J.; Radisky, D.C.; et al. Matrix metalloproteinase 3 is a mediator of pulmonary fibrosis. Am. J. Pathol. 2011, 179, 1733–1745. [Google Scholar] [CrossRef] [PubMed]
- Murray, G.I.; Duncan, M.E.; O’Neil, P.; Melvin, W.T.; Fothergill, J.E. Matrix metalloproteinase–1 is associated with poor prognosis in colorectal cancer. Nat. Med. 1996, 2, 461–462. [Google Scholar] [CrossRef]
- Zinzindohoué, F.; Lecomte, T.; Ferraz, J.-M.; Houllier, A.-M.; Cugnenc, P.-H.; Berger, A.; Blons, H.; Laurent-Puig, P. Prognostic Significance of MMP-1 and MMP-3 Functional Promoter Polymorphisms in Colorectal Cancer. Clin. Cancer Res. 2005, 11, 594–599. [Google Scholar] [CrossRef]
- Midwood, K.S.; Chiquet, M.; Tucker, R.P.; Orend, G. Tenascin-C at a glance. J. Cell Sci. 2016, 129, 4321–4327. [Google Scholar] [CrossRef]
- Huang, W.; Chiquet-Ehrismann, R.; Moyano, J.V.; Garcia-Pardo, A.; Orend, G. Interference of tenascin-C with syndecan-4 binding to fibronectin blocks cell adhesion and stimulates tumor cell proliferation. Cancer Res. 2001, 61, 8586–8594. [Google Scholar] [PubMed]
- Nagaharu, K.; Zhang, X.; Yoshida, T.; Katoh, D.; Hanamura, N.; Kozuka, Y.; Ogawa, T.; Shiraishi, T.; Imanaka-Yoshida, K. Tenascin C Induces Epithelial-Mesenchymal Transition-Like Change Accompanied by SRC Activation and Focal Adhesion Kinase Phosphorylation in Human Breast Cancer Cells. Am. J. Pathol. 2011, 178, 754–763. [Google Scholar] [CrossRef] [PubMed]
- De Wever, O.; Nguyen, Q.-D.; Van Hoorde, L.; Bracke, M.; Bruyneel, E.; Gespach, C.; Mareel, M. Tenascin-C and SF/HGF produced by myofibroblasts in vitro provide convergent proinvasive signals to human colon cancer cells through RhoA and Rac. FASEB J. 2004, 18, 1016–1018. [Google Scholar] [CrossRef] [PubMed]
- Tsou, H.-K.; Chen, H.-T.; Hung, Y.-H.; Chang, C.-H.; Li, T.-M.; Fong, Y.-C.; Tang, C.-H. HGF and c-Met Interaction Promotes Migration in Human Chondrosarcoma Cells. PLoS ONE 2013, 8, e53974. [Google Scholar] [CrossRef]
- Yu, G.; Wang, Z.; Zeng, S.; Liu, S.; Zhu, C.; Xu, R.; Liu, R.-E. Paeoniflorin Inhibits Hepatocyte Growth Factor- (HGF-) Induced Migration and Invasion and Actin Rearrangement via Suppression of c-Met-Mediated RhoA/ROCK Signaling in Glioblastoma. BioMed Res. Int. 2019, 2019, 9053295. [Google Scholar] [CrossRef]
- Diao, B.; Yang, P. Comprehensive Analysis of the Expression and Prognosis for Laminin Genes in Ovarian Cancer. Pathol. Oncol. Res. 2021, 27, 1609855. [Google Scholar] [CrossRef]
- Cheng, J.-C.; Chang, H.-M.; Leung, P.C.K. Egr-1 mediates epidermal growth factor-induced downregulation of E-cadherin expression via Slug in human ovarian cancer cells. Oncogene 2013, 32, 1041–1049. [Google Scholar] [CrossRef]
- Zhang, X.; Yan, K.; Deng, L.; Liang, J.; Liang, H.; Feng, D.; Ling, B. Cyclooxygenase 2 Promotes Proliferation and Invasion in Ovarian Cancer Cells via the PGE2/NF-κB Pathway. Cell Transplant. 2019, 28, 1s–13s. [Google Scholar] [CrossRef]
- Chen, J.; Wang, M.; Xi, B.; Xue, J.; He, D.; Zhang, J.; Zhao, Y. SPARC is a key regulator of proliferation, apoptosis and invasion in human ovarian cancer. PLoS ONE 2012, 7, e42413. [Google Scholar] [CrossRef]
- Chen, C.; Gupta, P.; Parashar, D.; Nair, G.G.; George, J.; Geethadevi, A.; Wang, W.; Tsaih, S.-W.; Bradley, W.; Ramchandran, R.; et al. ERBB3-induced furin promotes the progression and metastasis of ovarian cancer via the IGF1R/STAT3 signaling axis. Oncogene 2020, 39, 2921–2933. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tedja, R.; Alvero, A.B.; Fox, A.; Cardenas, C.; Pitruzzello, M.; Chehade, H.; Bawa, T.; Adzibolosu, N.; Gogoi, R.; Mor, G. Generation of Stable Epithelial–Mesenchymal Hybrid Cancer Cells with Tumorigenic Potential. Cancers 2023, 15, 684. https://doi.org/10.3390/cancers15030684
Tedja R, Alvero AB, Fox A, Cardenas C, Pitruzzello M, Chehade H, Bawa T, Adzibolosu N, Gogoi R, Mor G. Generation of Stable Epithelial–Mesenchymal Hybrid Cancer Cells with Tumorigenic Potential. Cancers. 2023; 15(3):684. https://doi.org/10.3390/cancers15030684
Chicago/Turabian StyleTedja, Roslyn, Ayesha B. Alvero, Alexandra Fox, Carlos Cardenas, Mary Pitruzzello, Hussein Chehade, Tejeshwhar Bawa, Nicholas Adzibolosu, Radhika Gogoi, and Gil Mor. 2023. "Generation of Stable Epithelial–Mesenchymal Hybrid Cancer Cells with Tumorigenic Potential" Cancers 15, no. 3: 684. https://doi.org/10.3390/cancers15030684
APA StyleTedja, R., Alvero, A. B., Fox, A., Cardenas, C., Pitruzzello, M., Chehade, H., Bawa, T., Adzibolosu, N., Gogoi, R., & Mor, G. (2023). Generation of Stable Epithelial–Mesenchymal Hybrid Cancer Cells with Tumorigenic Potential. Cancers, 15(3), 684. https://doi.org/10.3390/cancers15030684






