EGFR Pathway Expression Persists in Recurrent Glioblastoma Independent of Amplification Status
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Clinical Information
2.2. FISH Evaluation of EGFR Amplification
2.3. Nucleic Acid Extraction, RNA Sequencing
2.4. RNA-Seq Data Post-Processing and Normalisation
2.5. Analysis of Tumour Subtypes
2.6. Differential Expression Analysis
2.7. Pathway Expression among Primary and Recurrent Glioblastoma Samples
2.8. Survival Analyses
2.9. Validation Cohort
3. Results
3.1. Cohort Characteristics
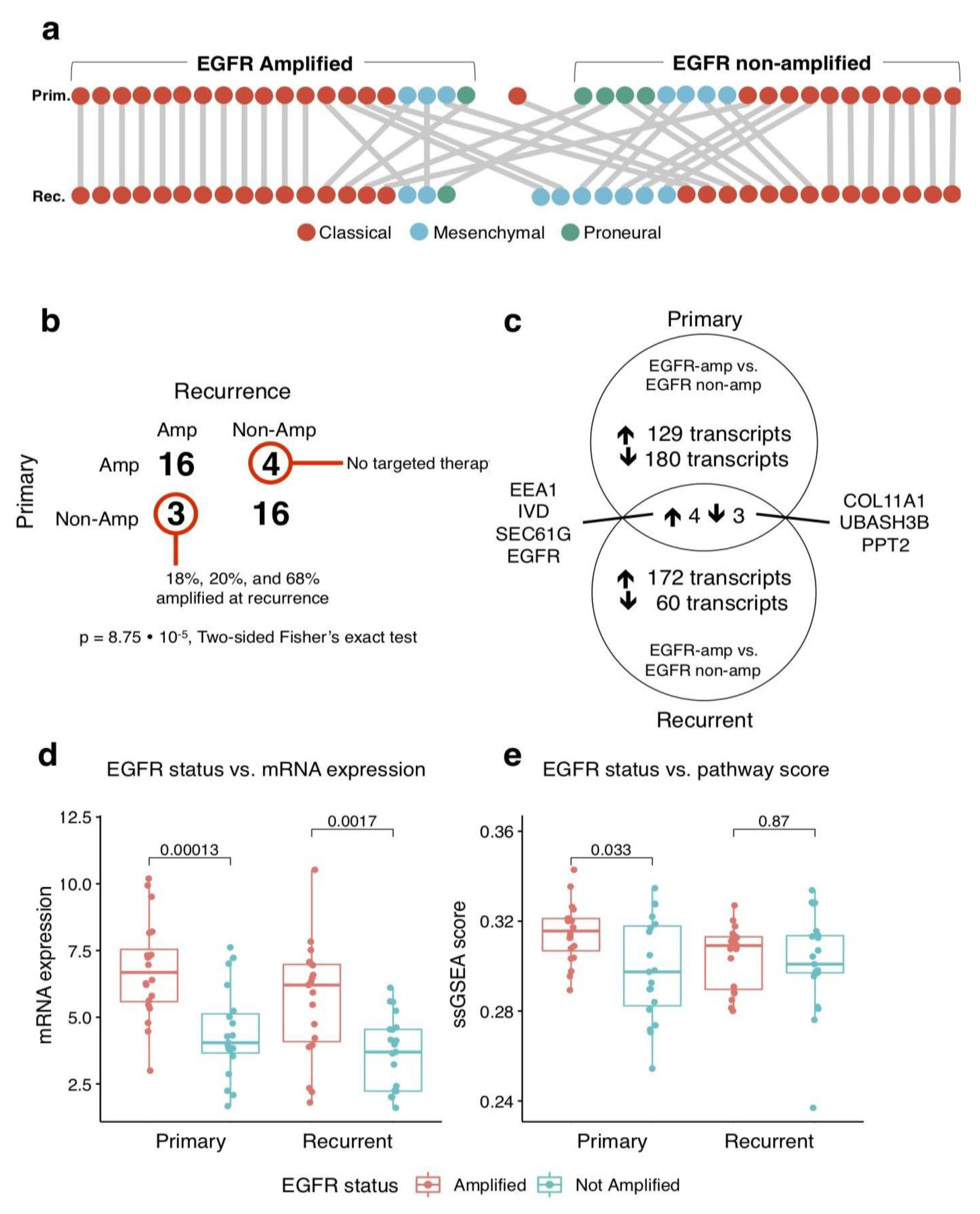
3.2. EGFR Amplification Remains Stable in Recurrence
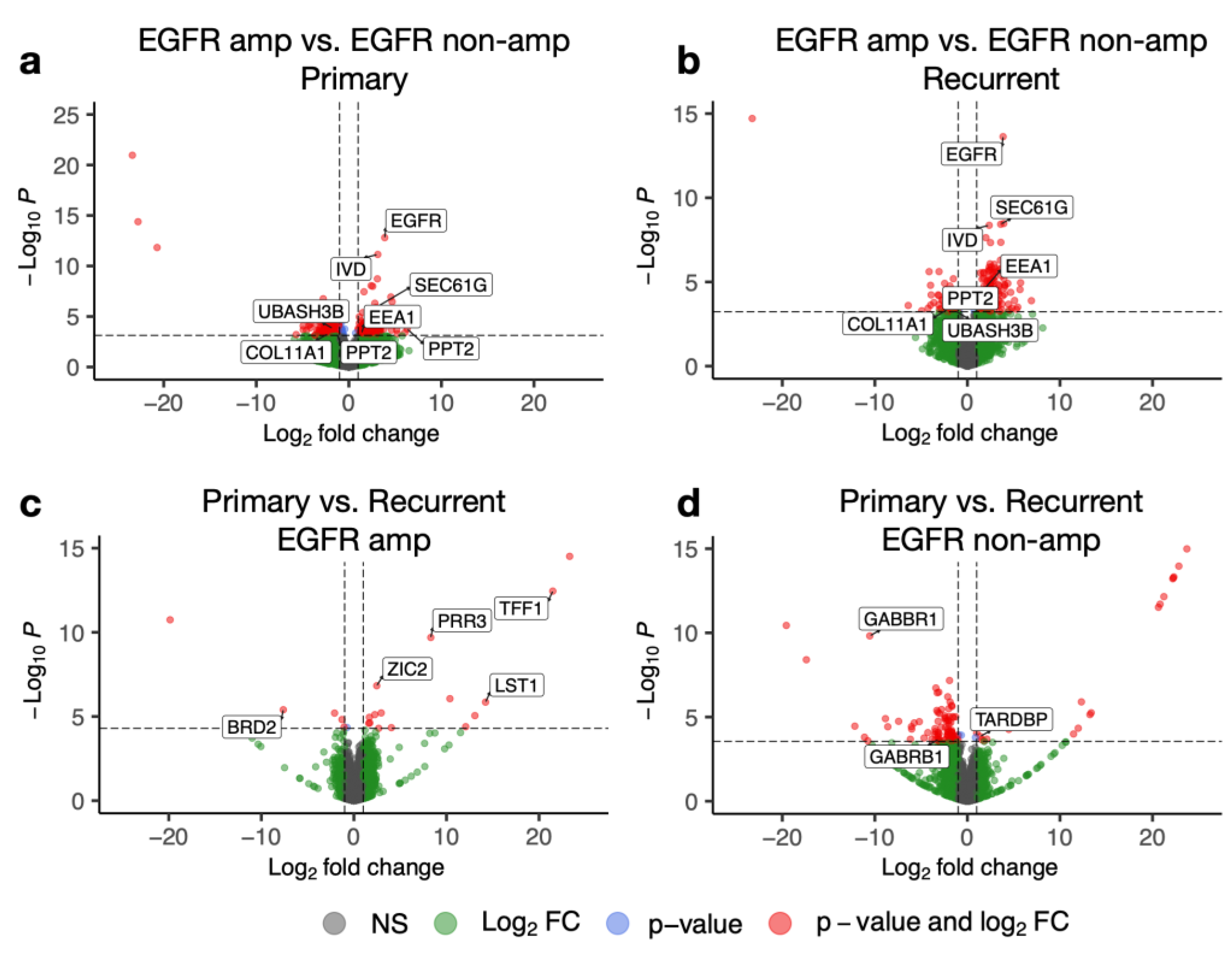
3.3. Recurrent Tumours Show Elevated EGFR Pathway Activity
3.4. Recurrent EGFR-Amplified Tumours Display a Distinct Gene Expression Profile with Increased BRD2 Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barthel, F.P.; Johnson, K.C.; Varn, F.S.; Moskalik, A.D.; Tanner, G.; Kocakavuk, E.; Anderson, K.J.; Abiola, O.; Aldape, K.; Alfaro, D.A.; et al. Longitudinal molecular trajectories of diffuse glioma in adults. Nature 2019, 576, 112–120. [Google Scholar] [CrossRef]
- Touat, M.; Li, Y.Y.; Boynton, A.N.; Spurr, L.F.; Iorgulescu, J.B.; Bohrson, C.L.; Cortes-Ciriano, I.; Birzu, C.; Geduldig, J.E.; Pelton, K.; et al. Mechanisms and therapeutic implications of hypermutation in gliomas. Nature 2020, 580, 517–523. [Google Scholar] [CrossRef]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- An, Z.; Aksoy, O.; Zheng, T.; Fan, Q.-W.; Weiss, W.A. Epidermal growth factor receptor and EGFRvIII in glioblastoma: Signaling pathways and targeted therapies. Oncogene 2018, 37, 1561–1575. [Google Scholar] [CrossRef]
- Gan, H.K.; Reardon, D.A.; Lassman, A.B.; Merrell, R.; Van Den Bent, M.; Butowski, N.; Lwin, Z.; Wheeler, H.; Fichtel, L.; Scott, A.M.; et al. Safety, pharmacokinetics, and antitumor response of depatuxizumab mafodotin as monotherapy or in combination with temozolomide in patients with glioblastoma. Neuro. Oncol. 2018, 20, 838–847. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.C.; Bady, P.; Kamoshima, Y.; Kouwenhoven, M.; Delorenzi, M.; Lambiv, W.L.; Hamou, M.F.; Matter, M.S.; Koch, A.; et al. Pathway analysis of glioblastoma tissue after preoperative treatment with the EGFR tyrosine kinase inhibitor gefitinib--a phase II trial. Mol. Cancer Ther. 2011, 10, 1102–1112. [Google Scholar] [CrossRef]
- Lassman, A.B.; Roberts-Rapp, L.; Sokolova, I.; Song, M.; Pestova, E.; Kular, R.; Mullen, C.; Zha, Z.; Lu, X.; Gomez, E.; et al. Comparison of Biomarker Assays for EGFR: Implications for Precision Medicine in Patients with Glioblastoma. Clin. Cancer Res. 2019, 25, 3259–3265. [Google Scholar] [CrossRef]
- Krueger, F. Trim Galore. 2022. Available online: https://github.com/FelixKrueger/TrimGalore (accessed on 18 June 2022).
- Andrews, S. FastQC. 2022. Available online: https://github.com/s-andrews/FastQC. (accessed on 18 June 2022).
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.I.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e472015. [Google Scholar] [CrossRef]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, D.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013, 14, 7. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2014–2018. Neuro-oncology 2021, 21, v1–v100. [Google Scholar] [CrossRef]
- Gosney, J.A.; Wilkey, D.W.; Merchant, M.L.; Ceresa, B.P. Proteomics reveals novel protein associations with early endosomes in an epidermal growth factor-dependent manner. J. Biol. Chem. 2018, 293, 5895–5908. [Google Scholar] [CrossRef]
- Rossi, M.R.; La Duca, J.; Matsui, S.-I.; Nowak, N.J.; Hawthorn, L.; Cowell, J.K. Novel amplicons on the short arm of chromosome 7 identified using high resolution array CGH contain over expressed genes in addition to EGFR in glioblastoma multiforme. Genes Chromosomes Cancer 2005, 44, 392–404. [Google Scholar] [CrossRef]
- Kowanetz, K.; Crosetto, N.; Haglund, K.; Schmidt, M.; Heldin, C.-H.; Dikic, I. Suppressors of T-cell receptor signaling Sts-1 and Sts-2 bind to Cbl and inhibit endocytosis of receptor tyrosine kinases. J. Biol. Chem. 2004, 279, 32786–32795. [Google Scholar] [CrossRef]
- Cheng, Z.; Gong, Y.; Ma, Y.; Lu, K.; Lu, X.; Pierce, L.A.; Thompson, R.C.; MullerKnapp, S.; Wang, J. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin. Cancer Res. 2013, 19, 1748–1759. [Google Scholar] [CrossRef]
- Pastori, C.; Daniel, M.; Penas, C.; Volmar, C.H.; Johnstone, A.L.; Brothers, S.P. BET bromodomain proteins are required for glioblastoma cell proliferation. Epigenetics 2014, 9, 611–620. [Google Scholar] [CrossRef]
- Labrakakis, C.; Patt, S.; Hartmann, J.; Kettenmann, H. Functional GABA(A) receptors on human glioma cells. Eur. J. Neurosci. 1998, 10, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Sephton, C.F.; Cenik, C.; Kucukural, A.; Dammer, E.B.; Cenik, B.; Han, Y. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J. Biol. Chem. 2011, 286, 1204–1215. [Google Scholar] [CrossRef] [PubMed]
- Yi, W.; Wang, J.; Yao, Z.; Kong, Q.; Zhang, N.; Mo, W.; Xu, L.; Li, X. The expression status of ZIC2 as a prognostic marker for nasopharyngeal carcinoma. Int. J. Clin. Exp. Pathol. 2018, 11, 4446–4460. [Google Scholar] [PubMed]
- Prest, S.J.; May, F.E.B.; Westley, B.R. The estrogen-regulated protein, TFF1, stimulates migration of human breast cancer cells. FASEB J. 2002, 16, 592–594. [Google Scholar] [CrossRef]
- Chen, J.-R.; Xu, H.-Z.; Yao, Y.; Qin, Z.-Y. Prognostic value of epidermal growth factor receptor amplification and EGFRvIII in glioblastoma: Meta-analysis. Acta Neurol. Scand. 2015, 132, 310–322. [Google Scholar] [CrossRef]
- Martinez, R.; Rohde, V.; Schackert, G. Different molecular patterns in glioblastoma multiforme subtypes upon recurrence. J. Neurooncol. 2010, 96, 321–329. [Google Scholar] [CrossRef]
- Kim, J.; Lee, I.H.; Cho, H.J.; Park, C.K.; Jung, Y.S.; Kim, Y.; Nam, S.H.; Kim, B.S.; Johnson, M.D.; Kong, S.K.; et al. Spatiotemporal Evolution of the Primary Glioblastoma Genome. Cancer Cell 2015, 28, 318–328. [Google Scholar] [CrossRef]

| Primary Tumour EGFR Status | EGFR-Amplified n = 20 | EGFR Non-Amplified/Indeterminate n = 20 |
|---|---|---|
| Age at diagnosis (y) | 56.9 (IQR 50–64.25) | 56.7 (IQR48.5–67.25) |
| Sex | Male (13/20) Female (7/20) | Male (15/20) Female (5/20) |
| KPS at time of diagnosis | 86.3 (IQR 80–90) | 86.1 (IQR 80–90) |
| KPS at time of recurrence | 76.3 (IQR 70–85) | 79.4 (IQR 77.5–82.5) |
| Extent of primary resection | Total (10/20) Near total (7/20) Subtotal (2/20) Partial (1/20) | Total (13/20) Near total (4/20) Subtotal (2/20) NA (1/20) |
| IDH1 mutation status | Wildtype (8/20) Mutant (1/20) NA (11/20) | Wildtype (5/20) Mutant (0/20) NA (15/20) |
| MGMT promoter methylation status | Unmethylated (6/20) Methylated (2/20) NA (12/20) | Unmethylated (2/20) Methylated (2/20) NA (16/20) |
| PFS (Median) | 433.9 days (IQR 134.25–520 days) | 320.2 days (IQR 101.5–394.75 days) |
| OS (Median) | 809.75 days (IQR 478.5–1094 days) | 697.1 days (IQR 308–824 days) |
| Laterality | Right (15/20) Left (5/20) | Right (13/20) Left (7/20) |
| Location | Temporal (7/20) Frontal (6/20) Parietal (6/20) Fronto-parietal (1/20) | Temporal (9/20) Frontal (6/20) Parietal (4/20) Parieto-occipital (1/20) |
| Targeted therapy received? | Yes (3/20) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhawan, A.; Manem, V.S.K.; Yeaney, G.; Lathia, J.D.; Ahluwalia, M.S. EGFR Pathway Expression Persists in Recurrent Glioblastoma Independent of Amplification Status. Cancers 2023, 15, 670. https://doi.org/10.3390/cancers15030670
Dhawan A, Manem VSK, Yeaney G, Lathia JD, Ahluwalia MS. EGFR Pathway Expression Persists in Recurrent Glioblastoma Independent of Amplification Status. Cancers. 2023; 15(3):670. https://doi.org/10.3390/cancers15030670
Chicago/Turabian StyleDhawan, Andrew, Venkata S. K. Manem, Gabrielle Yeaney, Justin D. Lathia, and Manmeet S. Ahluwalia. 2023. "EGFR Pathway Expression Persists in Recurrent Glioblastoma Independent of Amplification Status" Cancers 15, no. 3: 670. https://doi.org/10.3390/cancers15030670
APA StyleDhawan, A., Manem, V. S. K., Yeaney, G., Lathia, J. D., & Ahluwalia, M. S. (2023). EGFR Pathway Expression Persists in Recurrent Glioblastoma Independent of Amplification Status. Cancers, 15(3), 670. https://doi.org/10.3390/cancers15030670







