Novel Computed-Tomography-Based Transformer Models for the Noninvasive Prediction of PD-1 in Pre-Operative Settings
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort
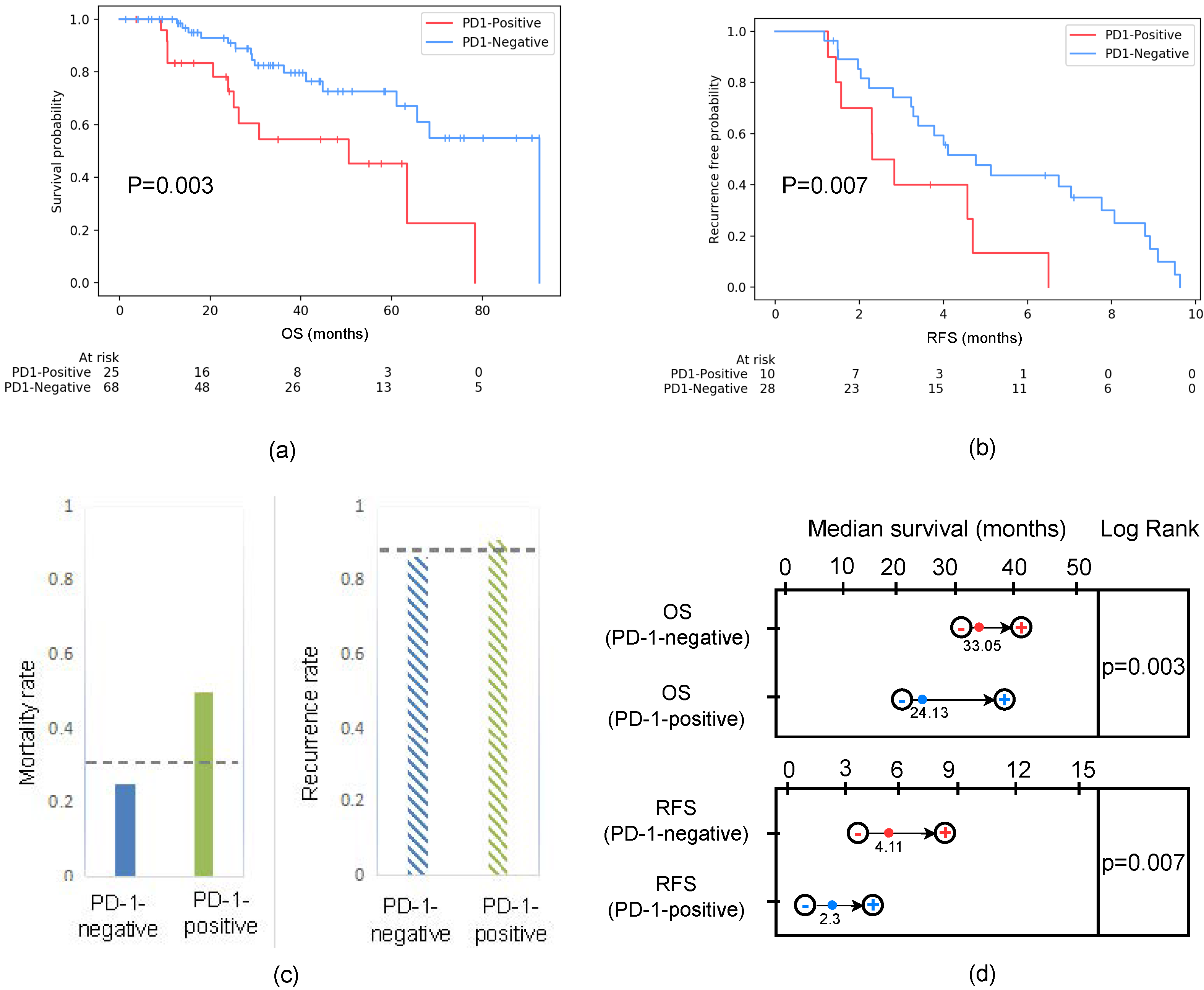
2.2. Patient Follow-Up
2.3. Immunohistochemistry Staining
2.4. CT Images
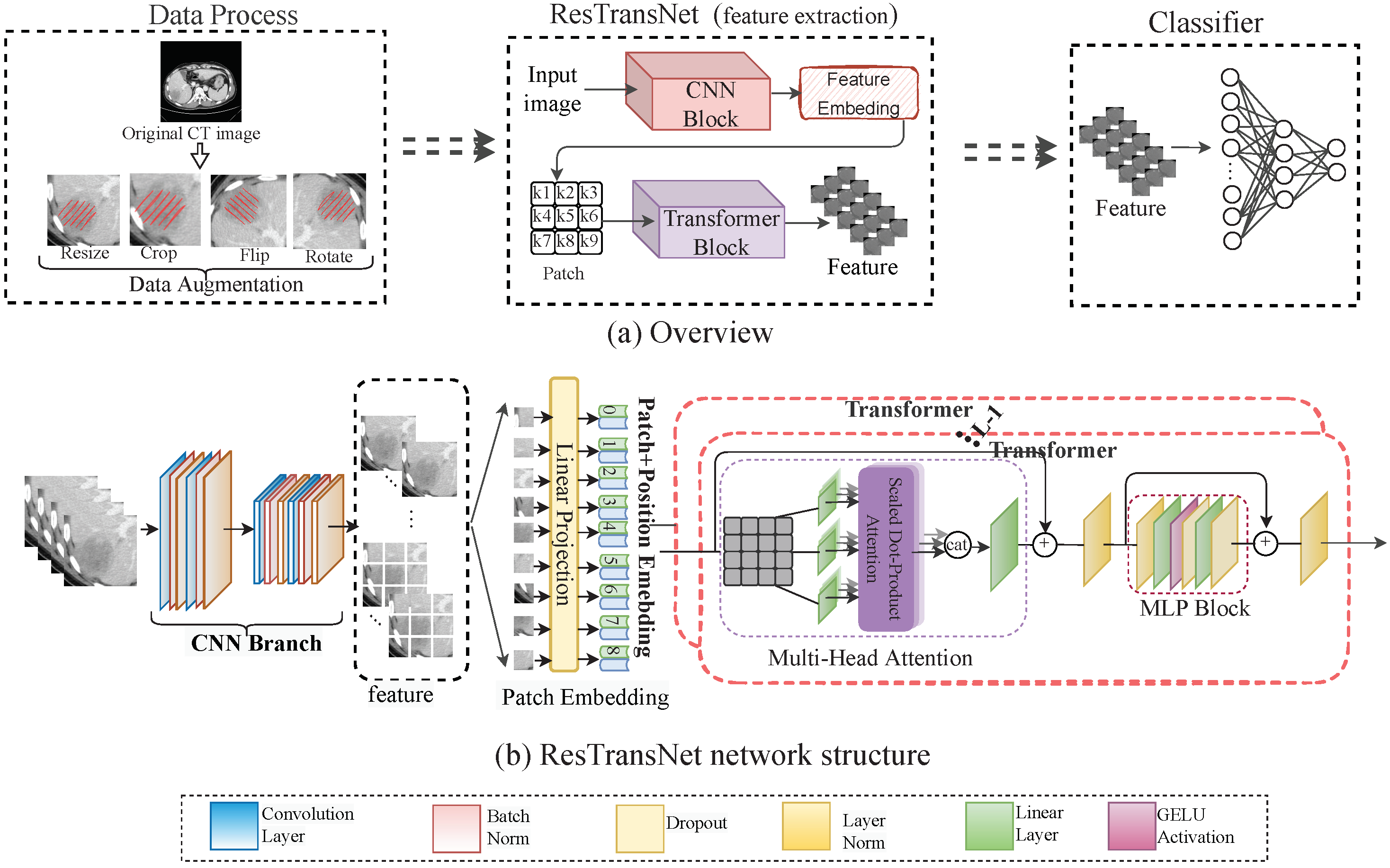
2.5. Prediction Network of PD-1
2.6. Gradient Penalty
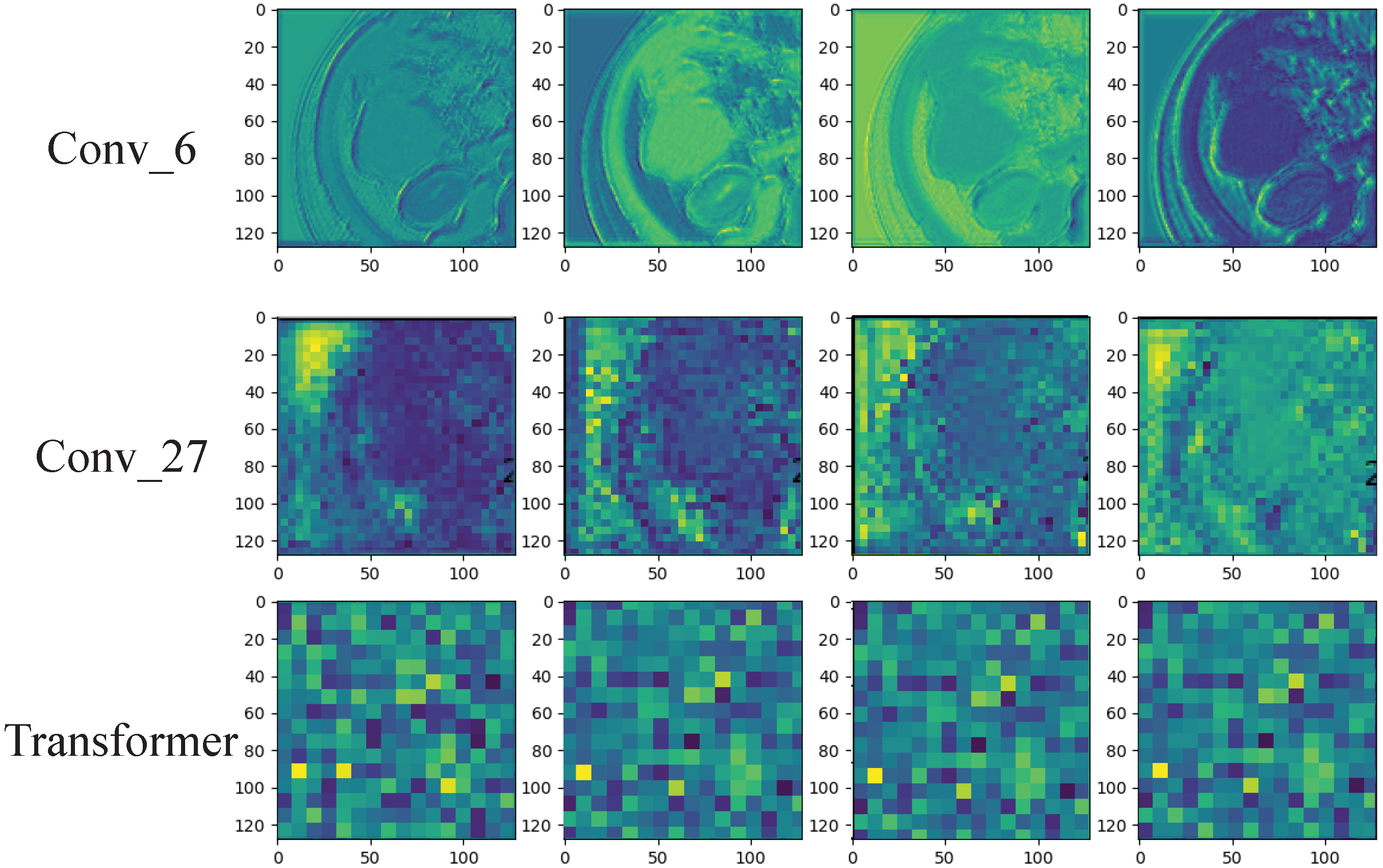
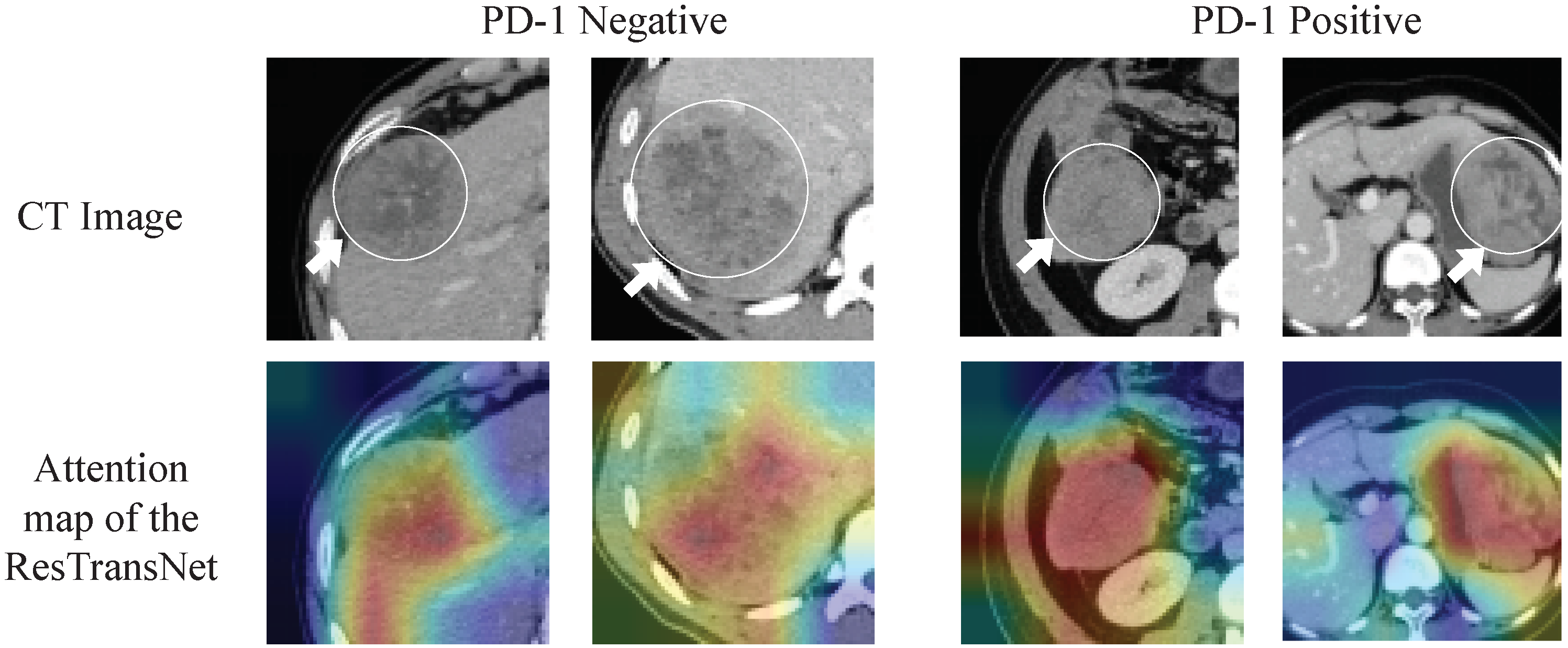
2.7. Visualization of the ResTransNet model
2.8. Statistical Analysis
3. Results
3.1. Patient Characteristics
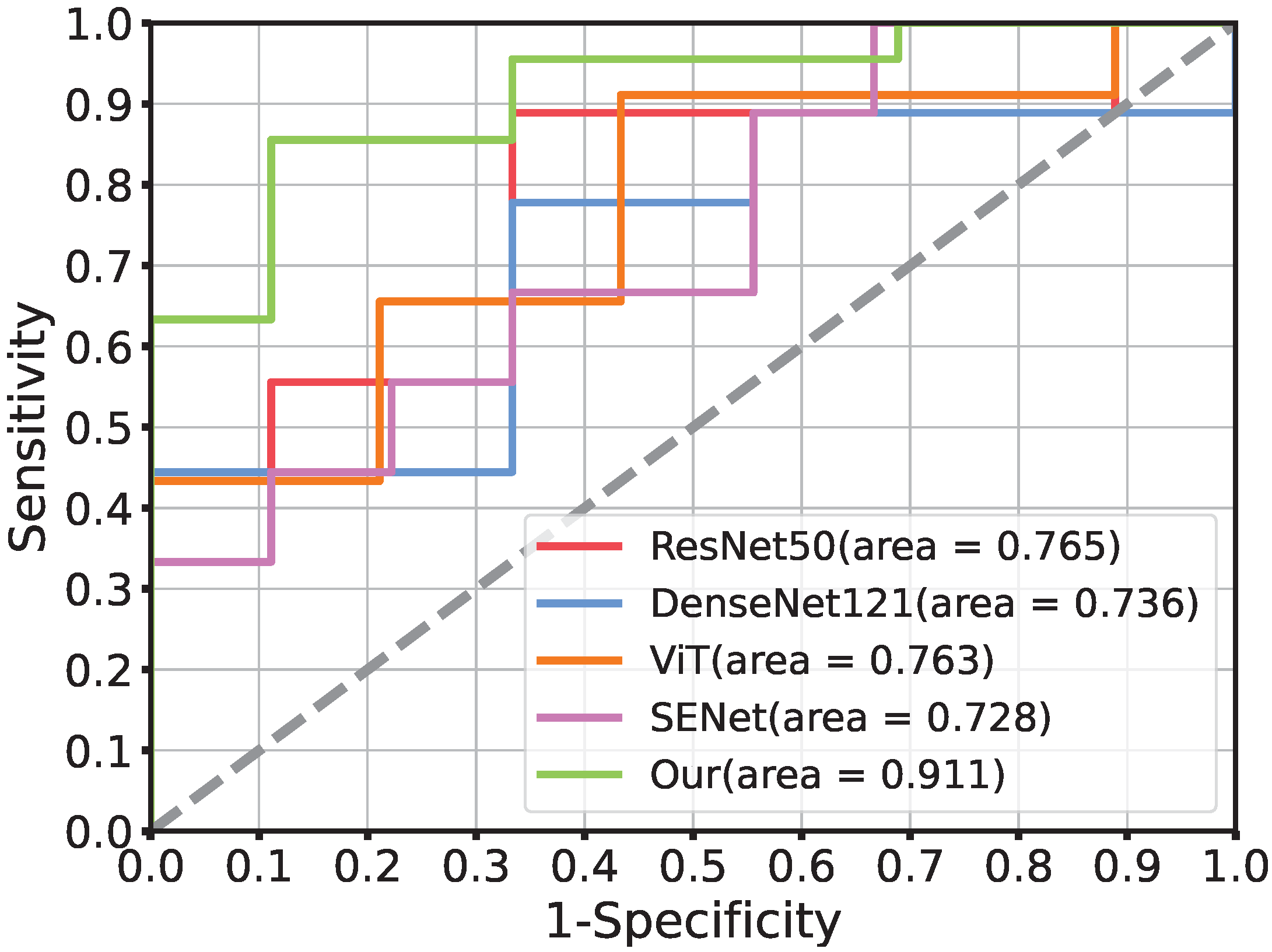
3.2. Performance of Prediction Model
3.3. Deep Learning Feature Analysis
3.4. Clinical Prognostic Validation of PD-1 in HCC Treatment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Tsilimigras, D.I.; Bagante, F.; Moris, D.; Hyer, J.M.; Pawlik, T.M. Recurrence patterns and outcomes after resection of hepatocellular carcinoma within and beyond the Barcelona Clinic liver cancer criteria. Ann. Surg. Oncol. 2020, 27, 2321–2331. [Google Scholar] [CrossRef] [PubMed]
- Heimbach, J.K.; Kulik, L.M.; Finn, R.S.; Sirlin, C.B.; Abecassis, M.M.; Roberts, L.R.; Zhu, A.X.; Murad, M.H.; Marrero, J.A. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018, 67, 358–380. [Google Scholar] [CrossRef] [PubMed]
- Ayuso, C.; Rimola, J.; Vilana, R.; Burrel, M.; Darnell, A.; García-Criado, Á.; Bianchi, L.; Belmonte, E.; Caparroz, C.; Barrufet, M.; et al. Diagnosis and staging of hepatocellular carcinoma (HCC): Current guidelines. Eur. J. Radiol. 2018, 101, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Sberna, A.; Bouillet, B.; Rouland, A.; Brindisi, M.; Nguyen, A.; Mouillot, T.; Duvillard, L.; Denimal, D.; Loffroy, R.; Vergès, B.; et al. European Association for the Study of the Liver (EASL), European Association for the Study of Diabetes (EASD) and European Association for the Study of Obesity (EASO) clinical practice recommendations for the management of non-alcoholic fatty liver disease: Evaluation of their application in people with Type 2 diabetes. Diabet. Med. 2018, 35, 368–375. [Google Scholar]
- Sangro, B.; Sarobe, P.; Hervás-Stubbs, S.; Melero, I. Advances in immunotherapy for hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 525–543. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, X.; Liang, J.; Liu, Y.; Hou, X.; Zhang, M.; Li, Y.; Jiang, X. Immunotherapy for hepatocellular carcinoma: Current status and future prospects. Front. Immunol. 2021, 12, 765101. [Google Scholar] [CrossRef]
- Yan, T.; Yu, L.; Zhang, N.; Peng, C.; Su, G.; Jing, Y.; Zhang, L.; Wu, T.; Cheng, J.; Guo, Q.; et al. The advanced development of molecular targeted therapy for hepatocellular carcinoma. Cancer Biol. Med. 2022, 19, 802. [Google Scholar] [CrossRef] [PubMed]
- Sanmamed, M.F.; Chen, L. A paradigm shift in cancer immunotherapy: From enhancement to normalization. Cell 2018, 175, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Giraud, J.; Chalopin, D.; Blanc, J.F.; Saleh, M. Hepatocellular carcinoma immune landscape and the potential of immunotherapies. Front. Immunol. 2021, 12, 655697. [Google Scholar] [CrossRef]
- Llovet, J.M.; Castet, F.; Heikenwalder, M.; Maini, M.K.; Mazzaferro, V.; Pinato, D.J.; Pikarsky, E.; Zhu, A.X.; Finn, R.S. Immunotherapies for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2022, 19, 151–172. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Xiang, Y.; Sheng, J.; Zhang, D.; Yao, X.; Yang, Y.; Zhang, X. Immunotherapy for hepatocellular carcinoma: Current advances and future expectations. J. Immunol. Res. 2018, 2018, 8740976. [Google Scholar] [CrossRef]
- Lu, L.; Jiang, J.; Zhan, M.; Zhang, H.; Wang, Q.T.; Sun, S.N.; Guo, X.K.; Yin, H.; Wei, Y.; Li, S.Y.; et al. Targeting Tumor-Associated Antigens in Hepatocellular Carcinoma for Immunotherapy: Past Pitfalls and Future Strategies. Hepatology 2020, 73, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.C.; Levine, J.H.; Cogdill, A.P.; Zhao, Y.; Anang, N.A.A.; Andrews, M.C.; Sharma, P.; Wang, J.; Wargo, J.A.; Pe’er, D.; et al. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell 2017, 170, 1120–1133. [Google Scholar] [CrossRef]
- Pinter, M.; Jain, R.K.; Duda, D.G. The current landscape of immune checkpoint blockade in hepatocellular carcinoma: A review. JAMA Oncol. 2021, 7, 113–123. [Google Scholar] [CrossRef]
- Gordan, J.D.; Kennedy, E.B.; Abou-Alfa, G.K.; Beg, M.S.; Brower, S.T.; Gade, T.P.; Goff, L.; Gupta, S.; Guy, J.; Harris, W.P.; et al. Systemic therapy for advanced hepatocellular carcinoma: ASCO guideline. J. Clin. Oncol. 2020, 38, 4317–4345. [Google Scholar] [CrossRef] [PubMed]
- Sonbol, M.B.; Riaz, I.B.; Naqvi, S.A.A.; Almquist, D.R.; Mina, S.; Almasri, J.; Shah, S.; Almader-Douglas, D.; Junior, P.L.S.U.; Mahipal, A.; et al. Systemic therapy and sequencing options in advanced hepatocellular carcinoma: A systematic review and network meta-analysis. JAMA Oncol. 2020, 6, e204930. [Google Scholar] [CrossRef] [PubMed]
- El-Khoueiry, A.B.; Sangro, B.; Yau, T.; Crocenzi, T.S.; Kudo, M.; Hsu, C.; Kim, T.Y.; Choo, S.P.; Trojan, J.; Welling, T.H., 3rd; et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): An open-label, non-comparative, phase? dose escalation and expansion trial. Lancet Lond. 2017, 389, 2492–2502. [Google Scholar] [CrossRef]
- Finn, R.; Ryoo, B.; Merle, P.; Kudo, M.; Bouattour, M.; Lim, H.; Breder, V.; Edeline, J.; Chao, Y.; Ogasawara, S.; et al. Pembrolizumab As Second-Line Therapy in Patients With Advanced Hepatocellular Carcinoma in KEYNOTE-240: A Randomized, Double-Blind, Phase III Trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019, 38, 193–202. [Google Scholar] [CrossRef]
- Ruiz de Galarreta, M.; Bresnahan, E.; Molina-Sánchez, P.; Lindblad, K.E.; Maier, B.; Sia, D.; Puigvehi, M.; Miguela, V.; Casanova-Acebes, M.; Dhainaut, M.; et al. β-Catenin Activation Promotes Immune Escape and Resistance to Anti–PD-1 Therapy in Hepatocellular Carcinomaβ-Catenin Promotes Immune Resistance in Liver Cancer. Cancer Discov. 2019, 9, 1124–1141. [Google Scholar] [CrossRef]
- Wang, B.J.; Bao, J.J.; Wang, J.Z.; Wang, Y.; Jiang, M.; Xing, M.Y.; Zhang, W.G.; Qi, J.Y.; Roggendorf, M.; Lu, M.J.; et al. Immunostaining of PD-1/PD-Ls in liver tissues of patients with hepatitis and hepatocellular carcinoma. World J. Gastroenterol. WJG 2011, 17, 3322. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563. [Google Scholar] [CrossRef] [PubMed]
- Bi, W.L.; Hosny, A.; Schabath, M.B.; Giger, M.L.; Birkbak, N.J.; Mehrtash, A.; Allison, T.; Arnaout, O.; Abbosh, C.; Dunn, I.F.; et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J. Clin. 2019, 69, 127–157. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Gu, D.; Wei, J.; Ding, Y.; Yang, L.; Zhu, K.; Luo, R.; Rao, S.X.; Tian, J.; Zeng, M. A radiomics-based biomarker for cytokeratin 19 status of hepatocellular carcinoma with gadoxetic acid–enhanced MRI. Eur. Radiol. 2020, 30, 3004–3014. [Google Scholar] [CrossRef]
- Sun, L.; Mu, L.; Zhou, J.; Tang, W.; Zhang, L.; Xie, S.; Chen, J.; Wang, J. Imaging features of gadoxetic acid-enhanced MR imaging for evaluation of tumor-infiltrating CD8 cells and PD-L1 expression in hepatocellular carcinoma. Cancer Immunol. Immunother. 2022, 71, 25–38. [Google Scholar] [CrossRef]
- Hectors, S.J.; Lewis, S.; Besa, C.; King, M.J.; Said, D.; Putra, J.; Ward, S.; Higashi, T.; Thung, S.; Yao, S.; et al. MRI radiomics features predict immuno-oncological characteristics of hepatocellular carcinoma. Eur. Radiol. 2020, 30, 3759–3769. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, Z.; Zhang, X.; Liu, S.; Zhao, J.; Yuan, F.; Shi, Y.; Song, B. Machine learning: An approach to preoperatively predict PD-1/PD-L1 expression and outcome in intrahepatic cholangiocarcinoma using MRI biomarkers. ESMO Open 2020, 5, e000910. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. In Proceedings of the International Conference on Learning Representations, Addis Ababa, Ethiopia, 26–30 April 2020. [Google Scholar]
- Dai, Y.; Gao, Y.; Liu, F. Transmed: Transformers advance multi-modal medical image classification. Diagnostics 2021, 11, 1384. [Google Scholar] [CrossRef]
- Valanarasu, J.M.J.; Oza, P.; Hacihaliloglu, I.; Patel, V.M. Medical Transformer: Gated Axial-Attention for Medical Image Segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; pp. 36–46. [Google Scholar]
- Han, C.; Rundo, L.; Murao, K.; Noguchi, T.; Shimahara, Y.; Milacski, Z.Á.; Koshino, S.; Sala, E.; Nakayama, H.; Satoh, S. MADGAN: Unsupervised medical anomaly detection GAN using multiple adjacent brain MRI slice reconstruction. BMC Bioinform. 2021, 22, 31. [Google Scholar] [CrossRef]
- Wu, Y.; Hatipoglu, S.; Alonso-Álvarez, D.; Gatehouse, P.; Li, B.; Gao, Y.; Firmin, D.; Keegan, J.; Yang, G. Fast and automated segmentation for the three-directional multi-slice cine myocardial velocity mapping. Diagnostics 2021, 11, 346. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Gidaris, S.; Singh, P.; Komodakis, N. Unsupervised Representation Learning by Predicting Image Rotations. In Proceedings of the International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Kim, J.H. Estimating classification error rate: Repeated cross-validation, repeated hold-out and bootstrap. Comput. Stat. Data Anal. 2009, 53, 3735–3745. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Cui, Y.; Jia, M.; Lin, T.Y.; Song, Y.; Belongie, S. Class-balanced loss based on effective number of samples. In Proceedings of the Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 9268–9277. [Google Scholar]
- Cao, K.; Wei, C.; Gaidon, A.; Arechiga, N.; Ma, T. Learning imbalanced datasets with label-distribution-aware margin loss. In Proceedings of the 33rd Conference on Neural Information Processing Systems (NeurIPS 2019), Vancouver, BC, Canada, 8–14 December 2019. [Google Scholar]
- Khan, S.; Hayat, M.; Zamir, S.W.; Shen, J.; Shao, L. Striking the right balance with uncertainty. In Proceedings of the CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 103–112. [Google Scholar]
- Yi, M.; Jiao, D.; Xu, H.; Liu, Q.; Zhao, W.; Han, X.; Wu, K. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol. Cancer 2018, 17, 129. [Google Scholar] [CrossRef]
- Wu, C.C.; Wang, Y.A.; Livingston, J.A.; Zhang, J.; Futreal, P.A. Prediction of biomarkers and therapeutic combinations for anti-PD-1 immunotherapy using the global gene network association. Nat. Commun. 2022, 13, 42. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Jiang, L.; Zhang, J.; Shi, Y.; Gray, J.E.; Tunali, I.; Gao, C.; Sun, Y.; Tian, J.; Zhao, X.; et al. Non-invasive decision support for NSCLC treatment using PET/CT radiomics. Nat. Commun. 2020, 11, 5228. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Dong, Y.; Xiao, T.; Zhang, S.; Yu, J.; Li, L.; Zhang, Q.; Wang, Y.; Xiao, Y.; Wang, W. Prediction of programmed cell death protein 1 in hepatocellular carcinoma patients using radiomics analysis with radiofrequency-based ultrasound multifeature maps. BioMed. Eng. Online 2022, 21, 24. [Google Scholar] [CrossRef]
- Mu, W.; Jiang, L.; Shi, Y.; Tunali, I.; Gray, J.E.; Katsoulakis, E.; Tian, J.; Gillies, R.J.; Schabath, M.B. Non-invasive measurement of PD-L1 status and prediction of immunotherapy response using deep learning of PET/CT images. J. Immunother. Cancer 2021, 9, e002118. [Google Scholar] [CrossRef] [PubMed]
- Kalathil, S.G.; Lugade, A.A.; Miller, A.; Iyer, R.; Thanavala, Y. PD-1+ and Foxp3+ T cell reduction correlates with survival of HCC patients after sorafenib therapy. JCI Insight 2016, 1, e86182. [Google Scholar] [CrossRef]
- Yuan, G.; Song, Y.; Li, Q.; Hu, X.; Zang, M.; Dai, W.; Cheng, X.; Huang, W.; Yu, W.; Chen, M.; et al. Development and validation of a contrast-enhanced CT-based radiomics nomogram for prediction of therapeutic efficacy of anti-PD-1 antibodies in advanced HCC patients. Front. Immunol. 2021, 11, 613946. [Google Scholar] [CrossRef]


| Attribute | Training Cohort | Testing Cohort | ||||
|---|---|---|---|---|---|---|
| PD-1-Positive | PD-1-Negative | p Value | PD-1-Positive | PD-1-Negative | p Value | |
| Age | 0.219 | 1.000 | ||||
| Mean (SD) | 47.25 (13.5) | 52.88 (12.62) | 50 (10.54) | 52.5 (11.11) | ||
| Gender | 0.227 | 0.725 | ||||
| Male (%) | 13 (81.25%) | 54 (91.5%) | 8 (88.9%) | 8 (88.9%) | ||
| FeMale | 3 (18.75%) | 5 (8.5%) | 1 (11.1%) | 1 (11.1%) | ||
| HBV | 0.786 | 0.317 | ||||
| + (%) | 13 (81.25%) | 54 (91.5%) | 8 (88.9%) | 9 (100%) | ||
| − (%) | 3 (18.75%) | 5 (8.5%) | 1 (11.1%) | 0 (0%) | ||
| ALT | 0.655 | 0.307 | ||||
| >40 (%) | 8 (50%) | 26 (44.06%) | 5 (55.6%) | 1 (11.1%) | ||
| AST | 0.522 | 0.910 | ||||
| >35 (%) | 11 (68.7%) | 30 (50.8%) | 7 (77.8%) | 5 (55.6%) | ||
| AFP | 0.137 | 0.070 | ||||
| >400 (%) | 2 (12.5%) | 9 (15.25%) | 1 (11.1%) | 1 (11.1%) | ||
| CEA | 0.693 | 0.226 | ||||
| >3.4 (%) | 4 (25%) | 11 (18.6%) | 3 (33.3%) | 1 (11.1%) | ||
| TB | 0.248 | 0.226 | ||||
| Mean (SD) | 18.53 (9.56) | 19.04 (14.04) | 12.77 (4.38) | 18.17 (9.63) | ||
| PLT | 0.349 | 0.226 | ||||
| Mean (SD) | 203.7 (138.58) | 150.8 (63.47) | 126.2 (43.51) | 164.7 (72.39) | ||
| ALB | 0.298 | 0.520 | ||||
| Mean (SD) | 60.58 (67.12) | 41.9 (8.53) | 45.32 (4.72) | 43.53 (6.91) | ||
| GGT | 0.509 | 0.344 | ||||
| Mean (SD) | 69 (47.26) | 90 (85.04) | 108.8 (84.43) | 85.1 (92.39) | ||
| Creatinine | 0.013 | 1.000 | ||||
| Mean (SD) | 62.38 (17.75) | 76.6 (19.31) | 70.7 (12.39) | 75.9 (27.36) | ||
| MVI | 0.549 | 0.687 | ||||
| Absent (%) | 8 (50%) | 29 (49.1%) | 7 (77.7%) | 6 (66.7%) | ||
| Present (%) | 8 (50%) | 30 (50.9%) | 2 (22.3%) | 3 (33.3%) | ||
| BCLC | 0.445 | 0.768 | ||||
| 0–A (%) | 4 (25%) | 11 (18.6%) | 1 (11.1%) | 1 (11.1%) | ||
| B (%) | 8 (50%) | 31 (52.5%) | 3 (33.3%) | 4 (36.4%) | ||
| C (%) | 4 (25%) | 17 (28.9%) | 5 (55.6%) | 4 (52.5%) | ||
| Model | Training Cohort | Testing Cohort | ||||||
|---|---|---|---|---|---|---|---|---|
| ACC | AUC | SEN | SPEC | ACC | AUC | SEN | SPEC | |
| ResNet50 | 97.4% | 96.5% | 94.3% | 98.8% | 77.7% | 76.5% | 66.7% | 88.9% |
| DenseNet121 | 96.4% | 93.7% | 92.5% | 97.8% | 76.4% | 73.6% | 55.5% | 87.5% |
| SENet154 | 95.5% | 92.8% | 93.1% | 91.6% | 66.7% | 72.8% | 77.8% | 55.6% |
| Vit | 97.8% | 95.5% | 94.1% | 98.7% | 77.8% | 76.3% | 66.7% | 88.9% |
| ResTransNet(Our) | 99.2% | 99.8% | 98.9% | 98.9% | 88.2% | 91.1% | 88.9% | 88.5% |
| Strategy | Training Cohort | Testing Cohort | ||||||
|---|---|---|---|---|---|---|---|---|
| ACC | AUC | SEN | SPEC | ACC | AUC | SEN | SPEC | |
| Oversampling | 96.6% | 94.6% | 93.5% | 97.8% | 76.4% | 73.6% | 55.5% | 87.5% |
| Loss re-weighting | 98.8% | 95.2% | 98.8% | 96.7% | 83.3% | 82.2% | 88.8% | 70.5% |
| Gradient penalty | 99.8% | 99.7% | 98.5% | 98.5% | 88.5% | 91.1% | 88.9% | 88.5% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Y.; Yang, M.; Xu, L.; Liu, M.; Zhang, F.; Xie, T.; Cheng, X.; Wang, X.; Che, F.; Li, Q.; et al. Novel Computed-Tomography-Based Transformer Models for the Noninvasive Prediction of PD-1 in Pre-Operative Settings. Cancers 2023, 15, 658. https://doi.org/10.3390/cancers15030658
Wei Y, Yang M, Xu L, Liu M, Zhang F, Xie T, Cheng X, Wang X, Che F, Li Q, et al. Novel Computed-Tomography-Based Transformer Models for the Noninvasive Prediction of PD-1 in Pre-Operative Settings. Cancers. 2023; 15(3):658. https://doi.org/10.3390/cancers15030658
Chicago/Turabian StyleWei, Yi, Meiyi Yang, Lifeng Xu, Minghui Liu, Feng Zhang, Tianshu Xie, Xuan Cheng, Xiaomin Wang, Feng Che, Qian Li, and et al. 2023. "Novel Computed-Tomography-Based Transformer Models for the Noninvasive Prediction of PD-1 in Pre-Operative Settings" Cancers 15, no. 3: 658. https://doi.org/10.3390/cancers15030658
APA StyleWei, Y., Yang, M., Xu, L., Liu, M., Zhang, F., Xie, T., Cheng, X., Wang, X., Che, F., Li, Q., Xu, Q., Huang, Z., & Liu, M. (2023). Novel Computed-Tomography-Based Transformer Models for the Noninvasive Prediction of PD-1 in Pre-Operative Settings. Cancers, 15(3), 658. https://doi.org/10.3390/cancers15030658









