Prediction of Neoadjuvant Chemoradiotherapy Response in Rectal Cancer Patients Using Harmonized Radiomics of Multcenter 18F-FDG-PET Image
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort
2.2. Image Feature Extraction
2.3. Harmonization Methodology
2.4. Deep Learning and Machine Learning
3. Results
3.1. Patients Cohort
3.2. Evaluation of Deep Learning Model
3.3. Image Feature Extraction and Harmonization
3.4. Evaluation of Machine Learning Model
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Z.; Zhang, X.-Y.; Shi, Y.-J.; Wang, L.; Zhu, H.-T.; Tang, Z.; Wang, S.; Li, X.-T.; Tian, J.; Sun, Y.-S.; et al. Radiomics analysis for evaluation of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Clin. Cancer Res. 2017, 23, 7253–7262. [Google Scholar] [CrossRef] [PubMed]
- Valentini, V.; Coco, C.; Cellini, N.; Picciocchi, A.; Genovesi, D.; Mantini, G.; Barbaro, B.; Cogliandolo, S.; Mattana, C.; Ambesiimpiombato, F.; et al. Preoperative chemoradiation for extraperitoneal T3 rectal cancer: Acute toxicity, tumor response, and sphincter preservation. Int. J. Radiat. Oncol. Biol. Phys. 1998, 40, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.T.; Liney, G.P.; Wong, K.; Barton, M.B. Functional MRI for quantitative treatment response prediction in locally advanced rectal cancer. Br. J. Radiol. 2017, 90, 20151078. [Google Scholar] [CrossRef] [PubMed]
- Maas, M.; Nelemans, P.J.; Valentini, V.; Das, P.; Rödel, C.; Kuo, L.J.; Beets, G.L. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: A pooled analysis of individual patient data. Lancet Oncol. 2010, 11, 835–844. [Google Scholar] [CrossRef]
- Maffione, A.; Chondrogiannis, S.; Capirci, C.; Galeotti, F.; Fornasiero, A.; Crepaldi, G.; Grassetto, G.; Rampin, L.; Marzola, M.; Rubello, D. Early prediction of response by 18F-FDG PET/CT during preoperative therapy in locally advanced rectal cancer: A systematic review. Eur. J. Surg. Oncol. (EJSO) 2014, 40, 1186–1194. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, K.W.; Yang, R.; Geng, L.S. Review of deep learning based automatic segmentation for lung cancer radiotherapy. Front. Oncol. 2021, 11, 717039. [Google Scholar] [CrossRef]
- Zhong, Y.; She, Y.; Deng, J.; Chen, S.; Wang, T.; Yang, M.; Ma, M.; Song, Y.; Qi, H.; Wang, Y.; et al. Multi-omics Classifier for Pulmonary Nodules (MISSION) Collaborative Group. Deep learning for prediction of N2 metastasis and survival for clinical stage I non–small cell lung cancer. Radiology 2022, 302, 200–211. [Google Scholar] [CrossRef]
- Kim, J.; Oh, J.E.; Lee, J.; Kim, M.J.; Hur, B.Y.; Sohn, D.K.; Lee, B. Rectal cancer: Toward fully automatic discrimination of T2 and T3 rectal cancers using deep convolutional neural network. Int. J. Imaging Syst. Technol. 2019, 29, 247–259. [Google Scholar] [CrossRef]
- Palma, P.; Conde-Muíño, R.; Rodríguez-Fernández, A.; Segura-Jiménez, I.; Sánchez-Sánchez, R.; Martín-Cano, J.; Gómez-Río, M.; Ferrón, J.A.; Llamas-Elvira, J.M. The value of metabolic imaging to predict tumour response after chemoradiation in locally advanced rectal cancer. Radiat. Oncol. 2010, 5, 1–8. [Google Scholar] [CrossRef]
- Martoni, A.A.; Di Fabio, F.; Pinto, C.; Castellucci, P.; Pini, S.; Ceccarelli, C.; Cuicchi, D.; Iacopino, B.; Di Tullio, P.; Giaquinta, S.; et al. Prospective study on the FDG–PET/CT predictive and prognostic values in patients treated with neoadjuvant chemoradiation therapy and radical surgery for locally advanced rectal cancer. Ann. Oncol. 2011, 22, 650–656. [Google Scholar] [CrossRef]
- Lovinfosse, P.; Polus, M.; Van Daele, D.; Martinive, P.; Daenen, F.; Hatt, M.; Hustinx, R. FDG PET/CT radiomics for predicting the outcome of locally advanced rectal cancer. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 365–375. [Google Scholar] [CrossRef]
- Sun, W.; Xu, J.; Hu, W.; Zhang, Z.; Shen, W. The role of sequential 18F-FDG PET/CT in predicting tumour response after preoperative chemoradiation for rectal cancer. Color. Dis. 2013, 15, e231–e238. [Google Scholar] [CrossRef] [PubMed]
- Joye, I.; Deroose, C.M.; Vandecaveye, V.; Haustermans, K. The role of diffusion-weighted MRI and 18F-FDG PET/CT in the prediction of pathologic complete response after radiochemotherapy for rectal cancer: A systematic review. Radiother. Oncol. 2014, 113, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Orlhac, F.; Eertink, J.J.; Cottereau, A.S.; Zijlstra, J.M.; Thieblemont, C.; Meignan, M.; Boellaard, R.; Buvat, I. A guide to ComBat harmonization of imaging biomarkers in multicenter studies. J. Nucl. Med. 2022, 63, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, I.W.; Tassi, E.; Bellani, M.; Benedetti, F.; Poletti, S.; Spalletta, G.; Piras, F.; Bianchi, A.M.; Brambilla, P.; Maggioni, E. Comparison of Multi-Site Neuroimaging Data Harmonization Techniques for Machine Learning Applications. In Proceedings of the IEEE EUROCON 2023-20th International Conference on Smart Technologies, Torino, Italy, 6–8 July 2023. [Google Scholar]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2017, 8, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Jovicich, J.; Czanner, S.; Greve, D.; Haley, E.; van Der Kouwe, A.; Gollub, R.; Kennedy, D.; Schmitt, F.; Brown, G.; MacFall, J.; et al. Reliability in multi-site structural MRI studies: Effects of gradient non-linearity correction on phantom and human data. Neuroimage 2006, 30, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, R.T.; Oh, J.; Nair, G.; Calabresi, P.A.; Davatzikos, C.; Doshi, J.; Henry, R.G.; Kim, G.; Linn, K.A.; Papinutto, N.; et al. Volumetric analysis from a harmonized multisite brain MRI study of a single subject with multiple sclerosis. Am. J. Neuroradiol. 2017, 38, 1501–1509. [Google Scholar] [CrossRef]
- Macaulay, B.O.; Aribisala, B.S.; Akande, S.A.; Akinnuwesi, B.A.; Olabanjo, O.A. Breast cancer risk prediction in African women using random forest classifier. Cancer Treat. Res. Commun. 2021, 28, 100396. [Google Scholar] [CrossRef]
- Kesler, S.R.; Rao, A.; Blayney, D.W.; Oakley-Girvan, I.A.; Karuturi, M.; Palesh, O. Predicting long-term cognitive outcome following breast cancer with pre-treatment resting state fMRI and random forest machine learning. Front. Hum. Neurosci. 2017, 11, 555. [Google Scholar] [CrossRef]
- Li, N.; Luo, P.; Li, C.; Hong, Y.; Zhang, M.; Chen, Z. Analysis of related factors of radiation pneumonia caused by precise radiotherapy of esophageal cancer based on random forest algorithm. Math. Biosci. Eng. 2021, 18, 4477–4490. [Google Scholar] [CrossRef]
- Bi, L.; Guo, Y. Development and Validation of the Random Forest Model via Combining CT-PET Image Features and Demographic Data for Distant Metastases among Lung Cancer Patients. J. Healthc. Eng. 2022, 2022, 7793533. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, X.; Zhao, Y.; Zhang, J.; Zhang, Z.; Wang, J.; Wang, Y.; Dai, M.; Han, J. Value of pre-therapy 18F-FDG PET/CT radiomics in predicting EGFR mutation status in patients with non-small cell lung cancer. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Kaira, K.; Higuchi, T.; Naruse, I.; Arisaka, Y.; Tokue, A.; Altan, B.; Suda, S.; Mogi, A.; Shimizu, K.; Sunaga, N.; et al. Metabolic activity by 18F-FDG-PET/CT is predictive of early response after nivolumab in previously treated NSCLC. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 56–66. [Google Scholar] [CrossRef]
- Mu, W.; Tunali, I.; Gray, J.E.; Qi, J.; Schabath, M.B.; Gillies, R.J. Radiomics of 18F-FDG PET/CT images predicts clinical benefit of advanced NSCLC patients to checkpoint blockade immunotherapy. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1168–1182. [Google Scholar] [CrossRef]
- Huh, J.W.; Min, J.J.; Lee, J.H.; Kim, H.R.; Kim, Y.J. The predictive role of sequential FDG-PET/CT in response of locally advanced rectal cancer to neoadjuvant chemoradiation. Am. J. Clin. Oncol. 2012, 35, 340–344. [Google Scholar] [CrossRef]
- Capirci, C.; Rubello, D.; Pasini, F.; Galeotti, F.; Bianchini, E.; Del Favero, G.; Panzavolta, R.; Crepaldi, G.; Rampin, L.; Facci, E.; et al. The role of dual-time combined 18-fluorodeoxyglucose positron emission tomography and computed tomography in the staging and restaging workup of locally advanced rectal cancer, treated with preoperative chemoradiation therapy and radical surgery. Int. J. Radiat. Oncol. Biol. Phys. 2009, 74, 1461–1469. [Google Scholar] [CrossRef]
- Melton, G.B.; Lavely, W.C.; Jacene, H.A.; Schulick, R.D.; Choti, M.A.; Wahl, R.L.; Gearhart, S.L. Efficacy of preoperative combined 18-fluorodeoxyglucose positron emission tomography and computed tomography for assessing primary rectal cancer response to neoadjuvant therapy. J. Gastrointest. Surg. 2007, 11, 961–969. [Google Scholar] [CrossRef]
- Guillem, J.G.; Puig-La Calle, J.; Akhurst, T.; Tickoo, S.; Ruo, L.; Minsky, B.D.; Gollub, M.J.; Klimstra, D.S.; Mazumdar, M.; Paty, P.B.; et al. Prospective assessment of primary rectal cancer response to preoperative radiation and chemotherapy using 18-fluorodeoxyglucose positron emission tomography. Dis. Colon Rectum 2000, 43, 18–24. [Google Scholar] [CrossRef]
- Nyúl, L.G.; Udupa, J.K. On standardizing the MR image intensity scale. Magn. Reson. Med. Off. J. Int. Soc. Magn. Reson. Med. 1999, 42, 1072–1081. [Google Scholar] [CrossRef]
- Shinohara, R.T.; Sweeney, E.M.; Goldsmith, J.; Shiee, N.; Mateen, F.J.; Calabresi, P.A.; Jarso, S.; Pham, D.L.; Reich, D.S.; Crainiceanu, C.M. Statistical normalization techniques for magnetic resonance imaging. NeuroImage Clin. 2014, 6, 9–19. [Google Scholar] [CrossRef]
- Gullo, G.; Petousis, S.; Papatheodorou, A.; Panagiotidis, Y.; Margioula-Siarkou, C.; Prapas, N.; D’Anna, R.; Perino, A.; Cucinella, G.; Prapas, Y. Closed vs. Open oocyte vitrification methods are equally effective for blastocyst embryo transfers: Prospective study from a sibling oocyte donation program. Gynecol. Obstet. Investig. 2020, 85, 206–212. [Google Scholar] [CrossRef]
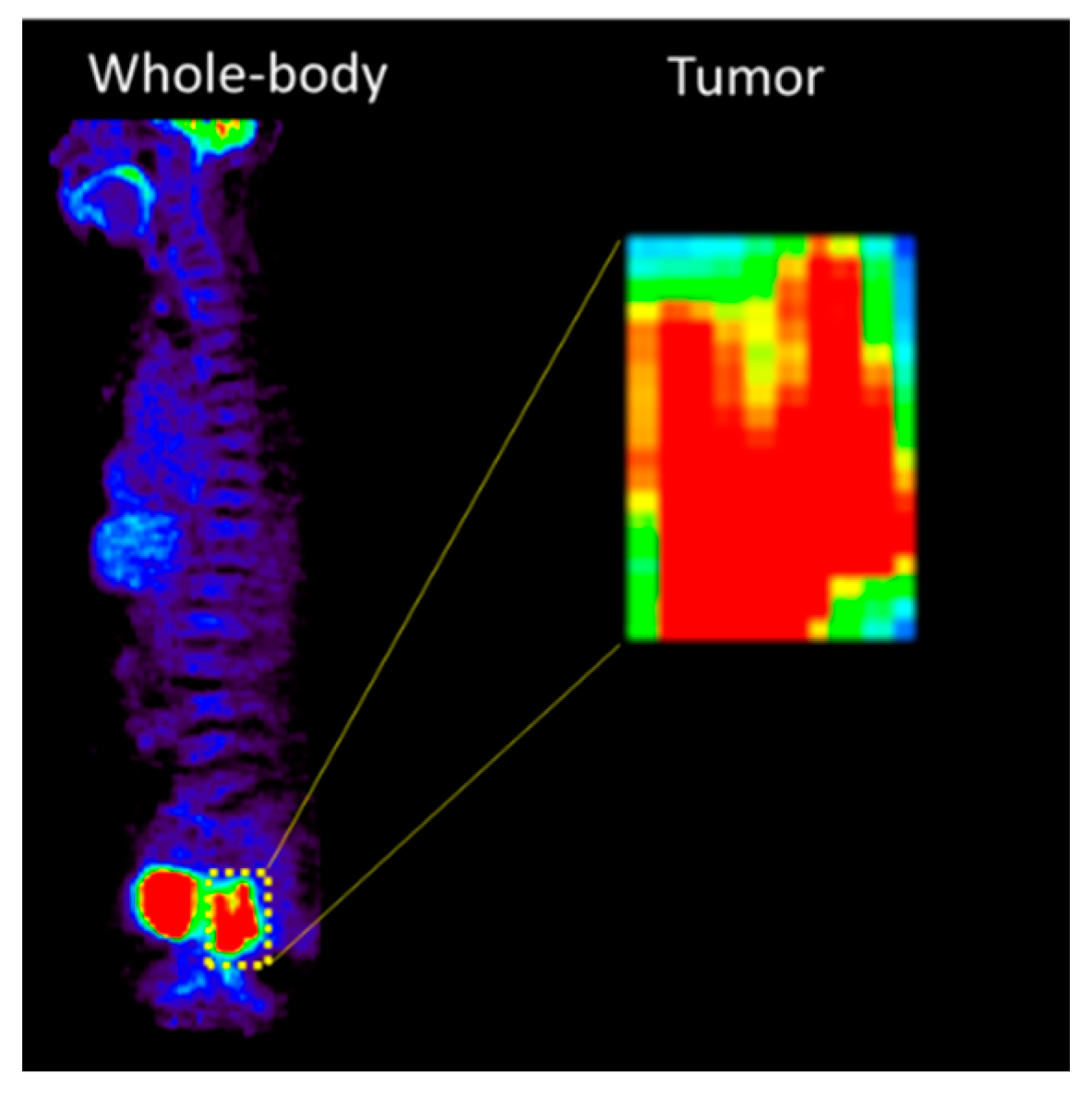
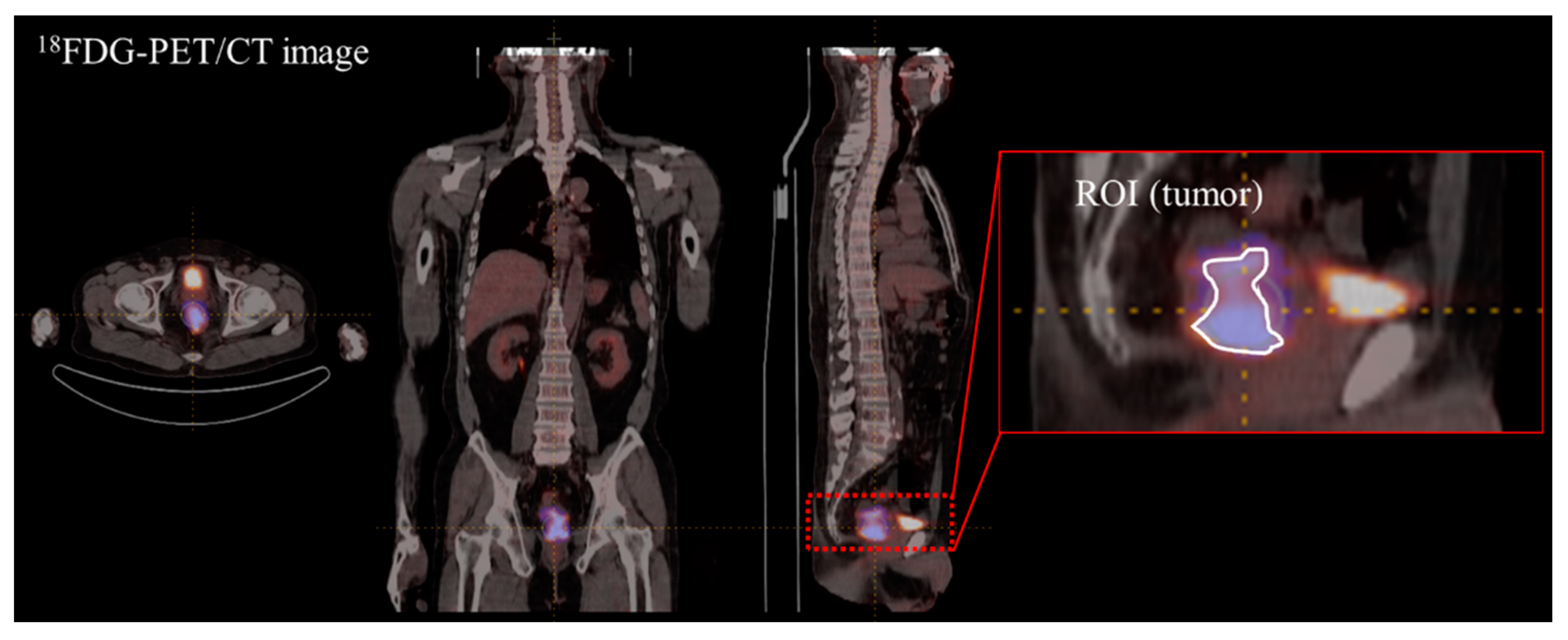
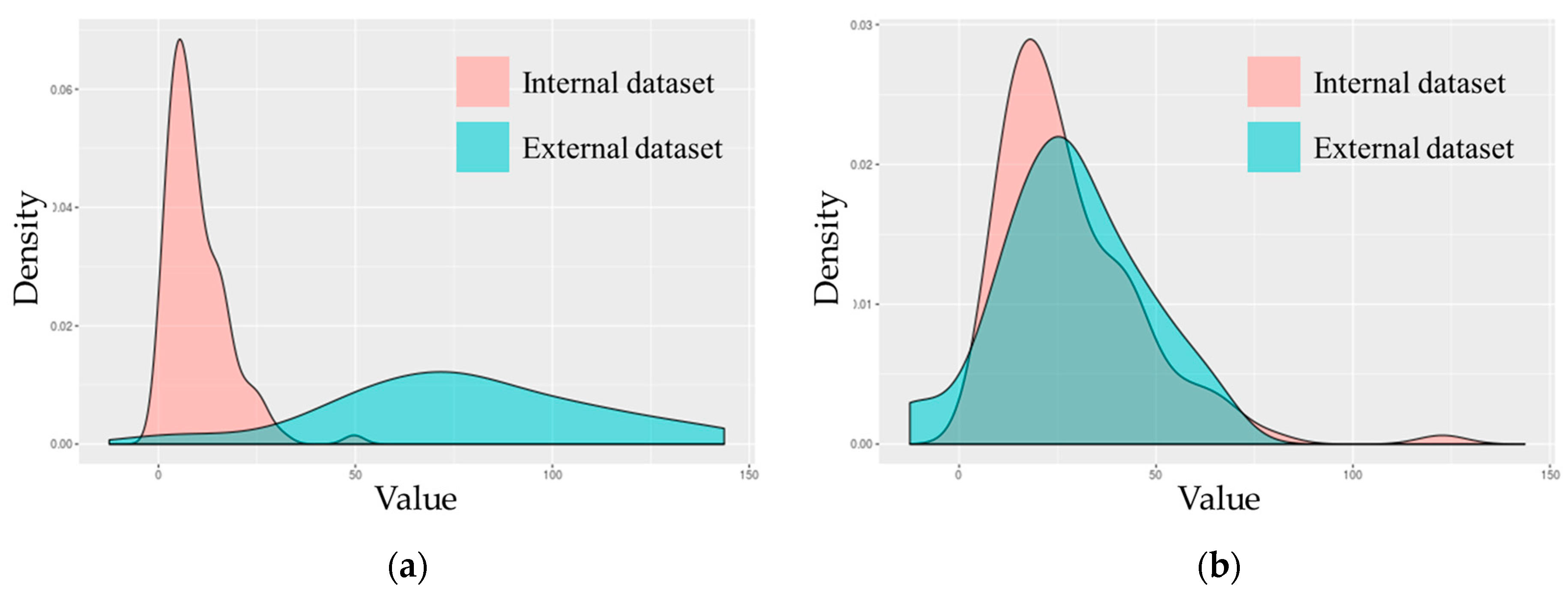
| Characteristics | Internal Dataset (n = 116) | External Dataset (n = 40) |
|---|---|---|
| Chemoradiotherapy response (%) | ||
| pCR | 21 (18.1) | 6 (15) |
| non-pCR | 95 (81.9) | 34 (85) |
| Age (%) | ||
| <65 | 69 (59.48) | 23 (57.5) |
| ≥65 | 47 (40.52) | 17 (42.5) |
| Mean age (y) | 61.85 | 59.88 |
| Sex (%) | ||
| Male | 75 (64.66) | 27 (67.5) |
| Female | 41 (35.34) | 13 (32.5) |
| Clinical T-stage, n (%) | ||
| T3 | 116 (100) | 40 |
| Clinical N stage (%) | ||
| N0 | 19 (16.38) | 5 (12.5) |
| N1 | 31 (26.72) | 8 (20) |
| N1a | 2 (1.72) | |
| N1b | 13 (11.21) | 1 (2.5) |
| N2 | 37 (31.9) | 6 (15) |
| N2a | 13 (11.21) | 12 (30) |
| N2b | 1 (0.86) | 8 (20) |
| Clinical M stage (%) | ||
| M0 | 106 (91.38) | 32 (80) |
| M1 | 6 (5.17) | |
| M1a | 3 (2.59) | 8 (50) |
| M1b | 1 (0.86) | |
| Clinical stage (%) | ||
| IIA | 5 (12.5) | |
| IIB | 18 (15.52) | |
| IIC | ||
| IIIA | 42 (36.21) | 21 (52.5) |
| IIIB | 46 (39.66) | 6 (15) |
| IIIC | 8 (20) | |
| IVA | 10 (8.62) |
| Number of Data | Efficiency Evaluation | |||||
|---|---|---|---|---|---|---|
| Data Set | pCR | Non-pCR | Accuracy | Precision | Sensitivity | AUC (95% CI) |
| Imbalanced | 21 | 21 | 0.867 | 0.871 | 0.871 | 0.903 (0.856–0.949) |
| Balanced | 84 | 95 | 0.789 | 0.843 | 0.677 | 0.835 (0.804–0.866) |
| Number of Data | Efficiency Evaluation | |||||
|---|---|---|---|---|---|---|
| Data Set | pCR | Non-pCR | Accuracy | Precision | Sensitivity | AUC (95% CI) |
| Imbalanced | 6 | 6 | 0.557 | 0.542 | 0.495 | 0.498 (0.412–0.583) |
| Balanced | 24 | 25 | 0.355 | 0.241 | 0.475 | 0.443 (0.378–0.509) |
| First-Order Image Feature | |||
|---|---|---|---|
| 18F-FDG PET | AUC | CT | AUC |
| SHAPE Sphericity | 0.715 | Uniformity | 0.663 |
| SUVQ1 | 0.707 | Entropy log10 | 0.659 |
| SUVmean | 0.694 | Entropy log2 | 0.659 |
| SUVQ3 | 0.692 | SHAPE Compacity | 0.618 |
| SUVQ2 | 0.69 | SHAPE Volume | 0.604 |
| Uniformity | 0.681 | SUVstd | 0.6 |
| Entropy log10 | 0.677 | SUVmax | 0.593 |
| Entropy log2 | 0.677 | SUVQ3 | 0.589 |
| SUVstd | 0.667 | Kurtosis | 0.582 |
| SUVmin | 0.65 | ExcessKurtosis | 0.582 |
| Volume | 0.663 | ||
| Sphericity | 0.579 | ||
| Skewness | 0.578 | ||
| TLG | 0.563 | ||
| Second-Order Image Feature | |||
|---|---|---|---|
| 18F-FDG PET | AUC | CT | AUC |
| GLZLM LZLGE | 0.766 | NGLDM Contrast | 0.704 |
| GLZLM LZE | 0.765 | GLZLM ZP | 0.698 |
| GLRLM GLNU | 0.763 | GLRLM LRE | 0.69 |
| GLRLM SRE | 0.756 | GLRLM RP | 0.69 |
| GLRLM RP | 0.755 | GLRLM SRE | 0.689 |
| GLRLM LRE | 0.753 | GLZLM LZLGE | 0.689 |
| NGLDM Contrast | 0.74 | GLCM Homogeneity | 0.689 |
| GLZLM ZP | 0.74 | GLZLM LZE | 0.685 |
| GLZLM LZHGE | 0.74 | GLZLM LZHGE | 0.683 |
| GLCM Homogeneity | 0.734 | GLCM Energy | 0.683 |
| NGLDM Busyness | 0.732 | GLCM Entropy log10 | 0.667 |
| GLRLM LRLGE | 0.731 | GLCM Entropy log2 | 0.667 |
| GLCM Dissimilarity | 0.71 | GLCM Dissimilarity | 0.661 |
| GLCM Contrast | 0.702 | GLRLM GLNU | 0.647 |
| GLRLM LGRE | 0.701 | GLRLM LRHGE | 0.633 |
| NGLDM Busyness | 0.628 | ||
| GLRLM SRHGE | 0.617 | ||
| GLCM Contrast | 0.613 | ||
| GLRLM LRLGE | 0.613 | ||
| Image Feature | Value | Without Harmonization | Without Harmonization | With Harmonization | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Internal Test | External Test | External Test | ||||||||
| CT | PET | PET/CT | CT | PET | PET/CT | CT | PET | PET/CT | ||
| First order | Accuracy | 0.54 | 0.62 | 0.56 | 0.55 | 0.7 | 0.525 | 0.6 | 0.646 | 0.771 |
| Precision | 0.524 | 0.575 | 0.615 | 0.227 | 0.2 | 0.19 | 0.222 | 0.769 | 0.882 | |
| Sensitivity | 0.88 | 0.92 | 0.32 | 0.833 | 0.333 | 0.667 | 0.667 | 0.417 | 0.625 | |
| AUC | 0.54 | 0.62 | 0.56 | 0.667 | 0.549 | 0.583 | 0.627 | 0.646 | 0.771 | |
| 95% CI for AUC | - | - | - | 0.412–0.921 | 0.291–0.807 | 0.325–0.842 | 0.37–0.885 | 0.469–0.962 | 0.429–0.934 | |
| Second order | Accuracy | 0.52 | 0.64 | 0.7 | 0.425 | 0.525 | 0.7 | 0.65 | 0.583 | 0.675 |
| Precision | 0.516 | 0.63 | 0.727 | 0.185 | 0.19 | 0.25 | 0.25 | 0.7 | 0.632 | |
| Sensitivity | 0.64 | 0.68 | 0.64 | 0.833 | 0.667 | 0.5 | 0.667 | 0.292 | 0.5 | |
| AUC | 0.52 | 0.64 | 0.7 | 0.593 | 0.583 | 0.618 | 0.657 | 0.583 | 0.603 | |
| 95% CI for AUC | - | - | - | 0.334–0.852 | 0.325–0.842 | 0.36–0.876 | 0.402–0.912 | 0.562–1 | 0.344–0.862 | |
| All | Accuracy | 0.68 | 0.76 | 0.7 | 0.65 | 0.675 | 0.775 | 0.425 | 0.875 | 0.725 |
| Precision | 0.765 | 0.81 | 0.639 | 0.214 | 0.267 | 0.333 | 0.185 | 0.952 | 0.333 | |
| Sensitivity | 0.52 | 0.68 | 0.92 | 0.5 | 0.667 | 0.5 | 0.833 | 0.833 | 0.833 | |
| AUC | 0.68 | 0.76 | 0.7 | 0.588 | 0.672 | 0.662 | 0.593 | 0.896 | 0.77 | |
| 95% CI for AUC | - | - | - | 0.329–0.847 | 0.418–0.925 | 0.556–1 | 0.334–0.852 | 0.562–1 | 0.536–1 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ju, H.-M.; Yang, J.; Park, J.-M.; Choi, J.-H.; Song, H.; Kim, B.-I.; Shin, U.-S.; Moon, S.M.; Cho, S.; Woo, S.-K. Prediction of Neoadjuvant Chemoradiotherapy Response in Rectal Cancer Patients Using Harmonized Radiomics of Multcenter 18F-FDG-PET Image. Cancers 2023, 15, 5662. https://doi.org/10.3390/cancers15235662
Ju H-M, Yang J, Park J-M, Choi J-H, Song H, Kim B-I, Shin U-S, Moon SM, Cho S, Woo S-K. Prediction of Neoadjuvant Chemoradiotherapy Response in Rectal Cancer Patients Using Harmonized Radiomics of Multcenter 18F-FDG-PET Image. Cancers. 2023; 15(23):5662. https://doi.org/10.3390/cancers15235662
Chicago/Turabian StyleJu, Hye-Min, Jingyu Yang, Jung-Mi Park, Joon-Ho Choi, Hyejin Song, Byung-Il Kim, Ui-Sup Shin, Sun Mi Moon, Sangsik Cho, and Sang-Keun Woo. 2023. "Prediction of Neoadjuvant Chemoradiotherapy Response in Rectal Cancer Patients Using Harmonized Radiomics of Multcenter 18F-FDG-PET Image" Cancers 15, no. 23: 5662. https://doi.org/10.3390/cancers15235662
APA StyleJu, H.-M., Yang, J., Park, J.-M., Choi, J.-H., Song, H., Kim, B.-I., Shin, U.-S., Moon, S. M., Cho, S., & Woo, S.-K. (2023). Prediction of Neoadjuvant Chemoradiotherapy Response in Rectal Cancer Patients Using Harmonized Radiomics of Multcenter 18F-FDG-PET Image. Cancers, 15(23), 5662. https://doi.org/10.3390/cancers15235662






