Concordance between Three Homologous Recombination Deficiency (HRD) Assays in Patients with High-Grade Epithelial Ovarian Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Tumor Tissue Processing and DNA Isolation
2.3. Targeted DNA NGS Analysis
2.4. AmoyDx® HRD Focus Panel
2.5. OncoScan™ (OncoScan Copy Number Variations (CNVs) Assay)
2.5.1. OncoScan Data Analysis
2.5.2. OncoScan GI Score Analysis Algorithm
2.6. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.1.1. Tumor Tissue Evaluation
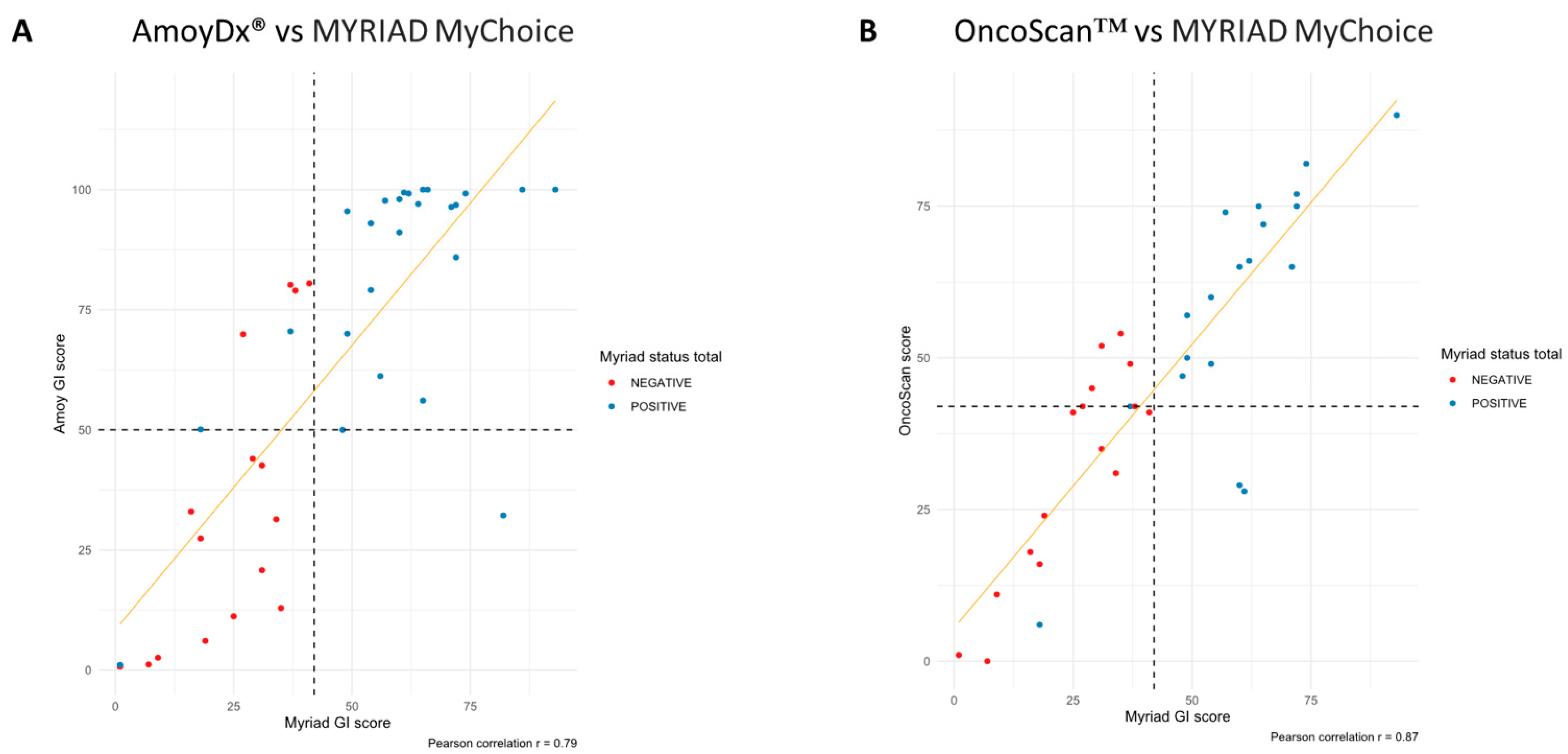
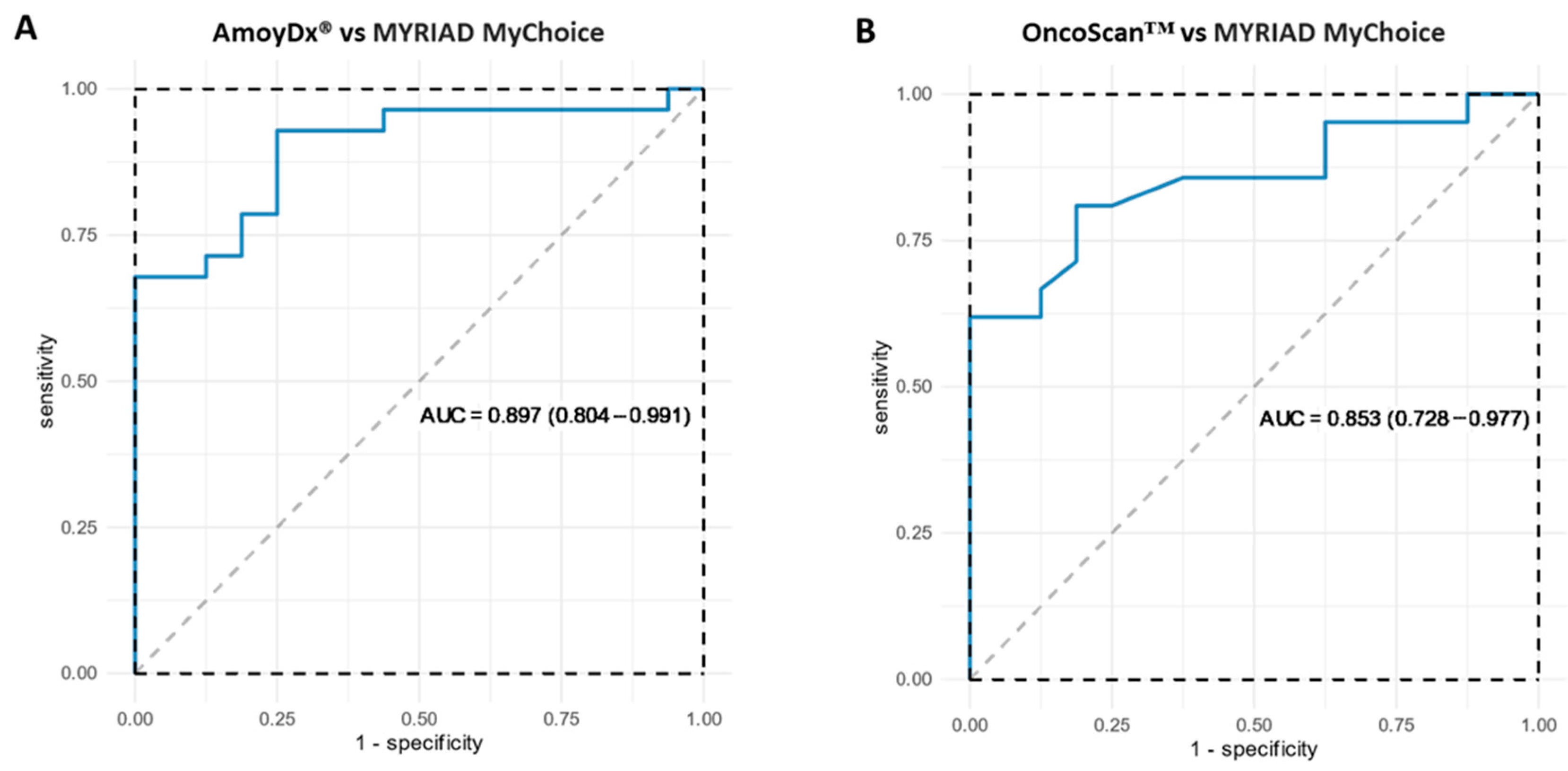
3.1.2. Concordance among Different HRD Assays
3.1.3. Concordance in Terms of GI Score
3.1.4. Concordance in Terms of GI Status
3.1.5. Mutations in BRCA1/2
3.1.6. Concordance in Terms of Final HRD Status
3.1.7. Mutations in HRR Genes
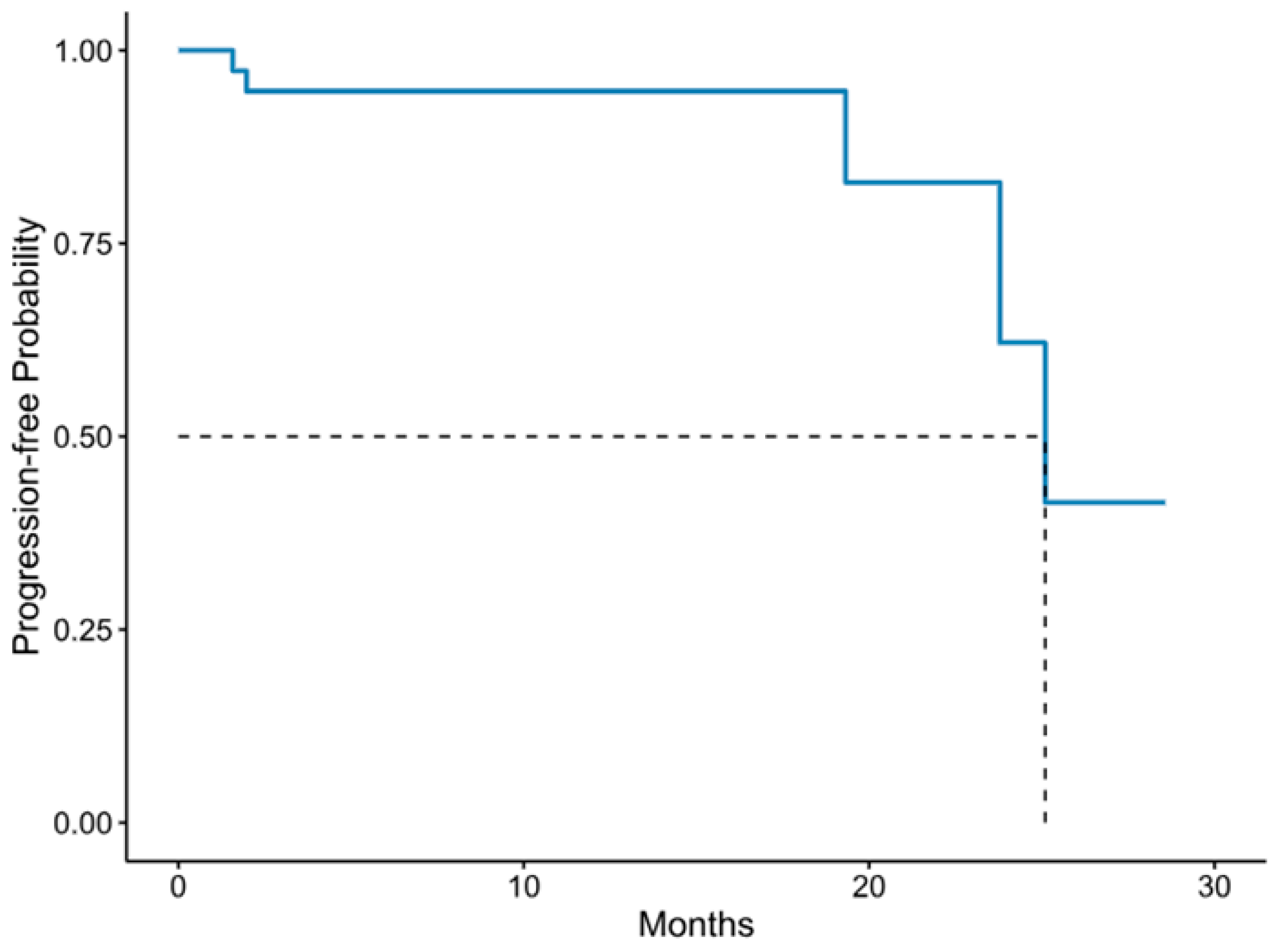
3.1.8. Patients’ Clinical Outcomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arora, T.; Mullangi, S.; Lekkala, M.R. Ovarian Cancer; StatPearls: Treasure Island, FL, USA, 2023. [Google Scholar]
- Chan, J.K.; Cheung, M.K.; Husain, A.; Teng, N.N.; West, D.; Whittemore, A.S.; Berek, J.S.; Osann, K. Patterns and progress in ovarian cancer over 14 years. Obstet. Gynecol. 2006, 108, 521–528. [Google Scholar] [CrossRef]
- Moore, K.; Colombo, N.; Scambia, G.; Kim, B.G.; Oaknin, A.; Friedlander, M.; Lisyanskaya, A.; Floquet, A.; Leary, A.; Sonke, G.S.; et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2018, 379, 2495–2505. [Google Scholar] [CrossRef]
- Penson, R.T.; Valencia, R.V.; Cibula, D.; Colombo, N.; Leath, C.A., 3rd; Bidzinski, M.; Kim, J.W.; Nam, J.H.; Madry, R.; Hernandez, C.; et al. Olaparib Versus Nonplatinum Chemotherapy in Patients With Platinum-Sensitive Relapsed Ovarian Cancer and a Germline BRCA1/2 Mutation (SOLO3): A Randomized Phase III Trial. J. Clin. Oncol. 2020, 38, 1164–1174. [Google Scholar] [CrossRef] [PubMed]
- González-Martín, A.; Pothuri, B.; Vergote, I.; DePont Christensen, R.; Graybill, W.; Mirza, M.R.; McCormick, C.; Lorusso, D.; Hoskins, P.; Freyer, G.; et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2391–2402. [Google Scholar] [CrossRef] [PubMed]
- Ray-Coquard, I.; Pautier, P.; Pignata, S.; Pérol, D.; González-Martín, A.; Berger, R.; Fujiwara, K.; Vergote, I.; Colombo, N.; Mäenpää, J.; et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2416–2428. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, D.; Mouret-Reynier, M.-A.; Harter, P.; Cropet, C.; Diaz, C.C.; Petru, E.; Satoh, T.; Vergote, I.B.; Parma, G.; Nøttrup, T.J.; et al. 5-year (y) overall survival (OS) with maintenance olaparib (ola) plus bevacizumab (bev) by clinical risk in patients (pts) with newly diagnosed advanced ovarian cancer (AOC) in the phase III PAOLA-1/ENGOT-ov25 trial. ESMO Open 2023, 8, 100812. [Google Scholar] [CrossRef]
- Trillsch, F.; Okamoto, A.; Reuss, A.; Kim, J.-W.; Rubio-Pérez, M.J.; Vardar, M.A.; Scambia, G.; Tredan, O.; Nyvang, G.-B.; Colombo, N.; et al. Durvalumab with paclitaxel/carboplatin (PC) and bevacizumab (bev), followed by maintenance durvalumab, bev, and olaparib in patients (pts) with newly diagnosed advanced ovarian cancer (AOC) without a tumor BRCA1/2 mutation (non-tBRCAm): Results from the randomized, placebo (pbo)-controlled phase III DUO-O trial. J. Clin. Oncol. 2023, 41, LBA5506. [Google Scholar] [CrossRef]
- Matulonis, U.; Herrstedt, J.; Oza, A.; Mahner, S.; Redondo, A.; Berton, D.; Berek, J.; Haslund, C.; Marmé, F.; González-Martín, A.; et al. Final overall survival and long-term safety in the ENGOT-OV16/NOVA phase III trial of niraparib in patients with recurrent ovarian cancer (LBA 6). Gynecol. Oncol. 2023, 176, S31–S32. [Google Scholar] [CrossRef]
- Konstantinopoulos, P.A.; Ceccaldi, R.; Shapiro, G.I.; D’Andrea, A.D. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov. 2015, 5, 1137–1154. [Google Scholar] [CrossRef]
- Blanc-Durand, F.; Tang, R.; Pommier, M.; Nashvi, M.; Cotteret, S.; Genestie, C.; Le Formal, A.; Pautier, P.; Michels, J.; Kfoury, M.; et al. Clinical relevance of BRCA1 promoter methylation testing in ovarian cancer patients. Clin. Cancer Res. 2023, 29, 3124–3129. [Google Scholar] [CrossRef]
- Lønning, P.E.; Nikolaienko, O.; Pan, K.; Kurian, A.W.; Eikesdal, H.P.; Pettinger, M.; Anderson, G.L.; Prentice, R.L.; Chlebowski, R.T.; Knappskog, S. Constitutional BRCA1 Methylation and Risk of Incident Triple-Negative Breast Cancer and High-grade Serous Ovarian Cancer. JAMA Oncol. 2022, 8, 1579–1587. [Google Scholar] [CrossRef] [PubMed]
- Hollis, R.L.; Churchman, M.; Michie, C.O.; Rye, T.; Knight, L.; McCavigan, A.; Perren, T.; Williams, A.R.W.; McCluggage, W.G.; Kaplan, R.S.; et al. High EMSY expression defines a BRCA-like subgroup of high-grade serous ovarian carcinoma with prolonged survival and hypersensitivity to platinum. Cancer 2019, 125, 2772–2781. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.H.; Balajee, A.S.; Wang, J.; Wu, H.; Eng, C.; Pandolfi, P.P.; Yin, Y. Essential Role for Nuclear PTEN in Maintaining Chromosomal Integrity. Cell 2007, 128, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Myriad Genetic Laboratories, Inc. Cancer-Related Germline Gene Mutation Detection System; Myriad Genetic Laboratories, Inc.: Salt Lake City, UT, USA, 2019. [Google Scholar]
- Cristescu, R.; Liu, X.Q.; Arreaza, G.; Chen, C.; Albright, A.; Qiu, P.; Marton, M.J. Concordance between single-nucleotide polymorphism-based genomic instability assays and a next-generation sequencing-based homologous recombination deficiency test. BMC Cancer 2022, 22, 1310. [Google Scholar] [CrossRef]
- Tsantikidi, A.; Papazisis, K.; Floros, T.; Gazouli, M.; Papadopoulou, E.; Tsaousis, G.; Nasioulas, G.; Mester, A.; Milan, K.P.; Gozman, B.; et al. RediScore: Prospective validation of a pipeline for homologous recombination deficiency analysis. Oncol. Lett. 2023, 26, 480. [Google Scholar] [CrossRef]
- Yuan, W.; Ni, J.; Wen, H.; Shi, W.; Chen, X.; Huang, H.; Zhang, X.; Lu, X.; Zhu, C.; Dong, H.; et al. Genomic Scar Score: A robust model predicting homologous recombination deficiency based on genomic instability. BJOG Int. J. Obstet. Gynaecol. 2022, 129 (Suppl. S2), 14–22. [Google Scholar] [CrossRef]
- Illumina. FoundationOne CDxTM. Available online: https://www.foundationmedicine.com/test/foundationone-cdx (accessed on 15 November 2023).
- Fountzilas, G.; Giannoulatou, E.; Alexopoulou, Z.; Zagouri, F.; Timotheadou, E.; Papadopoulou, K.; Lakis, S.; Bobos, M.; Poulios, C.; Sotiropoulou, M.; et al. TP53 mutations and protein immunopositivity may predict for poor outcome but also for trastuzumab benefit in patients with early breast cancer treated in the adjuvant setting. Oncotarget 2016, 7, 32731–32753. [Google Scholar] [CrossRef]
- Favero, F.; Joshi, T.; Marquard, A.M.; Birkbak, N.J.; Krzystanek, M.; Li, Q.; Szallasi, Z.; Eklund, A.C. Sequenza: Allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol. 2015, 26, 64–70. [Google Scholar] [CrossRef]
- Van Loo, P.; Nordgard, S.H.; Lingjaerde, O.C.; Russnes, H.G.; Rye, I.H.; Sun, W.; Weigman, V.J.; Marynen, P.; Zetterberg, A.; Naume, B.; et al. Allele-specific copy number analysis of tumors. Proc. Natl. Acad. Sci. USA 2010, 107, 16910–16915. [Google Scholar] [CrossRef]
- Ross, E.M.; Haase, K.; Van Loo, P.; Markowetz, F. Allele-specific multi-sample copy number segmentation in ASCAT. Bioinformatics 2021, 37, 1909–1911. [Google Scholar] [CrossRef]
- Imanishi, S.; Naoi, Y.; Shimazu, K.; Shimoda, M.; Kagara, N.; Tanei, T.; Miyake, T.; Kim, S.J.; Noguchi, S. Clinicopathological analysis of homologous recombination-deficient breast cancers with special reference to response to neoadjuvant paclitaxel followed by FEC. Breast Cancer Res. Treat. 2019, 174, 627–637. [Google Scholar] [CrossRef]
- Abkevich, V.; Timms, K.M.; Hennessy, B.T.; Potter, J.; Carey, M.S.; Meyer, L.A.; Smith-McCune, K.; Broaddus, R.; Lu, K.H.; Chen, J.; et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer 2012, 107, 1776–1782. [Google Scholar] [CrossRef] [PubMed]
- Birkbak, N.J.; Wang, Z.C.; Kim, J.Y.; Eklund, A.C.; Li, Q.; Tian, R.; Bowman-Colin, C.; Li, Y.; Greene-Colozzi, A.; Iglehart, J.D.; et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012, 2, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Popova, T.; Manie, E.; Rieunier, G.; Caux-Moncoutier, V.; Tirapo, C.; Dubois, T.; Delattre, O.; Sigal-Zafrani, B.; Bollet, M.; Longy, M.; et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012, 72, 5454–5462. [Google Scholar] [CrossRef] [PubMed]
- Timms, K.M.; Abkevich, V.; Hughes, E.; Neff, C.; Reid, J.; Morris, B.; Kalva, S.; Potter, J.; Tran, T.V.; Chen, J.; et al. Association of BRCA1/2 defects with genomic scores predictive of DNA damage repair deficiency among breast cancer subtypes. Breast Cancer Res. 2014, 16, 475. [Google Scholar] [CrossRef]
- Ruopp, M.D.; Perkins, N.J.; Whitcomb, B.W.; Schisterman, E.F. Youden Index and optimal cut-point estimated from observations affected by a lower limit of detection. Biom. J. Biom. Z. 2008, 50, 419–430. [Google Scholar] [CrossRef]
- Weichert, W.; Bartels, S.; Baretton, G.; Braicu, E.; Demes, M.; Endris, V.; Herold, S.; Heukamp, L.; Hummel, M.; Lehmann, U.; et al. 758P Concordance between multiple HRD assays is substantial in high-grade ovarian cancer. Ann. Oncol. 2021, 32, S747. [Google Scholar] [CrossRef]
- Fumagalli, C.; Betella, I.; Ranghiero, A.; Guerini-Rocco, E.; Bonaldo, G.; Rappa, A.; Vacirca, D.; Colombo, N.; Barberis, M. In-house testing for homologous recombination repair deficiency (HRD) testing in ovarian carcinoma: A feasibility study comparing AmoyDx HRD Focus panel with Myriad myChoiceCDx assay. Pathologica 2022, 114, 288–294. [Google Scholar] [CrossRef]
- Magliacane, G.; Brunetto, E.; Calzavara, S.; Bergamini, A.; Pipitone, G.B.; Marra, G.; Redegalli, M.; Grassini, G.; Rabaiotti, E.; Taccagni, G.; et al. Locally Performed HRD Testing for Ovarian Cancer? Yes, We Can! Cancers 2022, 15, 43. [Google Scholar] [CrossRef]
- Illumina. TruSight Oncology 500. Available online: https://emea.illumina.com/products/by-type/clinical-research-products/trusight-oncology-500.html (accessed on 15 November 2023).
- Weichert, W.; Qiu, P.; Lunceford, J.; Wehn, A.; Yarunin, A.; Cristescu, R.; Liu, L.; Roessler, K.; Timms, K.; Marton, M.J. Assessing homologous recombination deficiency (HRD) in ovarian cancer: Optimizing concordance of the regulatory-approved companion diagnostic and a next-generation sequencing (NGS) assay kit. J. Clin. Oncol. 2022, 40, e1757. [Google Scholar] [CrossRef]
- Loverix, L.; Vergote, I.; Busschaert, P.; Vanderstichele, A.; Boeckx, B.; Venken, T.; Harter, P.; Brems, H.; Nieuwenhuysen, E.V.; Pignata, S.; et al. Predictive value of the Leuven HRD test compared with Myriad myChoice PLUS on 468 ovarian cancer samples from the PAOLA-1/ENGOT-ov25 trial (LBA 6). Gynecol. Oncol. 2022, 166, S51–S52. [Google Scholar] [CrossRef]
- Zhao, M.; Wang, Q.; Wang, Q.; Jia, P.; Zhao, Z. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: Features and perspectives. BMC Bioinform. 2013, 14 (Suppl. 11), S1. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Morrison, C.D.; Johnson, C.S.; Trump, D.L.; Qin, M.; Conroy, J.C.; Wang, J.; Liu, S. Computational methods for detecting copy number variations in cancer genome using next generation sequencing: Principles and challenges. Oncotarget 2013, 4, 1868–1881. [Google Scholar] [CrossRef] [PubMed]
- Zare, F.; Dow, M.; Monteleone, N.; Hosny, A.; Nabavi, S. An evaluation of copy number variation detection tools for cancer using whole exome sequencing data. BMC Bioinform. 2017, 18, 286. [Google Scholar] [CrossRef] [PubMed]
- Konstantopoulou, I.; Tsitlaidou, M.; Fostira, F.; Pertesi, M.; Stavropoulou, A.V.; Triantafyllidou, O.; Tsotra, E.; Tsiftsoglou, A.P.; Tsionou, C.; Droufakou, S.; et al. High prevalence of BRCA1 founder mutations in Greek breast/ovarian families. Clin. Genet. 2014, 85, 36–42. [Google Scholar] [CrossRef]
- Apostolou, P.; Fostira, F.; Kouroussis, C.; Kalfakakou, D.; Delimitsou, A.; Agelaki, S.; Androulakis, N.; Christodoulou, C.; Kalbakis, K.; Kalykaki, A.; et al. BRCA1 and BRCA2 germline testing in Cretan isolates reveals novel and strong founder effects. Int. J. Cancer 2020, 147, 1334–1342. [Google Scholar] [CrossRef]
- Fostira, F.; Kostantopoulou, I.; Apostolou, P.; Papamentzelopoulou, M.S.; Papadimitriou, C.; Faliakou, E.; Christodoulou, C.; Boukovinas, I.; Razis, E.; Tryfonopoulos, D.; et al. One in three highly selected Greek patients with breast cancer carries a loss-of-function variant in a cancer susceptibility gene. J. Med. Genet. 2020, 57, 53–61. [Google Scholar] [CrossRef]
- Fostira, F.; Tsitlaidou, M.; Papadimitriou, C.; Pertesi, M.; Timotheadou, E.; Stavropoulou, A.V.; Glentis, S.; Bournakis, E.; Bobos, M.; Pectasides, D.; et al. Prevalence of BRCA1 mutations among 403 women with triple-negative breast cancer: Implications for genetic screening selection criteria: A Hellenic Cooperative Oncology Group Study. Breast Cancer Res. Treat. 2012, 134, 353–362. [Google Scholar] [CrossRef]
- Apessos, A.; Agiannitopoulos, K.; Pepe, G.; Tsaousis, G.N.; Papadopoulou, E.; Metaxa-Mariatou, V.; Tsirigoti, A.; Efstathiadou, C.; Markopoulos, C.; Xepapadakis, G.; et al. Comprehensive BRCA mutation analysis in the Greek population. Experience from a single clinical diagnostic center. Cancer Genet. 2018, 220, 1–12. [Google Scholar] [CrossRef]
- Tsaousis, G.N.; Papadopoulou, E.; Apessos, A.; Agiannitopoulos, K.; Pepe, G.; Kampouri, S.; Diamantopoulos, N.; Floros, T.; Iosifidou, R.; Katopodi, O.; et al. Analysis of hereditary cancer syndromes by using a panel of genes: Novel and multiple pathogenic mutations. BMC Cancer 2019, 19, 535. [Google Scholar] [CrossRef]
- Agiannitopoulos, K.; Pepe, G.; Tsaousis, G.N.; Potska, K.; Bouzarelou, D.; Katseli, A.; Ntogka, C.; Meintani, A.; Tsoulos, N.; Giassas, S.; et al. Copy Number Variations (CNVs) Account for 10.8% of Pathogenic Variants in Patients Referred for Hereditary Cancer Testing. Cancer Genom. Proteom. 2023, 20, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Giannini, A.; Di Dio, C.; Di Donato, V.; D’Oria, O.; Salerno, M.G.; Capalbo, G.; Cuccu, I.; Perniola, G.; Muzii, L.; Bogani, G. PARP Inhibitors in Newly Diagnosed and Recurrent Ovarian Cancer. Am. J. Clin. Oncol. 2023, 46, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Soung, Y.H.; Chung, J. Combination Treatment Strategies to Overcome PARP Inhibitor Resistance. Biomolecules 2023, 13, 1480. [Google Scholar] [CrossRef]
| Characteristic | Total n = 50 |
|---|---|
| Age at diagnosis | |
| Median (min, max) | 60 (38–84) |
| Histological subtype (n = 50) | n (%) |
| Serous | 45 (90.0) |
| Clear cell | 3 (6.0) |
| Endometrioid | 2 (4.0) |
| Family history cancer (n = 47) | |
| No | 33 (70.2) |
| Yes | 14 (29.8) |
| Family history breast/ovarian cancer (n = 47) | |
| No | 42 (89.4) |
| Yes | 5 (10.6) |
| Performance status (n = 45) | |
| 0 | 36 (80) |
| 1 | 6 (13.3) |
| 2 | 3 (6.7) |
| 3 | 0 |
| 4 | 0 |
| Stage at diagnosis | |
| I | 3 |
| II | 7 |
| III | 30 |
| IV | 10 |
| Neoadjuvant treatment (n = 50) | |
| Yes | 14 (20.0) |
| No | 36 (80.0) |
| First-line treatment (n = 50) | |
| Yes | 39 (78.0) |
| No | 11 (22.0) |
| Maintenance treatment (n = 50) | |
| Yes | 33 (66.0) |
| No | 17 (34.0) |
| Response to first-line treatment (n = 39) | |
| Complete response/no evidence of disease | 17 (43.6) |
| Partial response | 12 (30.8) |
| Stable disease | 7 (17.9) |
| Disease progression | 3 (7.7) |
| Maintenance treatment (n = 33) | |
| PARP inhibitor | 9 (27.3) |
| Bevacizumab | 9 (27.3) |
| PARP inhibitor/bevacizumab | 15 (45.4) |
| Myriad MyChoice® CDx | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Metric | n | Result | + | − | OPA | Sensitivity | Specificity | PPV | NPV | |
| AmoyDx® HRD Focus | GI status | 42 | + | 22 | 6 | 83.3% (68.6–93.3%) | 95.7% (78.1–99.9) | 68.4% (43.4–87.4) | 78.6% | 92.9% |
| − | 1 | 13 | ||||||||
| HRD | 44 | + | 28 | 5 | 88.6% (75.4–96.2) | 100% (87.7–100) | 68.8% (41.3–89) | 84.8% | 100% | |
| − | 0 | 11 | ||||||||
| OncoScan™ | GI status | 40 | + | 16 | 7 | 77.5% (61.5–89.2%) | 88.9% (65.3–98.6) | 68.1% (45.1–86.1) | 69.6% | 88.2% |
| − | 2 | 15 | ||||||||
| HRD | GI Status | BRCA1/2 Mutations | |||||
|---|---|---|---|---|---|---|---|
| ID | Myriad | Amoy | Myriad | Amoy | OncoScan | Myriad | Amoy |
| 1 | Positive | Positive | Positive | Positive | Positive | No | No |
| 2 | Positive | Positive | Positive | Negative | - | Yes | Yes |
| 3 | Negative | Negative | Negative | Negative | Negative | No | No |
| 4 | Negative | Negative | Negative | Negative | Negative | No | No |
| 5 | Positive | Positive | Positive | Positive | Positive | Yes | Yes |
| 6 | Positive | Positive | Positive | Positive | Negative | No | No |
| 7 | Negative | Negative | Negative | Negative | Positive | No | No |
| 8 | Positive | Positive | Positive | Positive | Positive | No | No |
| 9 | Positive | Positive | Positive | Positive | Positive | Yes | Yes |
| 10 | Negative | Negative | Negative | Negative | Negative | No | No |
| 11 | Negative | Negative * | Negative | Negative * | Negative | No | No |
| 12 | Positive | Positive | Positive | Positive | Positive | No | No |
| 13 | Positive | Positive | Positive | Positive | Negative | No | No |
| 14 | Negative | Negative | Negative | Negative | Positive | No | No |
| 15 | Positive | Positive | Positive | Positive | Positive | No | No |
| 16 | Positive | Positive | Negative | Positive | Positive | Yes | Yes |
| 17 | Inconclusive | Negative | Inconclusive | Negative | Negative | No | No |
| 18 | Negative | Positive | Negative | Negative | Negative | No | Yes |
| 19 | Positive | Positive | Positive | Positive | Positive | No | No |
| 20 | Positive | Positive | Positive | Positive | Positive | Yes | No |
| 21 | Positive | Positive | Positive | Positive | Positive | No | Yes |
| 22 | Negative | Negative | Negative | Negative | Positive | No | No |
| 23 | Positive | Positive | Positive | Positive | Positive | No | No |
| 24 | Positive | Positive | Positive | Positive | - | Yes | No |
| 25 | Positive | Positive | Positive | Positive | - | Yes | No |
| 26 | Positive | Positive | Positive | Positive | Positive | No | No |
| 27 | Positive | Positive | Positive | Positive | Positive | No | Yes |
| 28 | Positive | Positive | Positive | Positive | Positive | No | No |
| 29 | Inconclusive | Negative | Inconclusive | Negative | Negative | No | No |
| 30 | Negative | Negative * | Negative | Negative * | Negative | No | No |
| 31 | Negative | Negative * | Negative | Negative * | Negative | No | No |
| 32 | Positive | Positive | Positive | Positive | Positive | No | No |
| 33 | Negative | Negative | Negative | Negative | Negative | No | No |
| 34 | Negative | Negative | Negative | Negative | Negative | No | No |
| 35 | Negative | Positive | Negative | Positive | Negative | No | No |
| 36 | Positive | Positive | Positive | Positive | - | Yes | No |
| 37 | Negative | Positive | Negative | Positive | Positive | No | No |
| 38 | Positive | Positive | Positive | Positive | - | Yes | Yes |
| 39 | Positive | Positive | Negative | Negative | - | Yes | Yes |
| 40 | Negative | Negative * | Negative | Negative * | Negative | No | No |
| 41 | Positive | Positive | Inconclusive | Positive | - | Yes | Yes |
| 42 | Positive | Positive | Positive | Positive | Positive | No | No |
| 43 | Negative | Positive | Negative | Positive | Positive | No | No |
| 44 | Positive | Positive | Inconclusive | Positive | Positive | Yes | No |
| 45 | Negative | Negative | Negative | Negative | Negative | No | No |
| 46 | Positive | Positive | Negative | Positive | Negative | Yes | No |
| 47 | Negative | Negative | Negative | Negative | Negative | No | No |
| 48 | Negative | Positive | Negative | Positive | Positive | No | No |
| 49 | Negative | Negative | Negative | Negative | Negative | No | No |
| 50 | Positive | Positive | Positive | Positive | Positive | No | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fountzilas, E.; Papadopoulou, K.; Chatzikonstantinou, T.; Karakatsoulis, G.; Constantoulakis, P.; Tsantikidi, A.; Tsaousis, G.; Karageorgopoulou, S.; Koumarianou, A.; Mauri, D.; et al. Concordance between Three Homologous Recombination Deficiency (HRD) Assays in Patients with High-Grade Epithelial Ovarian Cancer. Cancers 2023, 15, 5525. https://doi.org/10.3390/cancers15235525
Fountzilas E, Papadopoulou K, Chatzikonstantinou T, Karakatsoulis G, Constantoulakis P, Tsantikidi A, Tsaousis G, Karageorgopoulou S, Koumarianou A, Mauri D, et al. Concordance between Three Homologous Recombination Deficiency (HRD) Assays in Patients with High-Grade Epithelial Ovarian Cancer. Cancers. 2023; 15(23):5525. https://doi.org/10.3390/cancers15235525
Chicago/Turabian StyleFountzilas, Elena, Kyriaki Papadopoulou, Thomas Chatzikonstantinou, Georgios Karakatsoulis, Pantelis Constantoulakis, Aikaterini Tsantikidi, Georgios Tsaousis, Sofia Karageorgopoulou, Anna Koumarianou, Davide Mauri, and et al. 2023. "Concordance between Three Homologous Recombination Deficiency (HRD) Assays in Patients with High-Grade Epithelial Ovarian Cancer" Cancers 15, no. 23: 5525. https://doi.org/10.3390/cancers15235525
APA StyleFountzilas, E., Papadopoulou, K., Chatzikonstantinou, T., Karakatsoulis, G., Constantoulakis, P., Tsantikidi, A., Tsaousis, G., Karageorgopoulou, S., Koumarianou, A., Mauri, D., Ntavatzikos, A., Saridaki, Z., Petrakis, G., Fostira, F., Fountzilas, G., & Liontos, M. (2023). Concordance between Three Homologous Recombination Deficiency (HRD) Assays in Patients with High-Grade Epithelial Ovarian Cancer. Cancers, 15(23), 5525. https://doi.org/10.3390/cancers15235525









