Nidogen-1/NID1 Function and Regulation during Progression and Metastasis of Colorectal Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture, Treatments, and Transfections
2.2. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qPCR) Assay
2.3. Chromatin Immunoprecipitation (ChIP) Assay
2.4. CellTiter GLO Cell Viability Assay
2.5. Metastases Formation in a Tail Vein Injection Xenograft Mouse Model
2.6. Bioinformatics Analysis of Public Datasets
2.7. Statistical Analysis
3. Results
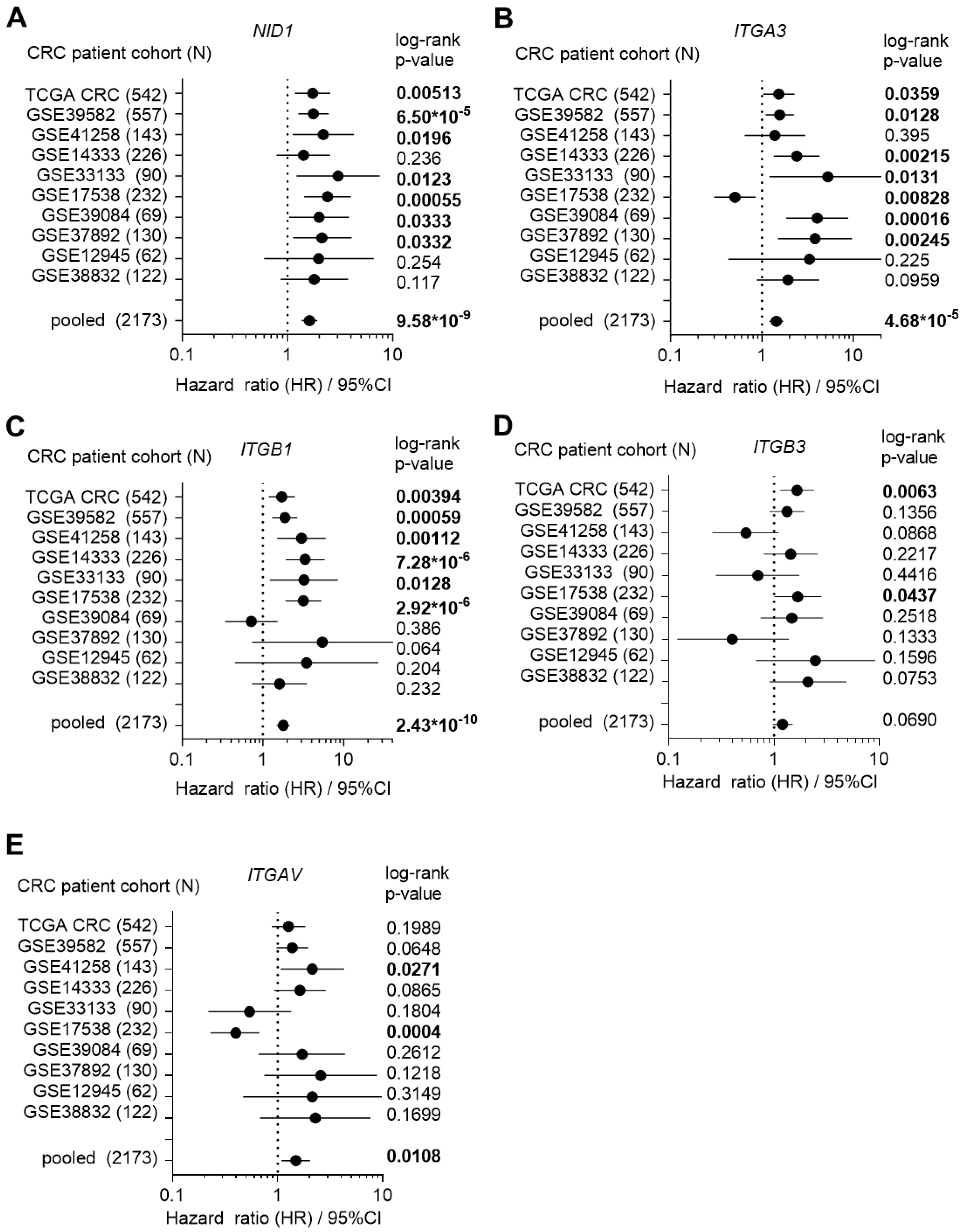
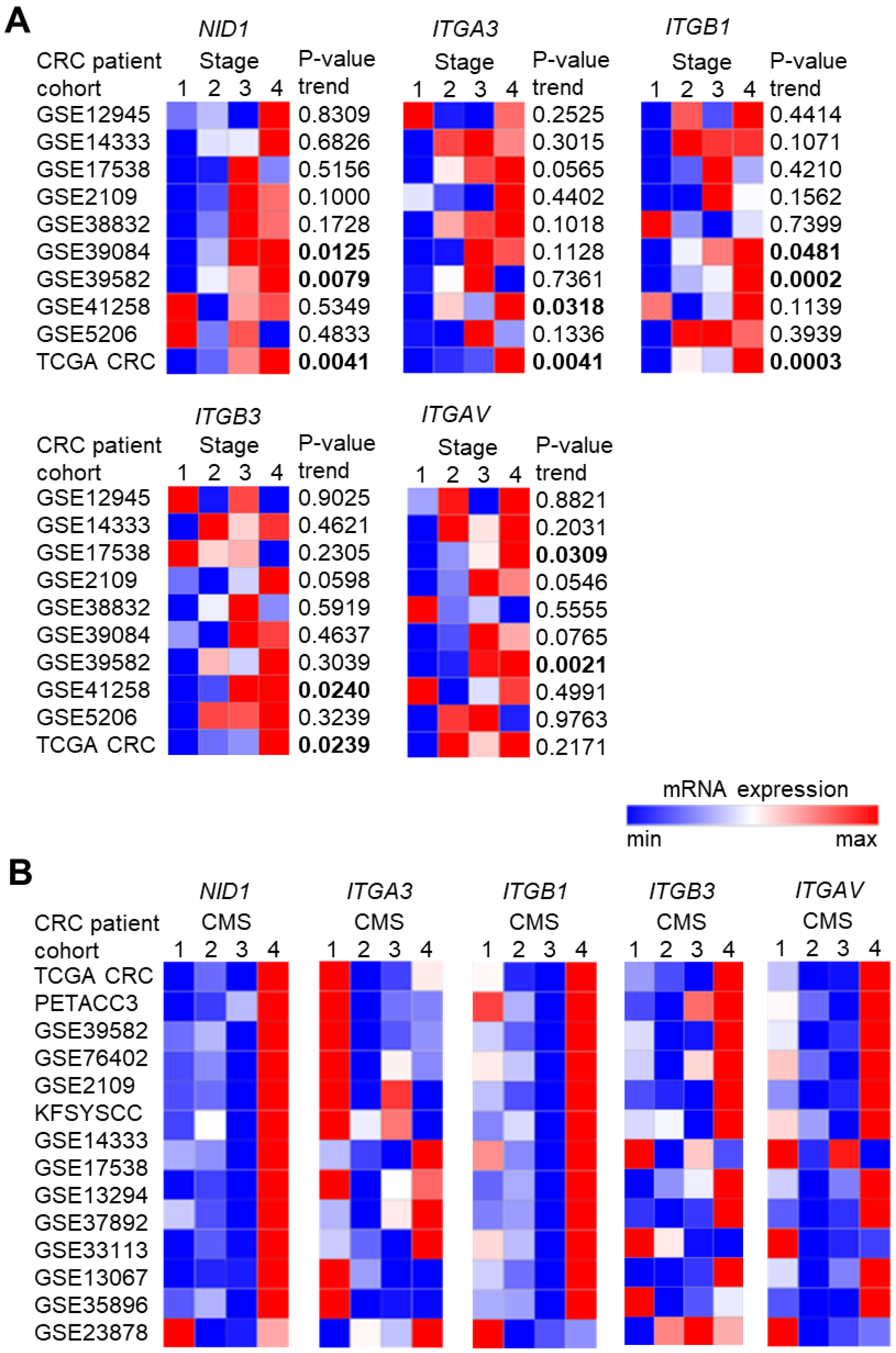
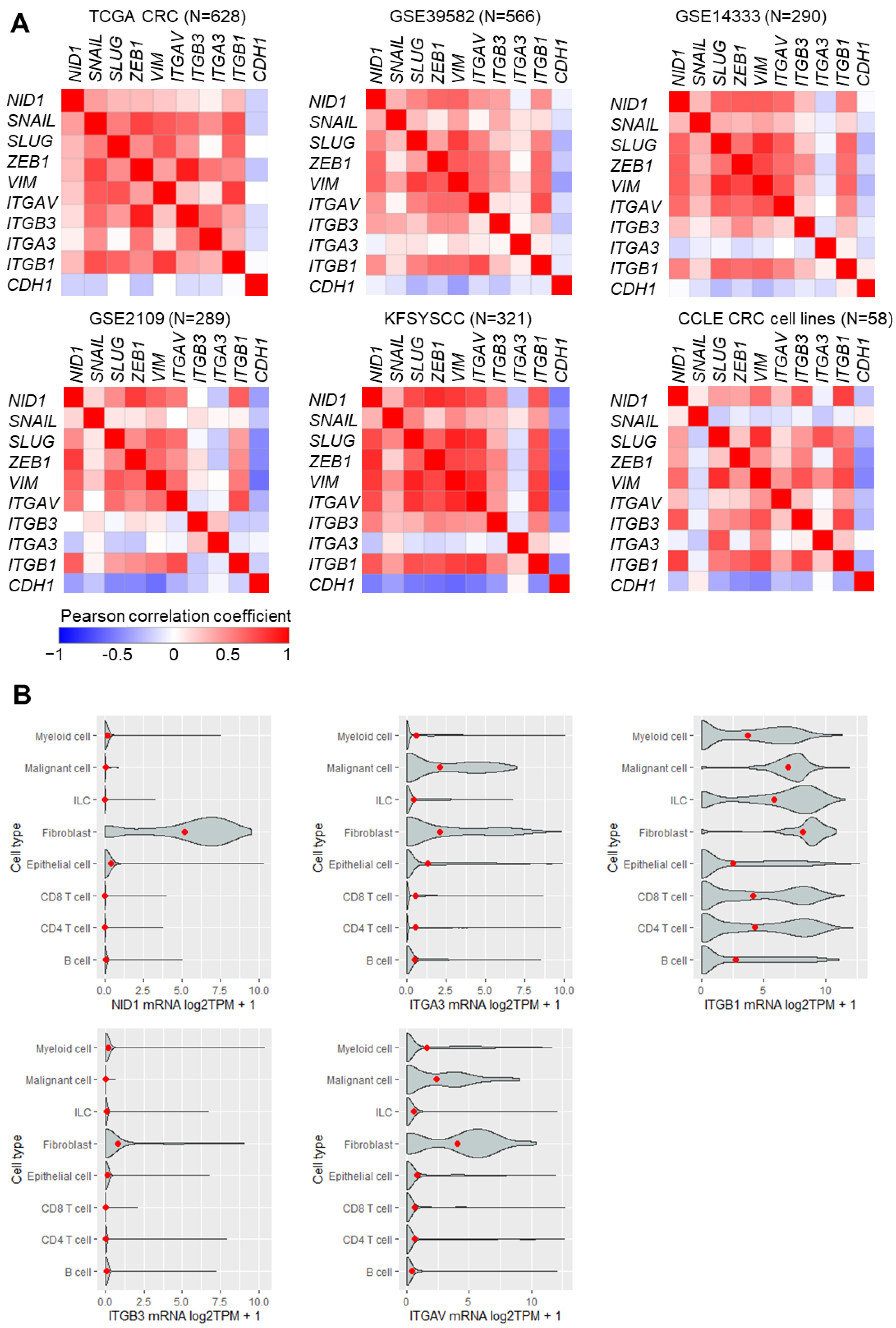
3.1. High Expression of NID1 and Its Receptors Is Associated with Poor Prognosis, Advanced Stage, and Mesenchymal Tumor Features in CRC
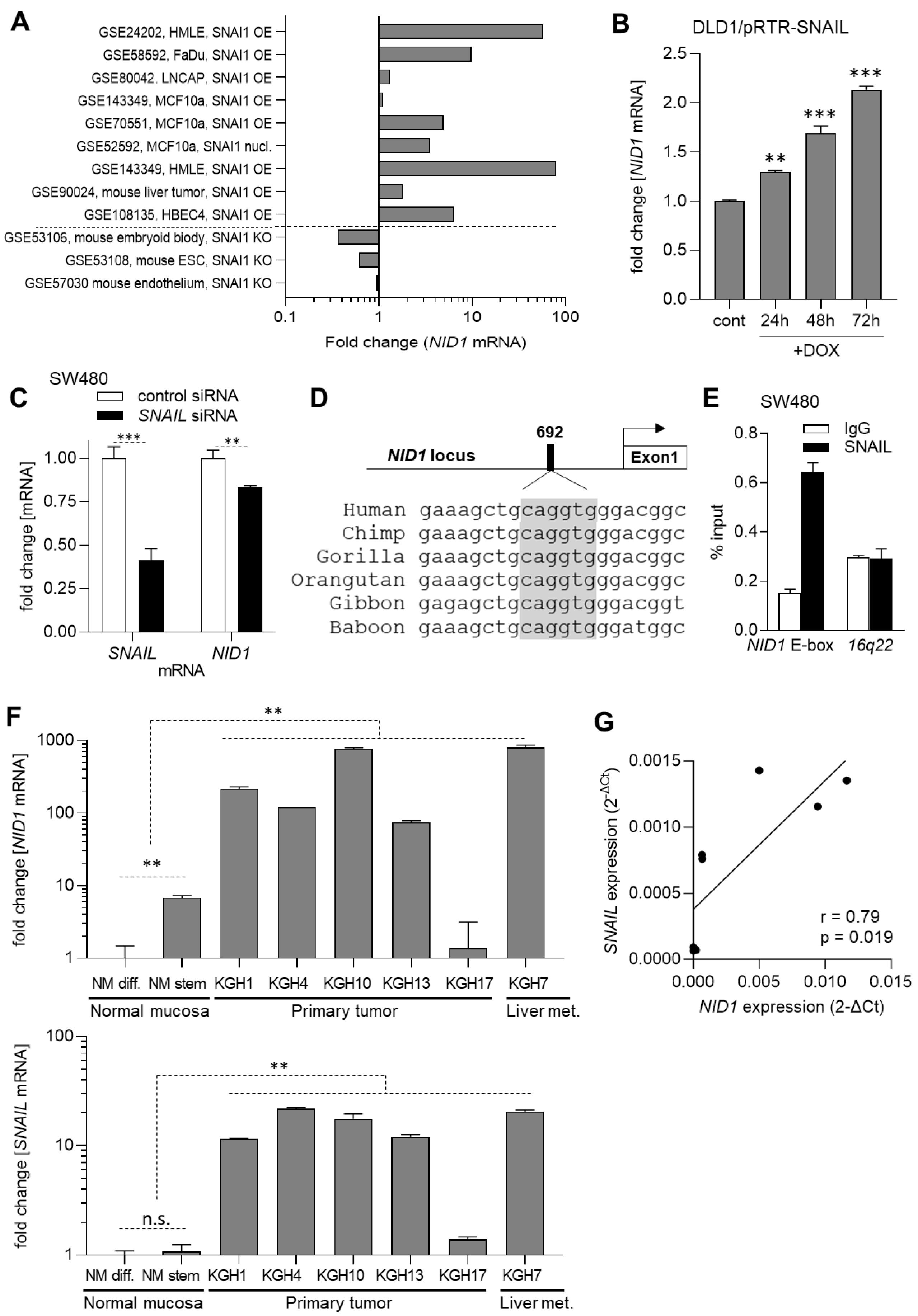
3.2. The Expression of NID1 Is Directly Induced by the EMT-TF SNAIL
3.3. Elevated Expression of NID1 in CRC Cells
3.4. NID1 Promotes Metastases Formation by CRC Cells
3.5. Suppression of ITGAV Decreases Cell Viability of CRC Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- GLOBOCAN Global Cancer Observatory (GCO): Cancer Today. Available online: https://gco.iarc.fr/today/ (accessed on 31 October 2023).
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef] [PubMed]
- Valastyan, S.; Weinberg, R.A. Tumor metastasis: Molecular insights and evolving paradigms. Cell 2011, 147, 275–292. [Google Scholar] [CrossRef]
- Gupta, P.B.; Pastushenko, I.; Skibinski, A.; Blanpain, C.; Kuperwasser, C. Phenotypic Plasticity: Driver of Cancer Initiation, Progression, and Therapy Resistance. Cell Stem Cell 2019, 24, 65–78. [Google Scholar] [CrossRef]
- Pastushenko, I.; Blanpain, C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019, 29, 212–226. [Google Scholar] [CrossRef]
- Tam, W.L.; Weinberg, R.A. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 2013, 19, 1438–1449. [Google Scholar] [CrossRef]
- Scheel, C.; Eaton, E.N.; Li, S.H.; Chaffer, C.L.; Reinhardt, F.; Kah, K.J.; Bell, G.; Guo, W.; Rubin, J.; Richardson, A.L.; et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 2011, 145, 926–940. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Bracken, C.P.; Smith, E.; Bert, A.G.; Wright, J.A.; Roslan, S.; Morris, M.; Wyatt, L.; Farshid, G.; Lim, Y.Y.; et al. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol. Biol. Cell 2011, 22, 1686–1698. [Google Scholar] [CrossRef]
- Rokavec, M.; Bouznad, N.; Hermeking, H. Paracrine Induction of Epithelial-Mesenchymal Transition Between Colorectal Cancer Cells and its Suppression by a p53/miR-192/215/NID1 Axis. Cell Mol. Gastroenterol. Hepatol. 2019, 7, 783–802. [Google Scholar] [CrossRef] [PubMed]
- Miosge, N.; Holzhausen, S.; Zelent, C.; Sprysch, P.; Herken, R. Nidogen-1 and nidogen-2 are found in basement membranes during human embryonic development. Histochem. J. 2001, 33, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, Y.; Li, N.; Feng, L.; Yao, H.; Zhang, R.; Li, B.; Li, X.; Han, N.; Gao, Y.; et al. Nidogen-1: A candidate biomarker for ovarian serous cancer. Jpn. J. Clin. Oncol. 2015, 45, 176–182. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, Y.; Fan, X.; Zhang, C.; Wang, Y.; Zhang, L.; Zhang, H.; Wen, T.; Zhang, K.; Huo, X.; et al. NID1, a new regulator of EMT required for metastasis and chemoresistance of ovarian cancer cells. Oncotarget 2017, 8, 33110–33121. [Google Scholar] [CrossRef]
- Aleckovic, M.; Wei, Y.; LeRoy, G.; Sidoli, S.; Liu, D.D.; Garcia, B.A.; Kang, Y. Identification of Nidogen 1 as a lung metastasis protein through secretome analysis. Genes Dev. 2017, 31, 1439–1455. [Google Scholar] [CrossRef] [PubMed]
- Pedrola, N.; Devis, L.; Llaurado, M.; Campoy, I.; Martinez-Garcia, E.; Garcia, M.; Muinelo-Romay, L.; Alonso-Alconada, L.; Abal, M.; Alameda, F.; et al. Nidogen 1 and Nuclear Protein 1: Novel targets of ETV5 transcription factor involved in endometrial cancer invasion. Clin. Exp. Metastasis 2015, 32, 467–478. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, B.; Liu, Y.; Yao, H.; Lu, N.; Li, B.; Gao, J.; Guo, S.; Han, N.; Qi, J.; et al. The ovarian cancer-derived secretory/releasing proteome: A repertoire of tumor markers. Proteomics 2012, 12, 1883–1891. [Google Scholar] [CrossRef]
- Dedhar, S.; Jewell, K.; Rojiani, M.; Gray, V. The receptor for the basement membrane glycoprotein entactin is the integrin alpha 3/beta 1. J. Biol. Chem. 1992, 267, 18908–18914. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.J.; Hsieh, J.C.; Chung, A.E. Two distinct cell attachment sites in entactin are revealed by amino acid substitutions and deletion of the RGD sequence in the cysteine-rich epidermal growth factor repeat 2. J. Biol. Chem. 1995, 270, 15838–15843. [Google Scholar] [CrossRef]
- Hamidi, H.; Ivaska, J. Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef]
- Mitra, S.K.; Schlaepfer, D.D. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr. Opin. Cell Biol. 2006, 18, 516–523. [Google Scholar] [CrossRef]
- Sheng, W.; Chen, C.; Dong, M.; Wang, G.; Zhou, J.; Song, H.; Li, Y.; Zhang, J.; Ding, S. Calreticulin promotes EGF-induced EMT in pancreatic cancer cells via Integrin/EGFR-ERK/MAPK signaling pathway. Cell Death Dis. 2017, 8, e3147. [Google Scholar] [CrossRef]
- Cooper, J.; Giancotti, F.G. Integrin Signaling in Cancer: Mechanotransduction, Stemness, Epithelial Plasticity, and Therapeutic Resistance. Cancer Cell 2019, 35, 347–367. [Google Scholar] [CrossRef] [PubMed]
- Attieh, Y.; Clark, A.G.; Grass, C.; Richon, S.; Pocard, M.; Mariani, P.; Elkhatib, N.; Betz, T.; Gurchenkov, B.; Vignjevic, D.M. Cancer-associated fibroblasts lead tumor invasion through integrin-beta3-dependent fibronectin assembly. J. Cell Biol. 2017, 216, 3509–3520. [Google Scholar] [CrossRef] [PubMed]
- Siemens, H.; Jackstadt, R.; Hunten, S.; Kaller, M.; Menssen, A.; Gotz, U.; Hermeking, H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle 2011, 10, 4256–4271. [Google Scholar] [CrossRef]
- Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Rokavec, M.; Kaller, M.; Horst, D.; Hermeking, H. Pan-cancer EMT-signature identifies RBM47 down-regulation during colorectal cancer progression. Sci. Rep. 2017, 7, 4687. [Google Scholar] [CrossRef]
- Loevenich, L.P.; Tschurtschenthaler, M.; Rokavec, M.; Silva, M.G.; Jesinghaus, M.; Kirchner, T.; Klauschen, F.; Saur, D.; Neumann, J.; Hermeking, H.; et al. SMAD4 Loss Induces c-MYC-Mediated NLE1 Upregulation to Support Protein Biosynthesis, Colorectal Cancer Growth, and Metastasis. Cancer Res. 2022, 82, 4604–4623. [Google Scholar] [CrossRef] [PubMed]
- Boos, S.L.; Loevenich, L.P.; Vosberg, S.; Engleitner, T.; Ollinger, R.; Kumbrink, J.; Rokavec, M.; Michl, M.; Greif, P.A.; Jung, A.; et al. Disease Modeling on Tumor Organoids Implicates AURKA as a Therapeutic Target in Liver Metastatic Colorectal Cancer. Cell. Mol. Gastroenterol. Hepatol. 2022, 13, 517–540. [Google Scholar] [CrossRef] [PubMed]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576.e516. [Google Scholar] [CrossRef]
- Chan, C.E.; Odde, D.J. Traction dynamics of filopodia on compliant substrates. Science 2008, 322, 1687–1691. [Google Scholar] [CrossRef] [PubMed]
- Vaes, N.; Schonkeren, S.L.; Rademakers, G.; Holland, A.M.; Koch, A.; Gijbels, M.J.; Keulers, T.G.; de Wit, M.; Moonen, L.; Van der Meer, J.R.M.; et al. Loss of enteric neuronal Ndrg4 promotes colorectal cancer via increased release of Nid1 and Fbln2. EMBO Rep. 2021, 22, e51913. [Google Scholar] [CrossRef]
- Elez, E.; Kocakova, I.; Hohler, T.; Martens, U.M.; Bokemeyer, C.; Van Cutsem, E.; Melichar, B.; Smakal, M.; Csoszi, T.; Topuzov, E.; et al. Abituzumab combined with cetuximab plus irinotecan versus cetuximab plus irinotecan alone for patients with KRAS wild-type metastatic colorectal cancer: The randomised phase I/II POSEIDON trial. Ann. Oncol. 2015, 26, 132–140. [Google Scholar] [CrossRef]
- Bates, R.C. Colorectal cancer progression: Integrin alphavbeta6 and the epithelial-mesenchymal transition (EMT). Cell Cycle 2005, 4, 1350–1352. [Google Scholar] [CrossRef]
- Bates, R.C.; Bellovin, D.I.; Brown, C.; Maynard, E.; Wu, B.; Kawakatsu, H.; Sheppard, D.; Oettgen, P.; Mercurio, A.M. Transcriptional activation of integrin beta6 during the epithelial-mesenchymal transition defines a novel prognostic indicator of aggressive colon carcinoma. J. Clin. Investig. 2005, 115, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reynies, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Ieguchi, K.; Fujita, M.; Ma, Z.; Davari, P.; Taniguchi, Y.; Sekiguchi, K.; Wang, B.; Takada, Y.K.; Takada, Y. Direct binding of the EGF-like domain of neuregulin-1 to integrins (alphavbeta3 and alpha6beta4) is involved in neuregulin-1/ErbB signaling. J. Biol. Chem. 2010, 285, 31388–31398. [Google Scholar] [CrossRef]
- Mori, S.; Wu, C.Y.; Yamaji, S.; Saegusa, J.; Shi, B.; Ma, Z.; Kuwabara, Y.; Lam, K.S.; Isseroff, R.R.; Takada, Y.K.; et al. Direct binding of integrin alphavbeta3 to FGF1 plays a role in FGF1 signaling. J. Biol. Chem. 2008, 283, 18066–18075. [Google Scholar] [CrossRef] [PubMed]
- Cedano Prieto, D.M.; Cheng, Y.; Chang, C.C.; Yu, J.; Takada, Y.K.; Takada, Y. Direct integrin binding to insulin-like growth factor-2 through the C-domain is required for insulin-like growth factor receptor type 1 (IGF1R) signaling. PLoS ONE 2017, 12, e0184285. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rokavec, M.; Jaeckel, S.; Hermeking, H. Nidogen-1/NID1 Function and Regulation during Progression and Metastasis of Colorectal Cancer. Cancers 2023, 15, 5316. https://doi.org/10.3390/cancers15225316
Rokavec M, Jaeckel S, Hermeking H. Nidogen-1/NID1 Function and Regulation during Progression and Metastasis of Colorectal Cancer. Cancers. 2023; 15(22):5316. https://doi.org/10.3390/cancers15225316
Chicago/Turabian StyleRokavec, Matjaz, Stephanie Jaeckel, and Heiko Hermeking. 2023. "Nidogen-1/NID1 Function and Regulation during Progression and Metastasis of Colorectal Cancer" Cancers 15, no. 22: 5316. https://doi.org/10.3390/cancers15225316
APA StyleRokavec, M., Jaeckel, S., & Hermeking, H. (2023). Nidogen-1/NID1 Function and Regulation during Progression and Metastasis of Colorectal Cancer. Cancers, 15(22), 5316. https://doi.org/10.3390/cancers15225316







