Image-Based Deep Learning Detection of High-Grade B-Cell Lymphomas Directly from Hematoxylin and Eosin Images
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Data Collection
2.3. Histopathological Analysis and Immunohistochemical (IHC) Staining
2.4. Fluorescence In Situ Hybridization (FISH) Analysis
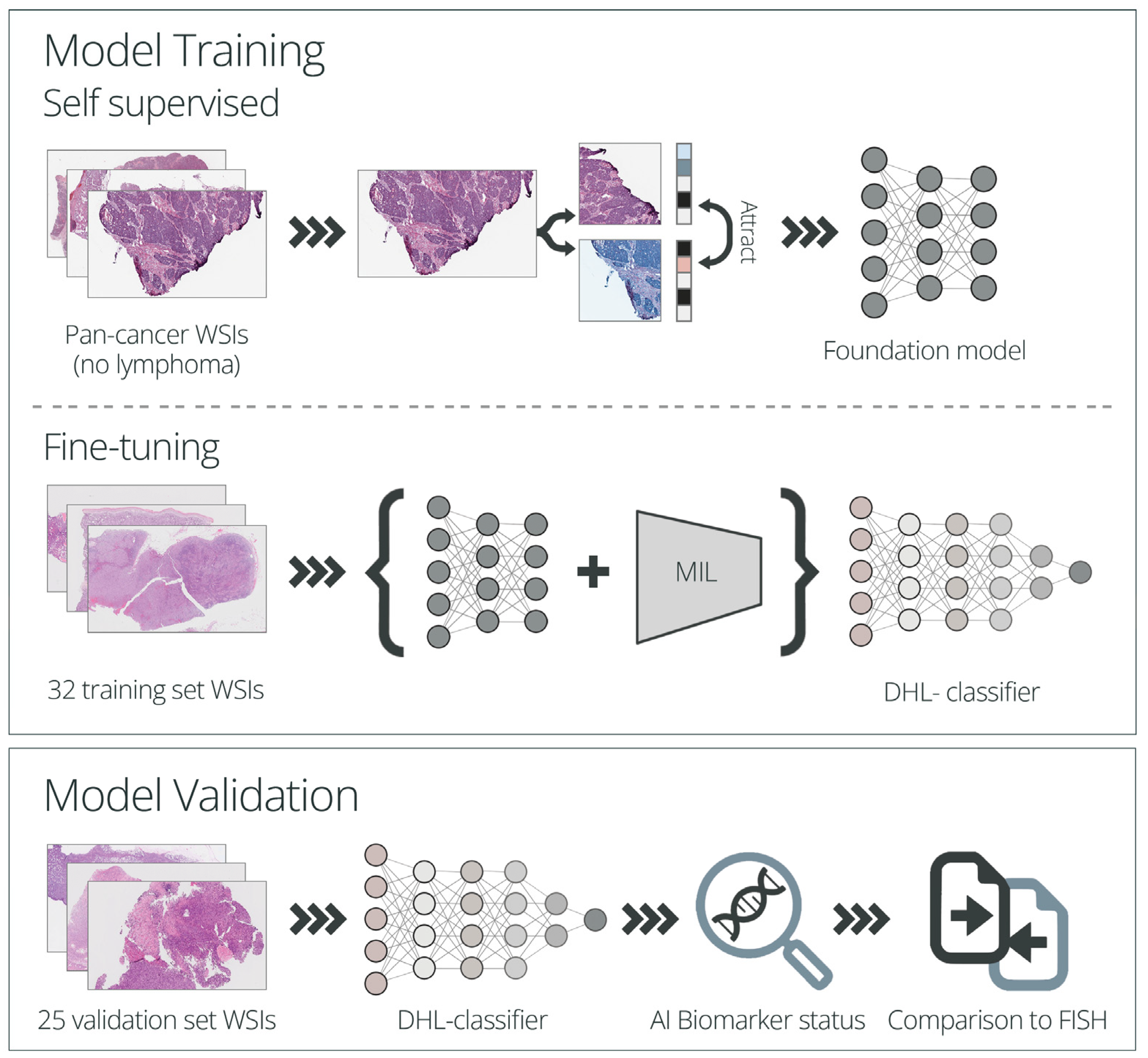
2.5. Algorithm Development and Application
2.5.1. Model Training
2.5.2. Model Performance Evaluation
3. Results
3.1. Patient Characteristics
3.2. Digital Imaging Analysis
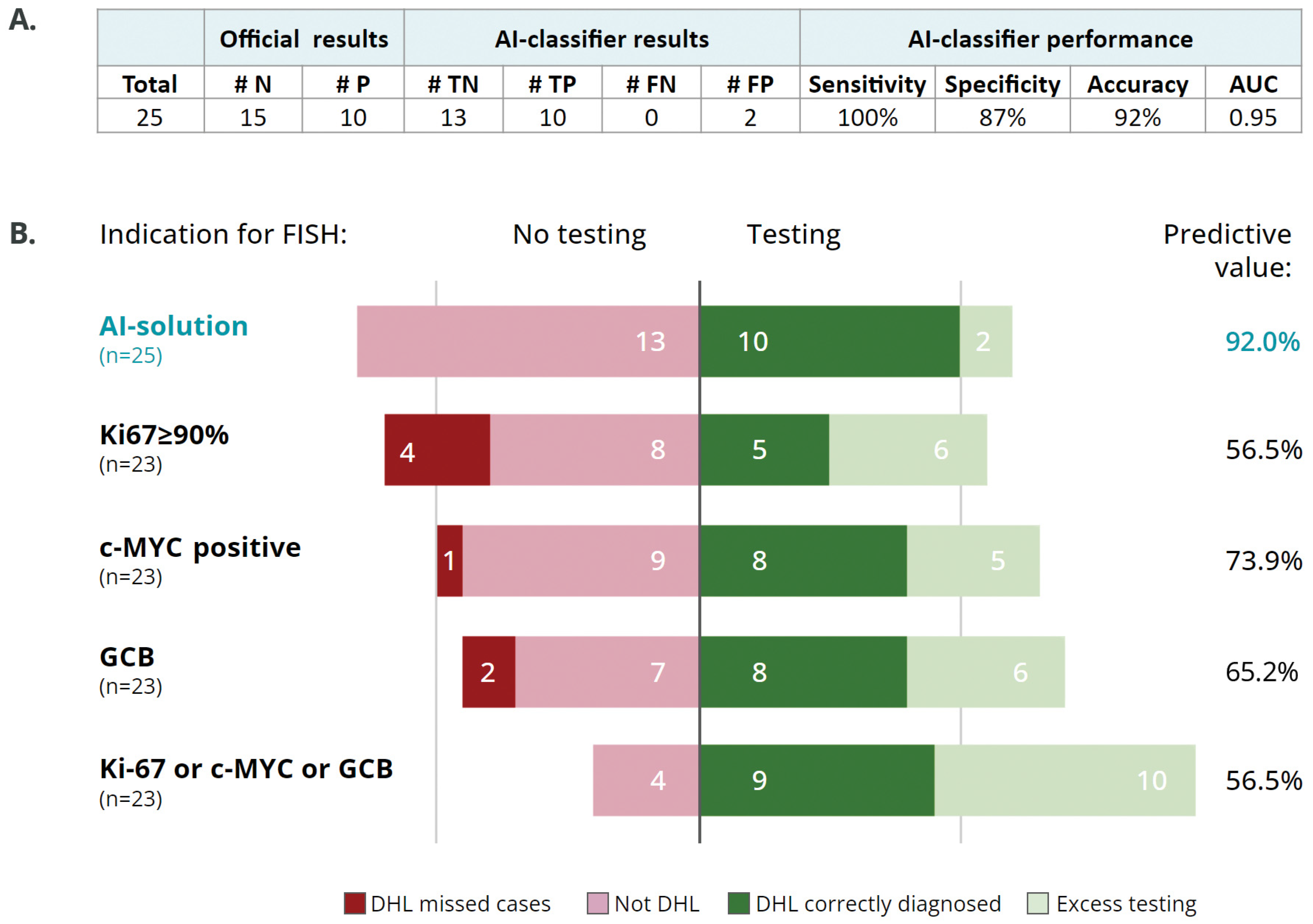
3.3. AI Classifier as a Screening Tool for Selecting Cases for FISH Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Rosenwald, A.; Wright, G.; Chan, W.C.; Connors, J.M.; Campo, E.; Fisher, R.I.; Gascoyne, R.D.; Muller-Hermelink, H.K.; Smeland, E.B.; Giltnane, J.M.; et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 2002, 346, 1937–1947. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; Papenhausen, P.; Shao, H. The Role of c-MYC in B-Cell Lymphomas: Diagnostic and Molecular Aspects. Genes 2017, 8, 116. [Google Scholar] [CrossRef] [PubMed]
- Barrans, S.; Crouch, S.; Smith, A.; Turner, K.; Owen, R.; Patmore, R.; Roman, E.; Jack, A. Rearrangement of MYC is associated with poor prognosis in patients with diffuse large B-cell lymphoma treated in the era of rituximab. J. Clin. Oncol. 2010, 28, 3360–3365. [Google Scholar] [CrossRef] [PubMed]
- Savage, K.J.; Johnson, N.A.; Ben-Neriah, S.; Connors, J.M.; Sehn, L.H.; Farinha, P.; Horsman, D.E.; Gascoyne, R.D. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood 2009, 114, 3533–3537. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [PubMed]
- Phuoc, V.; Sandoval-Sus, J.; Chavez, J.C. Drug therapy for double-hit lymphoma. Drugs Context 2019, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Dunleavy, K.; Fanale, M.A.; Abramson, J.S.; Noy, A.; Caimi, P.F.; Pittaluga, S.; Parekh, S.; Lacasce, A.; Hayslip, J.W.; Jagadeesh, D.; et al. Dose-adjusted EPOCH-R (etoposide, prednisone, vincristine, cyclophosphamide, doxorubicin, and rituximab) in untreated aggressive diffuse large B-cell lymphoma with MYC rearrangement: A prospective, multicentre, single-arm phase 2 study. Lancet Haematol. 2018, 5, e609–e617. [Google Scholar] [CrossRef]
- McMillan, A.K.; Phillips, E.H.; Kirkwood, A.A.; Barrans, S.; Burton, C.; Rule, S.; Patmore, R.; Pettengell, R.; Ardeshna, K.M.; Lawrie, A.; et al. Favourable outcomes for high-risk diffuse large B-cell lymphoma (IPI 3-5) treated with front-line R-CODOX-M/R-IVAC chemotherapy: Results of a phase 2 UK NCRI trial. Ann. Oncol. 2020, 31, 1251–1259. [Google Scholar] [CrossRef]
- Landsburg, D.J.; Falkiewicz, M.K.; Maly, J.; Blum, K.A.; Howlett, C.; Feldman, T.; Mato, A.R.; Hill, B.T.; Li, S.; Medeiros, L.J.; et al. Outcomes of Patients With Double-Hit Lymphoma Who Achieve First Complete Remission. J. Clin. Oncol. 2017, 35, 2260–2267. [Google Scholar] [CrossRef]
- Zhuang, Y.; Che, J.; Wu, M.; Guo, Y.; Xu, Y.; Dong, X.; Yang, H. Altered pathways and targeted therapy in double hit lymphoma. J. Hematol. Oncol. 2022, 15, 26. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H. Diagnosis of ‘double hit’ diffuse large B-cell lymphoma and B-cell lymphoma, unclassifiable, with features intermediate between DLBCL and Burkitt lymphoma: When and how, FISH versus IHC. Hematol. Am. Soc. Hematol. Educ. Program. 2014, 2014, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Thirunavukkarasu, B.; Bal, A.; Prakash, G.; Malhotra, P.; Singh, H.; Das, A. Screening Strategy for Detecting Double-Hit Lymphoma in a Resource-Limited Setting. Appl. Immunohistochem. Mol. Morphol. 2022, 30, 49–55. [Google Scholar] [CrossRef]
- Stephens, D.M.; Smith, S.M. Diffuse large B-cell lymphoma—Who should we FISH? Ann. Lymphoma 2018, 2, 8. [Google Scholar] [CrossRef]
- Shmatko, A.; Ghaffari Laleh, N.; Gerstung, M.; Kather, J.N. Artificial intelligence in histopathology: Enhancing cancer research and clinical oncology. Nat. Cancer 2022, 3, 1026–1038. [Google Scholar] [CrossRef]
- Baxi, V.; Edwards, R.; Montalto, M.; Saha, S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod. Pathol. 2022, 35, 23–32. [Google Scholar] [CrossRef]
- Laurent, C.; Baron, M.; Amara, N.; Haioun, C.; Dandoit, M.; Maynadie, M.; Parrens, M.; Vergier, B.; Copie-Bergman, C.; Fabiani, B.; et al. Impact of Expert Pathologic Review of Lymphoma Diagnosis: Study of Patients From the French Lymphopath Network. J. Clin. Oncol. 2017, 35, 2008–2017. [Google Scholar] [CrossRef] [PubMed]
- Bowen, J.M.; Perry, A.M.; Laurini, J.A.; Smith, L.M.; Klinetobe, K.; Bast, M.; Vose, J.M.; Aoun, P.; Fu, K.; Greiner, T.C.; et al. Lymphoma diagnosis at an academic centre: Rate of revision and impact on patient care. Br. J. Haematol. 2014, 166, 202–208. [Google Scholar] [CrossRef]
- Matasar, M.J.; Shi, W.; Silberstien, J.; Lin, O.; Busam, K.J.; Teruya-Feldstein, J.; Filippa, D.A.; Zelenetz, A.D.; Noy, A. Expert second-opinion pathology review of lymphoma in the era of the World Health Organization classification. Ann. Oncol. 2012, 23, 159–166. [Google Scholar] [CrossRef]
- Syrykh, C.; Abreu, A.; Amara, N.; Siegfried, A.; Maisongrosse, V.; Frenois, F.X.; Martin, L.; Rossi, C.; Laurent, C.; Brousset, P. Accurate diagnosis of lymphoma on whole-slide histopathology images using deep learning. NPJ Digit. Med. 2020, 3, 63. [Google Scholar] [CrossRef]
- Li, D.; Bledsoe, J.R.; Zeng, Y.; Liu, W.; Hu, Y.; Bi, K.; Liang, A.; Li, S. A deep learning diagnostic platform for diffuse large B-cell lymphoma with high accuracy across multiple hospitals. Nat. Commun. 2020, 11, 6004. [Google Scholar] [CrossRef]
- Steinbuss, G.; Kriegsmann, M.; Zgorzelski, C.; Brobeil, A.; Goeppert, B.; Dietrich, S.; Mechtersheimer, G.; Kriegsmann, K. Deep Learning for the Classification of Non-Hodgkin Lymphoma on Histopathological Images. Cancers 2021, 13, 2419. [Google Scholar] [CrossRef] [PubMed]
- Achi, H.E.; Belousova, T.; Chen, L.; Wahed, A.; Wang, I.; Hu, Z.; Kanaan, Z.; Rios, A.; Nguyen, A.N.D. Automated Diagnosis of Lymphoma with Digital Pathology Images Using Deep Learning. Ann. Clin. Lab. Sci. 2019, 49, 153–160. [Google Scholar] [PubMed]
- Miyoshi, H.; Sato, K.; Kabeya, Y.; Yonezawa, S.; Nakano, H.; Takeuchi, Y.; Ozawa, I.; Higo, S.; Yanagida, E.; Yamada, K.; et al. Deep learning shows the capability of high-level computer-aided diagnosis in malignant lymphoma. Lab. Investig. 2020, 100, 1300–1310. [Google Scholar] [CrossRef] [PubMed]
- Mohlman, J.S.; Leventhal, S.D.; Hansen, T.; Kohan, J.; Pascucci, V.; Salama, M.E. Improving Augmented Human Intelligence to Distinguish Burkitt Lymphoma From Diffuse Large B-Cell Lymphoma Cases. Am. J. Clin. Pathol. 2020, 153, 743–759. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.; Xiao, W.; Abdel-Wahab, O. Diagnosis and classification of hematologic malignancies on the basis of genetics. Blood 2017, 130, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Echle, A.; Rindtorff, N.T.; Brinker, T.J.; Luedde, T.; Pearson, A.T.; Kather, J.N. Deep learning in cancer pathology: A new generation of clinical biomarkers. Br. J. Cancer 2021, 124, 686–696. [Google Scholar] [CrossRef]
- Mayer, C.; Ofek, E.; Fridrich, D.E.; Molchanov, Y.; Yacobi, R.; Gazy, I.; Hayun, I.; Zalach, J.; Paz-Yaacov, N.; Barshack, I. Direct identification of ALK and ROS1 fusions in non-small cell lung cancer from hematoxylin and eosin-stained slides using deep learning algorithms. Mod. Pathol. 2022, 35, 1882–1887. [Google Scholar] [CrossRef] [PubMed]
- Rawat, R.R.; Ortega, I.; Roy, P.; Sha, F.; Shibata, D.; Ruderman, D.; Agus, D.B. Deep learned tissue "fingerprints" classify breast cancers by ER/PR/Her2 status from H&E images. Sci. Rep. 2020, 10, 7275. [Google Scholar] [CrossRef]
- Kather, J.N.; Heij, L.R.; Grabsch, H.I.; Loeffler, C.; Echle, A.; Muti, H.S.; Krause, J.; Niehues, J.M.; Sommer, K.A.J.; Bankhead, P.; et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat. Cancer 2020, 1, 789–799. [Google Scholar] [CrossRef]
- Cifci, D.; Foersch, S.; Kather, J.N. Artificial intelligence to identify genetic alterations in conventional histopathology. J. Pathol. 2022, 257, 430–444. [Google Scholar] [CrossRef] [PubMed]
- Swiderska-Chadaj, Z.; Hebeda, K.M.; van den Brand, M.; Litjens, G. Artificial intelligence to detect MYC translocation in slides of diffuse large B-cell lymphoma. Virchows Arch. 2021, 479, 617–621. [Google Scholar] [CrossRef] [PubMed]
- Hans, C.P.; Weisenburger, D.D.; Greiner, T.C.; Gascoyne, R.D.; Delabie, J.; Ott, G.; Muller-Hermelink, H.K.; Campo, E.; Braziel, R.M.; Jaffe, E.S.; et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004, 103, 275–282. [Google Scholar] [CrossRef]
- Chen, R.J.; Ding, T.; Lu, M.Y.; Williamson, D.F.K.; Jaume, G.; Chen, B.; Zhang, A.; Shao, D.; Song, A.H.; Shaban, M.; et al. A General-Purpose Self-Supervised Model for Computational Pathology. arXiv 2023, arXiv:2308.15474. [Google Scholar] [CrossRef]
- Saldanha, O.L.; Loeffler, C.M.L.; Niehues, J.M.; van Treeck, M.; Seraphin, T.P.; Hewitt, K.J.; Cifci, D.; Veldhuizen, G.P.; Ramesh, S.; Pearson, A.T.; et al. Self-supervised attention-based deep learning for pan-cancer mutation prediction from histopathology. NPJ Precis. Oncol. 2023, 7, 35. [Google Scholar] [CrossRef]
- Ilse, M.; Tomczak, J.M.; Welling, M. Attention-based Deep Multiple Instance Learning. arXiv 2018, arXiv:1802.04712. [Google Scholar] [CrossRef]
- Hilton, L.K.; Tang, J.; Ben-Neriah, S.; Alcaide, M.; Jiang, A.; Grande, B.M.; Rushton, C.K.; Boyle, M.; Meissner, B.; Scott, D.W.; et al. The double-hit signature identifies double-hit diffuse large B-cell lymphoma with genetic events cryptic to FISH. Blood 2019, 134, 1528–1532. [Google Scholar] [CrossRef]
- King, R.L.; McPhail, E.D.; Meyer, R.G.; Vasmatzis, G.; Pearce, K.; Smadbeck, J.B.; Ketterling, R.P.; Smoley, S.A.; Greipp, P.T.; Hoppman, N.L.; et al. False-negative rates for MYC fluorescence in situ hybridization probes in B-cell neoplasms. Haematologica 2019, 104, e248–e251. [Google Scholar] [CrossRef] [PubMed]
- Landsburg, D.J.; Petrich, A.M.; Abramson, J.S.; Sohani, A.R.; Press, O.; Cassaday, R.; Chavez, J.C.; Song, K.; Zelenetz, A.D.; Gandhi, M.; et al. Impact of oncogene rearrangement patterns on outcomes in patients with double-hit non-Hodgkin lymphoma. Cancer 2016, 122, 559–564. [Google Scholar] [CrossRef]
- Laude, M.C.; Lebras, L.; Sesques, P.; Ghesquieres, H.; Favre, S.; Bouabdallah, K.; Croizier, C.; Guieze, R.; Drieu La Rochelle, L.; Gyan, E.; et al. First-line treatment of double-hit and triple-hit lymphomas: Survival and tolerance data from a retrospective multicenter French study. Am. J. Hematol. 2021, 96, 302–311. [Google Scholar] [CrossRef]
- Howard, F.M.; Dolezal, J.; Kochanny, S.; Schulte, J.; Chen, H.; Heij, L.; Huo, D.; Nanda, R.; Olopade, O.I.; Kather, J.N.; et al. The impact of site-specific digital histology signatures on deep learning model accuracy and bias. Nat. Commun. 2021, 12, 4423. [Google Scholar] [CrossRef]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef]
| Entire Cohort (n = 57) | Training Set (n = 32) | Validation Set (n = 25) | |
|---|---|---|---|
| Male, n (%) | 32 (56.1) | 18 (56.3) | 14 (56) |
| Age (years), median (range) | 62 (8–84) | 66.5 (8–84) | 60 (17–77) |
| Tested tissue, n (%) | |||
| Lymph node | 22 (38.6) | 16 (50.0) | 6 (24.0) |
| Extra nodal | 35 (61.4) | 16 (50.0) | 19 (76.0) |
| Procedure, n (%) | |||
| Needle biopsy | 43 (75.4) | 24 (75.0) | 19 (76.0) |
| Excisional | 14 (24.6) | 8 (25.0) | 6 (24.0) |
| ECOG PS, n (%) * | |||
| 0/1 | 38 (82.6) | 24 (82.8) | 14 (82.4) |
| ≥2 | 8 (17.4) | 5 (17.2) | 3 (17.6) |
| Disease stage * | |||
| I/II | 13 (24.5) | 7 (23.3) | 6 (26.1) |
| III/IV | 40 (75.5) | 23 (76.7) | 17 (73.9) |
| LDH level * | |||
| Normal | 14 (28.0) | 10 (32.3) | 4 (21.1) |
| Increased | 36 (72.0) | 21 (67.7) | 15 (78.9) |
| COO, n (%) #* | |||
| GCB | 27 (52.9) | 13 (46.4) | 14 (60.9) |
| IHC | |||
| Ki67 * | |||
| Median % (range) | 80 (10–100) | 80 (10–100) | 85 (40–100) |
| Ki67 ≥ 90% | 23 (41.8) | 12 (37.5) | 11 (47.8) |
| c-MYC expression * | |||
| Positive/borderline positive | 33 (61.1) | 20 (64.5) | 13 (56.5) |
| Negative | 21 (38.9) | 11 (35.5) | 10 (43.5) |
| BCL2 expression * | |||
| Positive/borderline positive | 33 (62.3) | 21 (67.7) | 12 (54.5) |
| Negative | 20 (37.7) | 10 (32.3) | 10 (45.5) |
| BCL6 expression * | |||
| Positive/borderline positive | 49 (89.1) | 29 (90.6) | 20 (87.0) |
| Negative | 6 (10.9) | 3 (9.4) | 3 (13.0) |
| FISH | |||
| DHL/THL, n (%) | 15 (26.3) | 5 (15.6) | 10 (40) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perry, C.; Greenberg, O.; Haberman, S.; Herskovitz, N.; Gazy, I.; Avinoam, A.; Paz-Yaacov, N.; Hershkovitz, D.; Avivi, I. Image-Based Deep Learning Detection of High-Grade B-Cell Lymphomas Directly from Hematoxylin and Eosin Images. Cancers 2023, 15, 5205. https://doi.org/10.3390/cancers15215205
Perry C, Greenberg O, Haberman S, Herskovitz N, Gazy I, Avinoam A, Paz-Yaacov N, Hershkovitz D, Avivi I. Image-Based Deep Learning Detection of High-Grade B-Cell Lymphomas Directly from Hematoxylin and Eosin Images. Cancers. 2023; 15(21):5205. https://doi.org/10.3390/cancers15215205
Chicago/Turabian StylePerry, Chava, Orli Greenberg, Shira Haberman, Neta Herskovitz, Inbal Gazy, Assaf Avinoam, Nurit Paz-Yaacov, Dov Hershkovitz, and Irit Avivi. 2023. "Image-Based Deep Learning Detection of High-Grade B-Cell Lymphomas Directly from Hematoxylin and Eosin Images" Cancers 15, no. 21: 5205. https://doi.org/10.3390/cancers15215205
APA StylePerry, C., Greenberg, O., Haberman, S., Herskovitz, N., Gazy, I., Avinoam, A., Paz-Yaacov, N., Hershkovitz, D., & Avivi, I. (2023). Image-Based Deep Learning Detection of High-Grade B-Cell Lymphomas Directly from Hematoxylin and Eosin Images. Cancers, 15(21), 5205. https://doi.org/10.3390/cancers15215205








