Heparan-6-O-Endosulfatase 2 Promotes Invasiveness of Head and Neck Squamous Carcinoma Cell Lines in Co-Cultures with Cancer-Associated Fibroblasts
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. Generation of Fluorescently Labeled Cells
2.4. Generation of CRISPR/Cas9 Sulf-2 Knockout Cells
2.5. Spheroid Mono-Cultures and Co-Cultures
2.6. Quantification and Inhibition of Sulf-2 Activity
2.7. Disaccharide Analysis of Heparan Sulfate Desulfation
2.8. HfFucCS Treatment of the Spheroid Cell Cultures
2.9. Targeted MS Analysis of the Sulf-2 Protein
2.10. Statistical Analysis
3. Results
3.1. Generation of Sulf-2-Deficient HNSCC Cell Lines
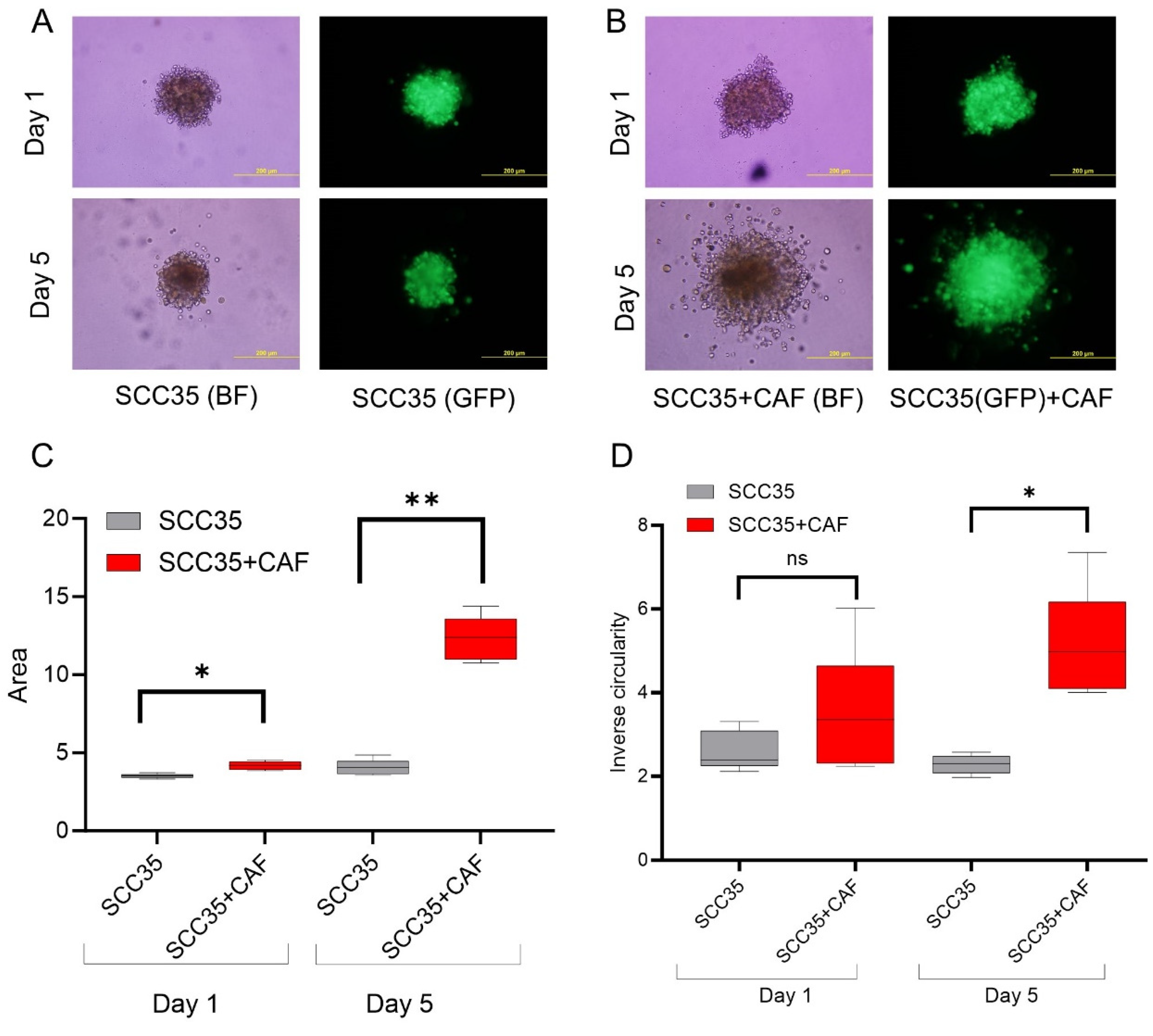
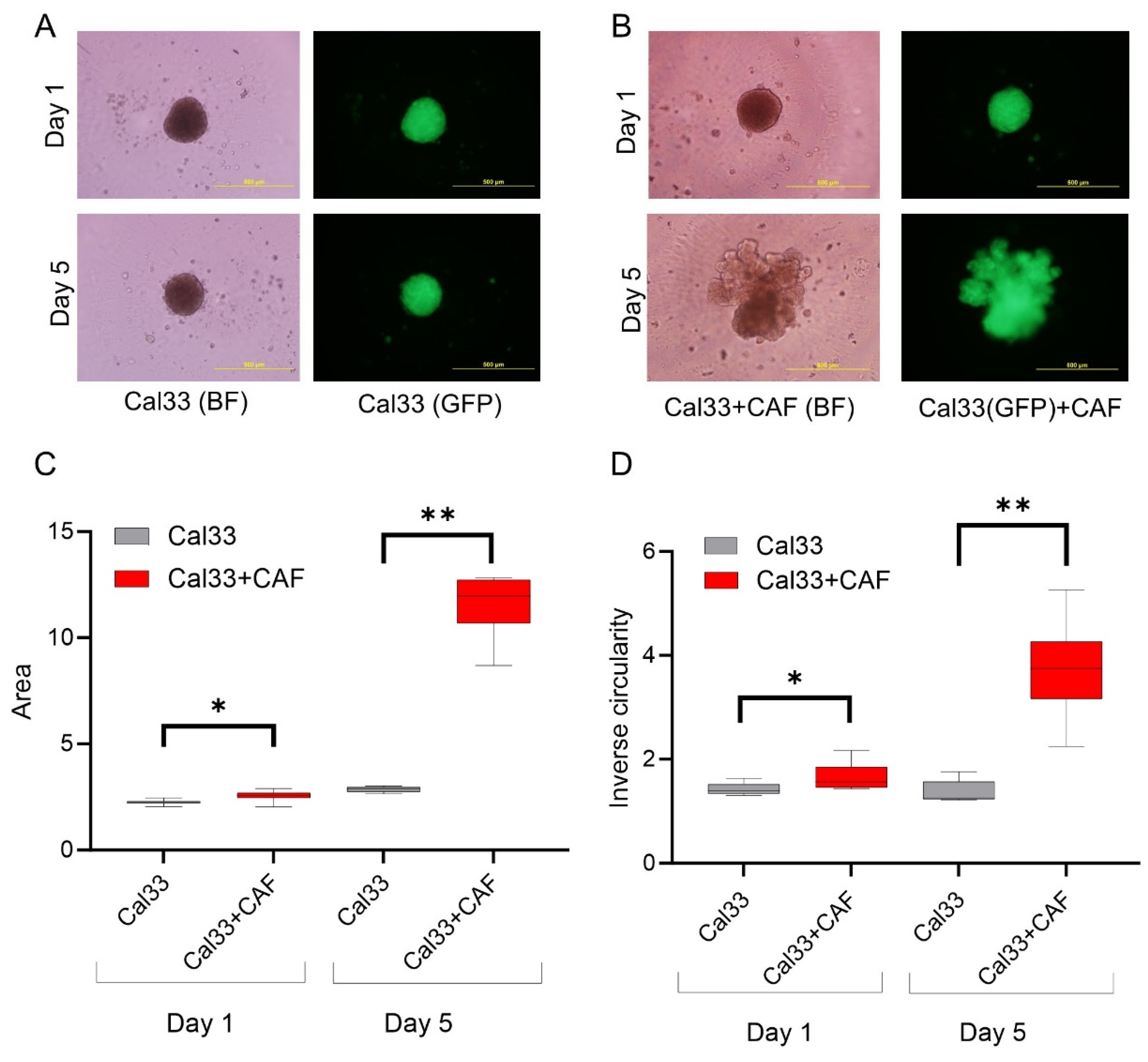
3.2. HNCAF37 Supports Invasion of HNSCC Cell Lines into Matrigel
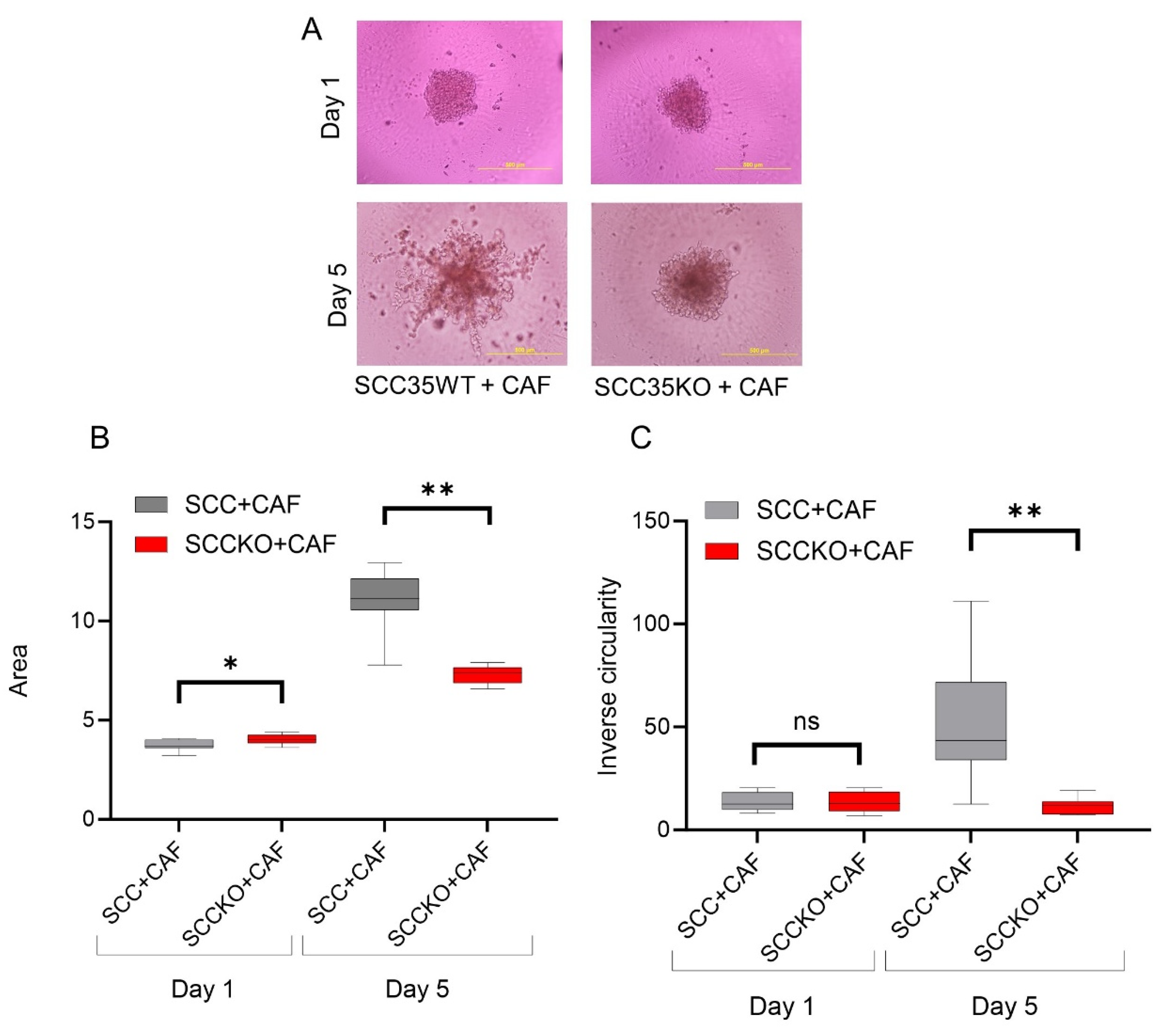
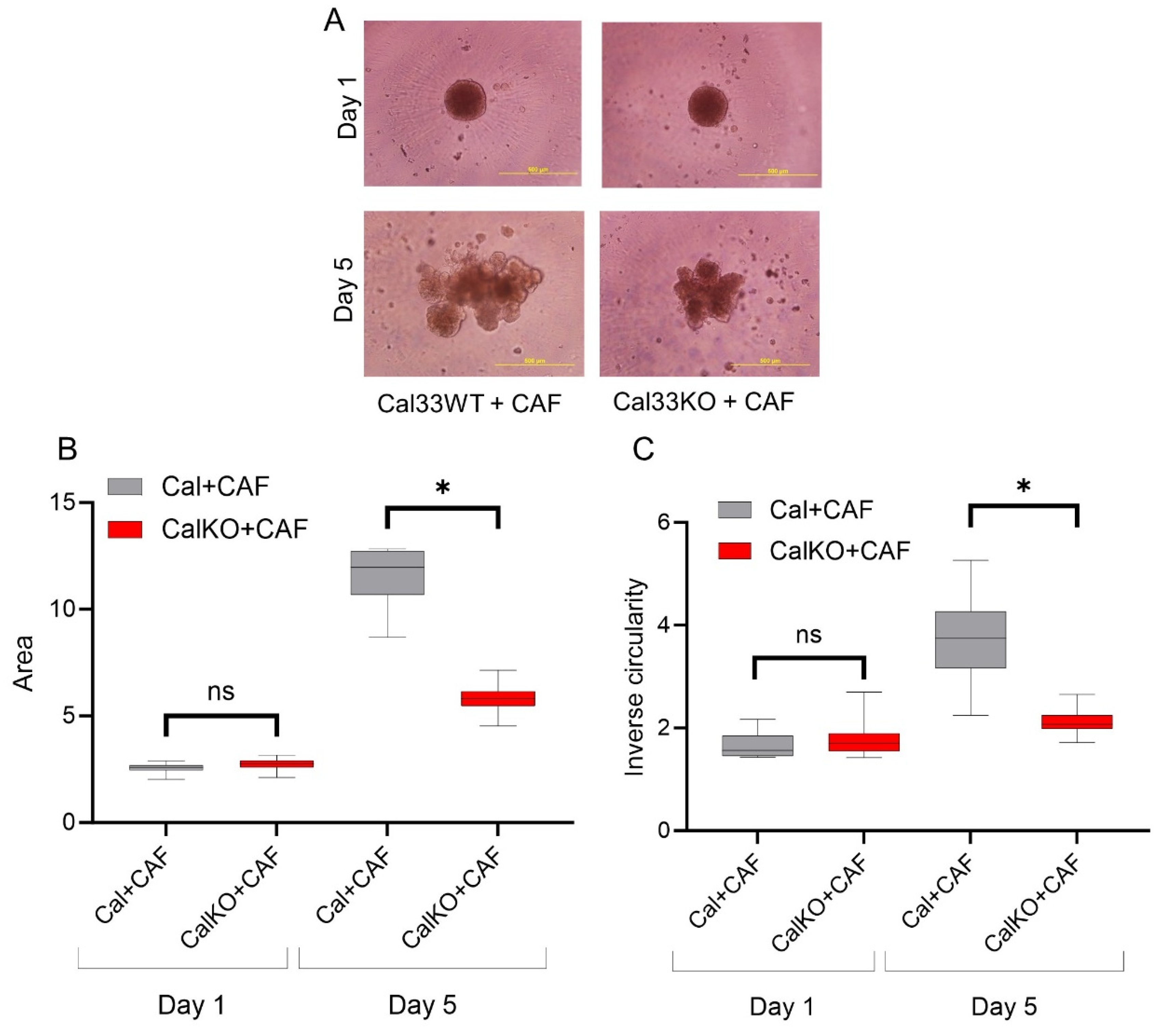
3.3. HNCAF37-Mediated Invasion of HNSCC Cell Lines Depends on Sulf-2
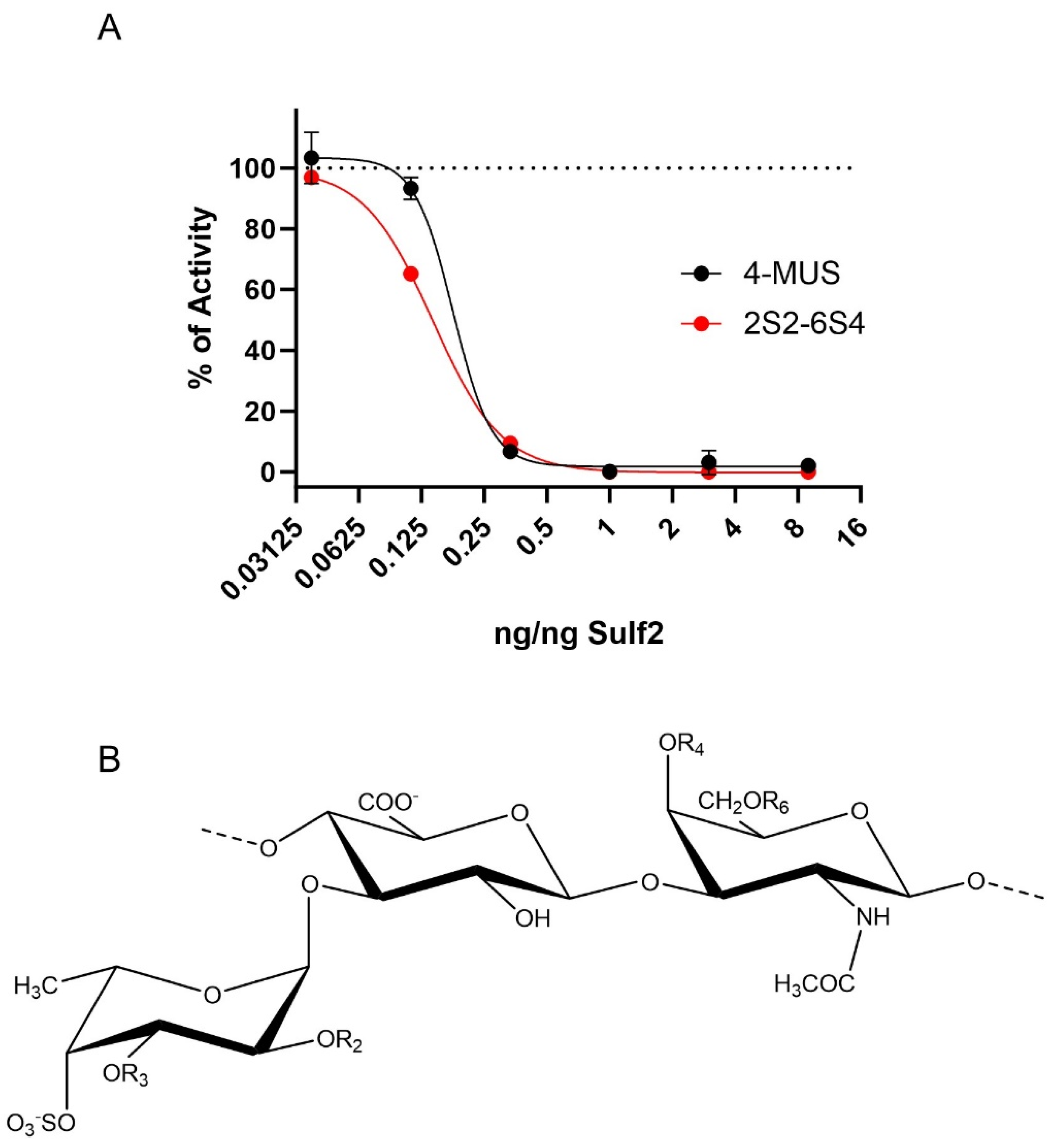
3.4. HfFucCS Blocks Activity of Sulf-2 In Vitro
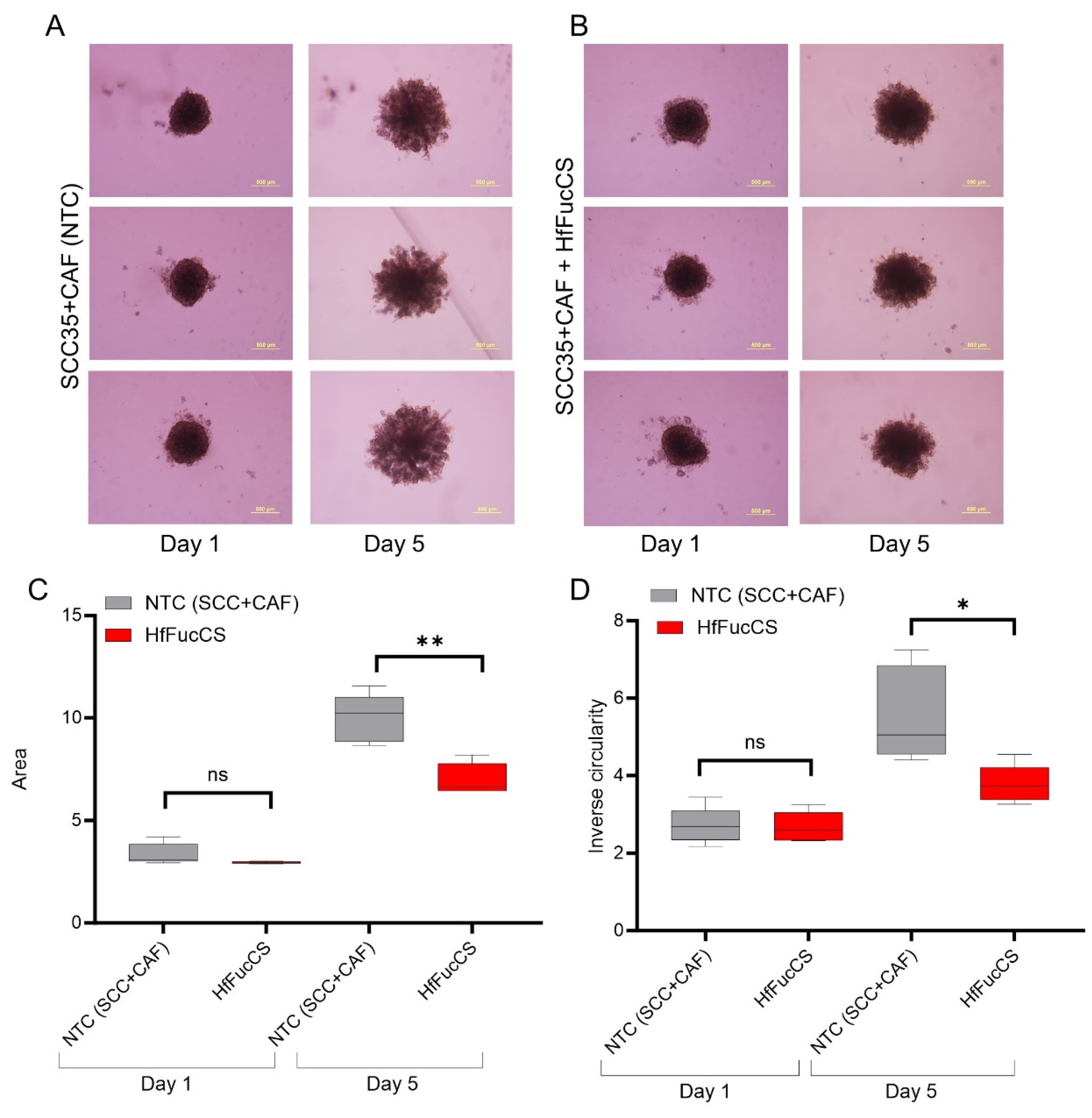
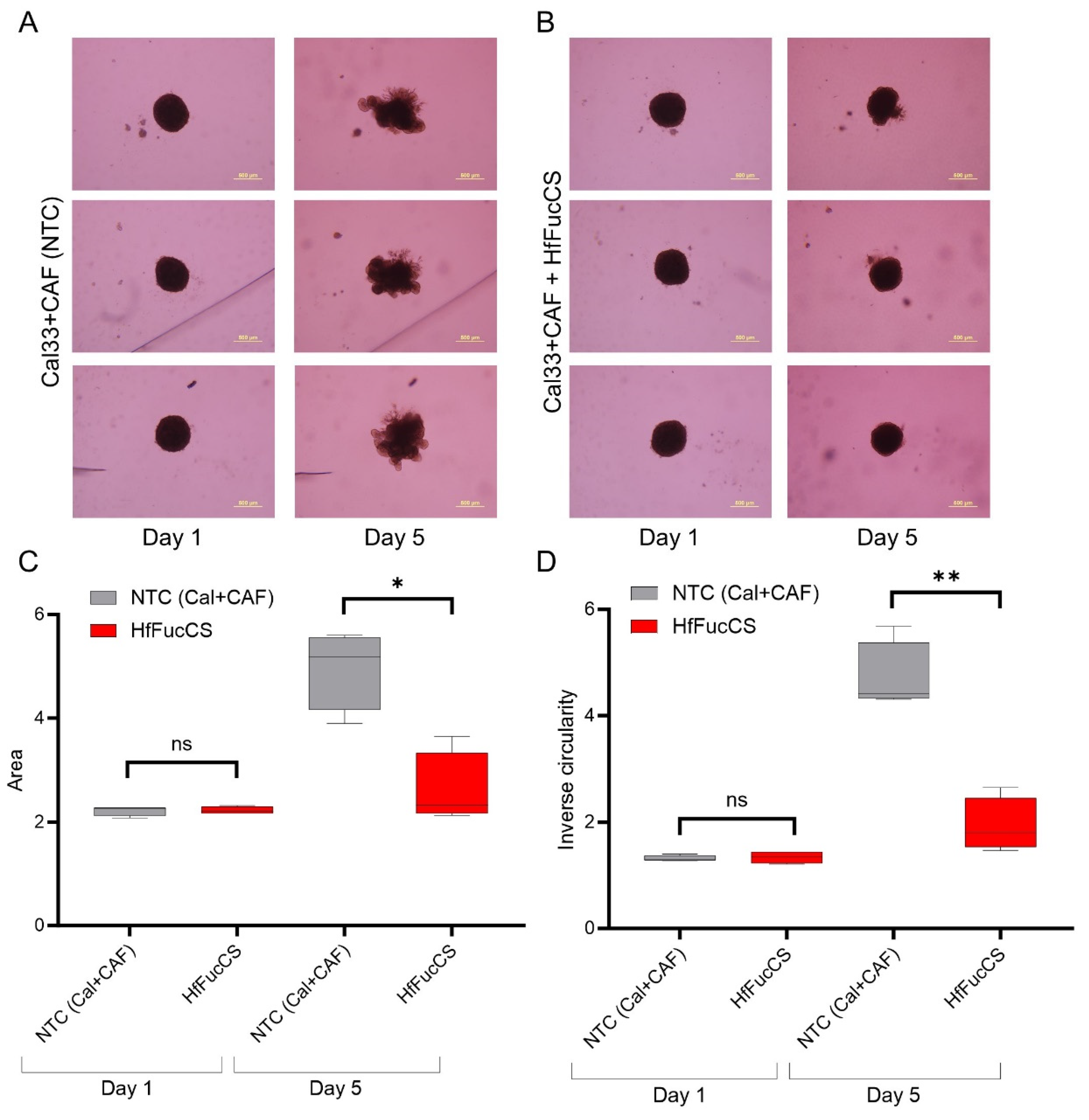
3.5. HfFucCS Suppresses the Invasion of HNSCC Co-Culture Spheroids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Jimenez, L.; Jayakar, S.K.; Ow, T.J.; Segall, J.E. Mechanisms of Invasion in Head and Neck Cancer. Arch. Pathol. Lab. Med. 2015, 139, 1334–1348. [Google Scholar] [CrossRef]
- Schmitd, L.B.; Scanlon, C.S.; D’Silva, N.J. Perineural Invasion in Head and Neck Cancer. J. Dent. Res. 2018, 97, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Banerjee, R.; Piao, S.; Costa de Medeiros, M.; Bellile, E.; Liu, M.; Damodaran Puthiya Veettil, D.; Schmitd, L.B.; Russo, N.; Danella, E.; et al. Squamous Cell Carcinoma Subverts Adjacent Histologically Normal Epithelium to Promote Lateral Invasion. J. Exp. Med. 2021, 218, e20200944. [Google Scholar] [CrossRef] [PubMed]
- Boyd, D.D.; Nicolson, G.L. Mechanisms of Invasion by Head and Neck Cancers. In Head and Neck Cancer: Basic and Clinical Aspects; Hong, W.K., Weber, R.S., Eds.; Cancer Treatment and Research; Springer US: Boston, MA, USA, 1995; pp. 117–130. ISBN 978-1-4615-2023-8. [Google Scholar]
- Friedl, P.; Alexander, S. Cancer Invasion and the Microenvironment: Plasticity and Reciprocity. Cell 2011, 147, 992–1009. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.A.; Ye, L.; Sanders, A.J.; Lane, J.; Jiang, W.G. Cancer Invasion and Metastasis: Molecular and Cellular Perspective. In Madame Curie Bioscience Database; Landes Bioscience: Austin, TX, USA, 2013. [Google Scholar]
- Chambers, A.F.; Groom, A.C.; MacDonald, I.C. Dissemination and Growth of Cancer Cells in Metastatic Sites. Nat. Rev. Cancer 2002, 2, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Krakhmal, N.V.; Zavyalova, M.V.; Denisov, E.V.; Vtorushin, S.V.; Perelmuter, V.M. Cancer Invasion: Patterns and Mechanisms. Acta Naturae 2015, 7, 17–28. [Google Scholar] [CrossRef]
- Poon, S.; Ailles, L.E. Modeling the Role of Cancer-Associated Fibroblasts in Tumor Cell Invasion. Cancers 2022, 14, 962. [Google Scholar] [CrossRef]
- Zhu, K.; Cai, L.; Cui, C.; de Los Toyos, J.R.; Anastassiou, D. Single-Cell Analysis Reveals the Pan-Cancer Invasiveness-Associated Transition of Adipose-Derived Stromal Cells into COL11A1-Expressing Cancer-Associated Fibroblasts. PLoS Comput. Biol. 2021, 17, e1009228. [Google Scholar] [CrossRef]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A Framework for Advancing Our Understanding of Cancer-Associated Fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef]
- Wright, K.; Ly, T.; Kriet, M.; Czirok, A.; Thomas, S.M. Cancer-Associated Fibroblasts: Master Tumor Microenvironment Modifiers. Cancers 2023, 15, 1899. [Google Scholar] [CrossRef]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and Neck Squamous Cell Carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Elmusrati, A.; Wang, J.; Wang, C.-Y. Tumor Microenvironment and Immune Evasion in Head and Neck Squamous Cell Carcinoma. Int. J. Oral. Sci. 2021, 13, 24. [Google Scholar] [CrossRef]
- Hassan, N.; Greve, B.; Espinoza-Sánchez, N.A.; Götte, M. Cell-Surface Heparan Sulfate Proteoglycans as Multifunctional Integrators of Signaling in Cancer. Cell Signal. 2021, 77, 109822. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, K.; Aquino, R.S.; Park, P.W. Coreceptor Functions of Cell Surface Heparan Sulfate Proteoglycans. Am. J. Physiol. Cell Physiol. 2022, 322, C896–C912. [Google Scholar] [CrossRef]
- Marques, C.; Reis, C.A.; Vivès, R.R.; Magalhães, A. Heparan Sulfate Biosynthesis and Sulfation Profiles as Modulators of Cancer Signalling and Progression. Front. Oncol. 2021, 11, 778752. [Google Scholar] [CrossRef]
- Soares da Costa, D.; Reis, R.L.; Pashkuleva, I. Sulfation of Glycosaminoglycans and Its Implications in Human Health and Disorders. Annu. Rev. Biomed. Eng. 2017, 19, 1–26. [Google Scholar] [CrossRef]
- Annaval, T.; Wild, R.; Crétinon, Y.; Sadir, R.; Vivès, R.R.; Lortat-Jacob, H. Heparan Sulfate Proteoglycans Biosynthesis and Post Synthesis Mechanisms Combine Few Enzymes and Few Core Proteins to Generate Extensive Structural and Functional Diversity. Molecules 2020, 25, 4215. [Google Scholar] [CrossRef]
- Lai, J.-P.; Sandhu, D.S.; Yu, C.; Han, T.; Moser, C.D.; Jackson, K.K.; Guerrero, R.B.; Aderca, I.; Isomoto, H.; Garrity-Park, M.M.; et al. Sulfatase 2 Up-Regulates Glypican 3, Promotes Fibroblast Growth Factor Signaling, and Decreases Survival in Hepatocellular Carcinoma. Hepatology 2008, 47, 1211–1222. [Google Scholar] [CrossRef]
- Li, J.; Kleeff, J.; Abiatari, I.; Kayed, H.; Giese, N.A.; Felix, K.; Giese, T.; Büchler, M.W.; Friess, H. Enhanced Levels of Hsulf-1 Interfere with Heparin-Binding Growth Factor Signaling in Pancreatic Cancer. Mol. Cancer 2005, 4, 14. [Google Scholar] [CrossRef]
- Hur, K.; Han, T.-S.; Jung, E.-J.; Yu, J.; Lee, H.-J.; Kim, W.H.; Goel, A.; Yang, H.-K. Up-Regulated Expression of Sulfatases (SULF1 and SULF2) as Prognostic and Metastasis Predictive Markers in Human Gastric Cancer. J. Pathol. 2012, 228, 88–98. [Google Scholar] [CrossRef]
- Zhu, C.; He, L.; Zhou, X.; Nie, X.; Gu, Y. Sulfatase 2 Promotes Breast Cancer Progression through Regulating Some Tumor-Related Factors. Oncol. Rep. 2016, 35, 1318–1328. [Google Scholar] [CrossRef]
- Jiang, T.; Chen, Z.-H.; Chen, Z.; Tan, D. SULF2 Promotes Tumorigenesis and Inhibits Apoptosis of Cervical Cancer Cells through the ERK/AKT Signaling Pathway. Braz. J. Med. Biol. Res. 2020, 53, e8901. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-Y.; Yeh, B.-W.; Chan, T.-C.; Yang, K.-F.; Li, W.-M.; Huang, C.-N.; Ke, H.-L.; Li, C.-C.; Yeh, H.-C.; Liang, P.-I.; et al. Sulfatase-1 Overexpression Indicates Poor Prognosis in Urothelial Carcinoma of the Urinary Bladder and Upper Tract. Oncotarget 2017, 8, 47216–47229. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.J.; Huillard, E.; Robinson, A.E.; Ward, A.; Lum, D.H.; Polley, M.-Y.; Rosen, S.D.; Rowitch, D.H.; Werb, Z. Heparan Sulfate Sulfatase SULF2 Regulates PDGFRα Signaling and Growth in Human and Mouse Malignant Glioma. J. Clin. Investig. 2012, 122, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Bret, C.; Moreaux, J.; Schved, J.-F.; Hose, D.; Klein, B. SULFs in Human Neoplasia: Implication as Progression and Prognosis Factors. J. Transl. Med. 2011, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ahn, J.; Edwards, N.J.; Benicky, J.; Rozeboom, A.M.; Davidson, B.; Karamboulas, C.; Nixon, K.C.J.; Ailles, L.; Goldman, R. Extracellular Heparan 6-O-Endosulfatases SULF1 and SULF2 in Head and Neck Squamous Cell Carcinoma and Other Malignancies. Cancers 2022, 14, 5553. [Google Scholar] [CrossRef]
- Flowers, S.A.; Zhou, X.; Wu, J.; Wang, Y.; Makambi, K.; Kallakury, B.V.; Singer, M.S.; Rosen, S.D.; Davidson, B.; Goldman, R. Expression of the Extracellular Sulfatase SULF2 Is Associated with Squamous Cell Carcinoma of the Head and Neck. Oncotarget 2016, 7, 43177–43187. [Google Scholar] [CrossRef]
- Yang, Y.; Ahn, J.; Raghunathan, R.; Kallakury, B.V.; Davidson, B.; Kennedy, Z.B.; Zaia, J.; Goldman, R. Expression of the Extracellular Sulfatase SULF2 Affects Survival of Head and Neck Squamous Cell Carcinoma Patients. Front. Oncol. 2021, 10, 582827. [Google Scholar] [CrossRef]
- Rosen, S.D.; Lemjabbar-Alaoui, H. Sulf-2: An Extracellular Modulator of Cell Signaling and a Cancer Target Candidate. Expert. Opin. Ther. Targets 2010, 14, 935–949. [Google Scholar] [CrossRef]
- Young, M.; Rodenhizer, D.; Dean, T.; D’Arcangelo, E.; Xu, B.; Ailles, L.; McGuigan, A.P. A TRACER 3D Co-Culture Tumour Model for Head and Neck Cancer. Biomaterials 2018, 164, 54–69. [Google Scholar] [CrossRef]
- Wu, N.C.; Cadavid, J.L.; Tan, X.; Latour, S.; Scaini, S.; Makhijani, P.; McGaha, T.L.; Ailles, L.; McGuigan, A.P. 3D Microgels to Quantify Tumor Cell Properties and Therapy Response Dynamics. Biomaterials 2022, 283, 121417. [Google Scholar] [CrossRef] [PubMed]
- Benicky, J.; Sanda, M.; Panigrahi, A.; Liu, J.; Wang, Z.; Pagadala, V.; Su, G.; Goldman, R. A 6-O-Endosulfatase Activity Assay Based on Synthetic Heparan Sulfate Oligomers. Glycobiology 2023, 33, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Farrag, M.; Dwivedi, R.; Sharma, P.; Kumar, D.; Tandon, R.; Pomin, V.H. Structural Requirements of Holothuria Floridana Fucosylated Chondroitin Sulfate Oligosaccharides in Anti-SARS-CoV-2 and Anticoagulant Activities. PLoS ONE 2023, 18, e0285539. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, R.; Sharma, P.; Farrag, M.; Kim, S.B.; Fassero, L.A.; Tandon, R.; Pomin, V.H. Inhibition of SARS-CoV-2 Wild-Type (Wuhan-Hu-1) and Delta (B.1.617.2) Strains by Marine Sulfated Glycans. Glycobiology 2022, 32, 849–854. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Arnold, K.; Xu, Y.; Pagadala, V.; Su, G.; Myatt, H.; Linhardt, R.J.; Liu, J. Quantitative Analysis of Heparan Sulfate Using Isotopically Labeled Calibrants. Commun. Biol. 2020, 3, 425. [Google Scholar] [CrossRef]
- Kamimura, K.; Koyama, T.; Habuchi, H.; Ueda, R.; Masu, M.; Kimata, K.; Nakato, H. Specific and Flexible Roles of Heparan Sulfate Modifications in Drosophila FGF Signaling. J. Cell Biol. 2006, 174, 773–778. [Google Scholar] [CrossRef]
- Kreuger, J.; Spillmann, D.; Li, J.; Lindahl, U. Interactions between Heparan Sulfate and Proteins: The Concept of Specificity. J. Cell Biol. 2006, 174, 323–327. [Google Scholar] [CrossRef]
- Nugent, M.A.; Zaia, J.; Spencer, J.L. Heparan Sulfate-Protein Binding Specificity. Biochemistry 2013, 78, 726–735. [Google Scholar] [CrossRef]
- Bishop, J.R.; Schuksz, M.; Esko, J.D. Heparan Sulphate Proteoglycans Fine-Tune Mammalian Physiology. Nature 2007, 446, 1030–1037. [Google Scholar] [CrossRef]
- Weiss, R.J.; Esko, J.D.; Tor, Y. Targeting Heparin– and Heparan Sulfate–Protein Interactions. Org. Biomol. Chem. 2017, 15, 5656–5668. [Google Scholar] [CrossRef]
- Esko, J.D.; Lindahl, U. Molecular Diversity of Heparan Sulfate. J. Clin. Investig. 2001, 108, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Sarrazin, S.; Lamanna, W.C.; Esko, J.D. Heparan Sulfate Proteoglycans. Cold Spring Harb. Perspect. Biol. 2011, 3, a004952. [Google Scholar] [CrossRef] [PubMed]
- Capila, I.; Linhardt, R.J. Heparin–Protein Interactions. Angew. Chem. Int. Ed. 2002, 41, 390–412. [Google Scholar] [CrossRef]
- Nagarajan, A.; Malvi, P.; Wajapeyee, N. Heparan Sulfate and Heparan Sulfate Proteoglycans in Cancer Initiation and Progression. Front. Endocrinol. 2018, 9, 483. [Google Scholar] [CrossRef]
- Hammond, E.; Khurana, A.; Shridhar, V.; Dredge, K. The Role of Heparanase and Sulfatases in the Modification of Heparan Sulfate Proteoglycans within the Tumor Microenvironment and Opportunities for Novel Cancer Therapeutics. Front. Oncol. 2014, 4, 195. [Google Scholar] [CrossRef]
- Uchimura, K.; Morimoto-Tomita, M.; Bistrup, A.; Li, J.; Lyon, M.; Gallagher, J.; Werb, Z.; Rosen, S.D. HSulf-2, an Extracellular Endoglucosamine-6-Sulfatase, Selectively Mobilizes Heparin-Bound Growth Factors and Chemokines: Effects on VEGF, FGF-1, and SDF-1. BMC Biochem. 2006, 7, 2. [Google Scholar] [CrossRef]
- Rajarathnam, K.; Desai, U.R. Structural Insights Into How Proteoglycans Determine Chemokine-CXCR1/CXCR2 Interactions: Progress and Challenges. Front. Immunol. 2020, 11, 660. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The Extracellular Matrix at a Glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef]
- Knelson, E.H.; Nee, J.C.; Blobe, G.C. Heparan Sulfate Signaling in Cancer. Trends Biochem. Sci. 2014, 39, 277–288. [Google Scholar] [CrossRef]
- Lai, J.-P.; Sandhu, D.S.; Shire, A.M.; Roberts, L.R. The Tumor Suppressor Function of Human Sulfatase 1 (SULF1) in Carcinogenesis. J. Gastrointest. Cancer 2008, 39, 149–158. [Google Scholar] [CrossRef]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624.e24. [Google Scholar] [CrossRef] [PubMed]
- Dean, T.; Li, N.T.; Cadavid, J.L.; Ailles, L.; McGuigan, A.P. A TRACER Culture Invasion Assay to Probe the Impact of Cancer Associated Fibroblasts on Head and Neck Squamous Cell Carcinoma Cell Invasiveness. Biomater. Sci. 2020, 8, 3078–3094. [Google Scholar] [CrossRef]
- Sun, L.-P.; Xu, K.; Cui, J.; Yuan, D.-Y.; Zou, B.; Li, J.; Liu, J.-L.; Li, K.-Y.; Meng, Z.; Zhang, B. Cancer-associated Fibroblast-derived Exosomal miR-382-5p Promotes the Migration and Invasion of Oral Squamous Cell Carcinoma. Oncol. Rep. 2019, 42, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- El Masri, R.; Seffouh, A.; Roelants, C.; Seffouh, I.; Gout, E.; Pérard, J.; Dalonneau, F.; Nishitsuji, K.; Noborn, F.; Nikpour, M.; et al. Extracellular Endosulfatase Sulf-2 Harbors a Chondroitin/Dermatan Sulfate Chain That Modulates Its Enzyme Activity. Cell Rep. 2022, 38, 110516. [Google Scholar] [CrossRef]
- Vasconcelos, A.A.; Pomin, V.H. The Sea as a Rich Source of Structurally Unique Glycosaminoglycans and Mimetics. Microorganisms 2017, 5, 51. [Google Scholar] [CrossRef]
- Zheng, X.; Gai, X.; Han, S.; Moser, C.D.; Hu, C.; Shire, A.M.; Floyd, R.A.; Roberts, L.R. The Human Sulfatase 2 Inhibitor 2,4-Disulfonylphenyl-Tert-Butylnitrone (OKN-007) Has an Antitumor Effect in Hepatocellular Carcinoma Mediated via Suppression of TGFB1/SMAD2 and Hedgehog/GLI1 Signaling. Genes. Chromosomes Cancer 2013, 52, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Chiu, L.-T.; Sabbavarapu, N.M.; Lin, W.-C.; Fan, C.-Y.; Wu, C.-C.; Cheng, T.-J.R.; Wong, C.-H.; Hung, S.-C. Trisaccharide Sulfate and Its Sulfonamide as an Effective Substrate and Inhibitor of Human Endo-O-Sulfatase-1. J. Am. Chem. Soc. 2020, 142, 5282–5292. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.M.; Hosono-Fukao, T.; Tang, R.; Sugaya, N.; van Kuppevelt, T.H.; Jenniskens, G.J.; Kimata, K.; Rosen, S.D.; Uchimura, K. Direct Detection of HSulf-1 and HSulf-2 Activities on Extracellular Heparan Sulfate and Their Inhibition by PI-88. Glycobiology 2010, 20, 175–186. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mukherjee, P.; Zhou, X.; Benicky, J.; Panigrahi, A.; Aljuhani, R.; Liu, J.; Ailles, L.; Pomin, V.H.; Wang, Z.; Goldman, R. Heparan-6-O-Endosulfatase 2 Promotes Invasiveness of Head and Neck Squamous Carcinoma Cell Lines in Co-Cultures with Cancer-Associated Fibroblasts. Cancers 2023, 15, 5168. https://doi.org/10.3390/cancers15215168
Mukherjee P, Zhou X, Benicky J, Panigrahi A, Aljuhani R, Liu J, Ailles L, Pomin VH, Wang Z, Goldman R. Heparan-6-O-Endosulfatase 2 Promotes Invasiveness of Head and Neck Squamous Carcinoma Cell Lines in Co-Cultures with Cancer-Associated Fibroblasts. Cancers. 2023; 15(21):5168. https://doi.org/10.3390/cancers15215168
Chicago/Turabian StyleMukherjee, Pritha, Xin Zhou, Julius Benicky, Aswini Panigrahi, Reem Aljuhani, Jian Liu, Laurie Ailles, Vitor H. Pomin, Zhangjie Wang, and Radoslav Goldman. 2023. "Heparan-6-O-Endosulfatase 2 Promotes Invasiveness of Head and Neck Squamous Carcinoma Cell Lines in Co-Cultures with Cancer-Associated Fibroblasts" Cancers 15, no. 21: 5168. https://doi.org/10.3390/cancers15215168
APA StyleMukherjee, P., Zhou, X., Benicky, J., Panigrahi, A., Aljuhani, R., Liu, J., Ailles, L., Pomin, V. H., Wang, Z., & Goldman, R. (2023). Heparan-6-O-Endosulfatase 2 Promotes Invasiveness of Head and Neck Squamous Carcinoma Cell Lines in Co-Cultures with Cancer-Associated Fibroblasts. Cancers, 15(21), 5168. https://doi.org/10.3390/cancers15215168




