AI Deployment on GBM Diagnosis: A Novel Approach to Analyze Histopathological Images Using Image Feature-Based Analysis
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. Morphology-Based Analysis
1.2. Texture-Based Analysis
1.3. Machine Learning in Cancer Diagnosis
2. Materials and Methods
2.1. Patient Dataset
2.2. Digitize Image from H&E Tissue Slide and Pre-Processing of Images
2.2.1. Digitize the H&E Tissue Slide from TCGA-GBM
2.2.2. Digitize the H&E Tissue Slide from Local Hospital
2.2.3. Standardization and Normalization of Images
2.3. Image Feature Extraction from Tissue Slide Images
2.4. Study Workflow
2.5. Machine Learning Algorithms
2.6. Ten-Fold Cross Validation to Minimize Overfitting
2.7. Data Analysis
2.8. Deployment of Models—Verified by Local Data
3. Results
3.1. Dataset Demographics
3.2. Image Features Selection
3.2.1. GLCM Image Features
Image Features Related to Local Intensity Variation
Image Features Related to Entropy
Image Features Related to Dissimilarity
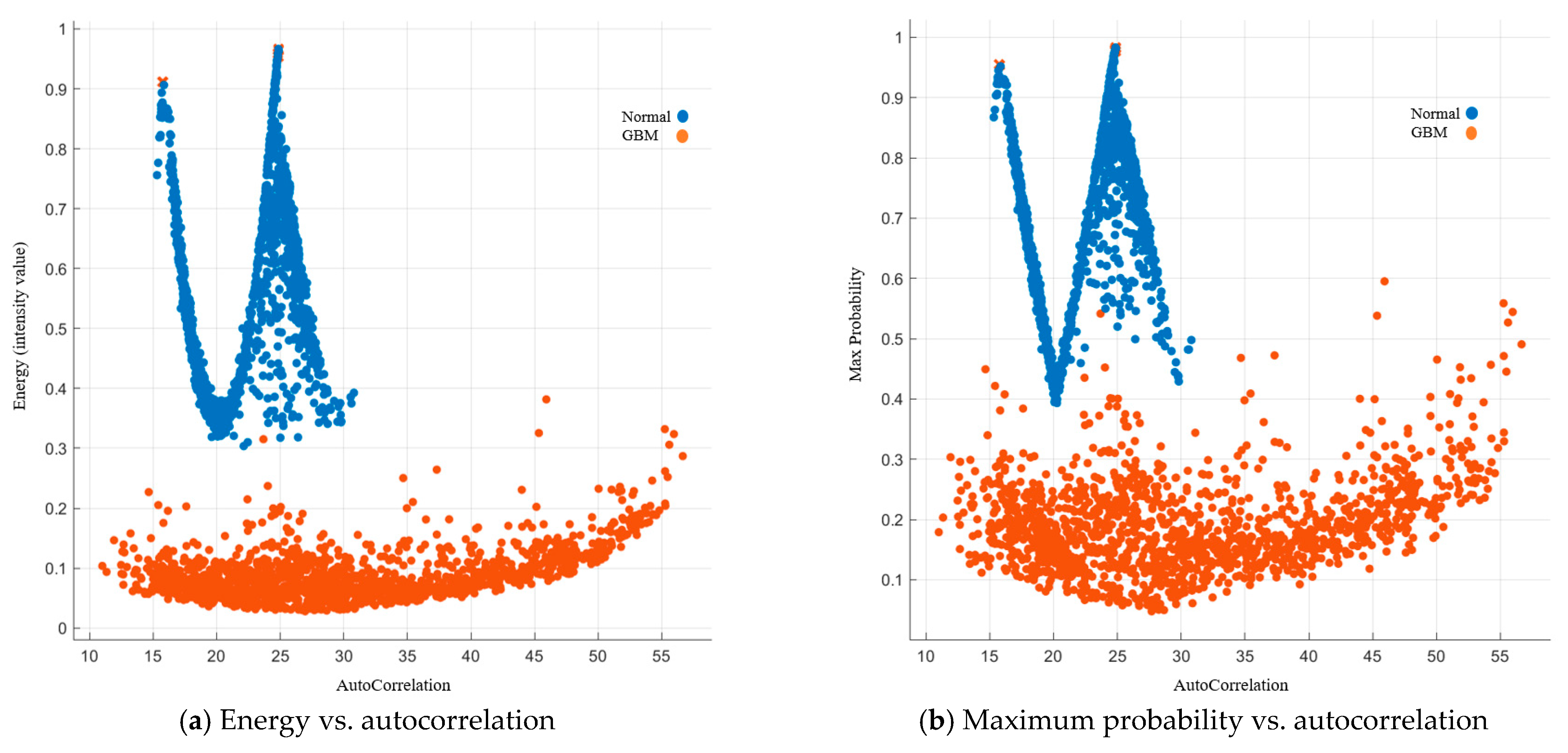
Image Features Related to Energy and Maximum Probability
Image Features Related to Homogeneity
3.2.2. GLRLM Image Features
3.3. Model Building and Validation Using Images from TCGA-GBM
3.4. Model Deployment Using Images from Local Hospital
4. Discussion
4.1. Significance of This Project
4.2. The Value of GLCM and GLRLM Image Features in H&E Images
4.3. Advantage of SVM in Classification
4.4. Study Limitations and Further Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bush, N.A.O.; Chang, S.M.; Berger, M.S. Current and Future Strategies for Treatment of Glioma. Neurosurg. Rev. 2017, 40, 1–14. [Google Scholar] [CrossRef] [PubMed]
- D’Alessio, A.; Proietti, G.; Sica, G.; Scicchitano, B.M. Pathological and Molecular Features of Glioblastoma and Its Peritumoral Tissue. Cancers 2019, 11, 469. [Google Scholar] [CrossRef] [PubMed]
- Cheung, E.Y.W.; Lee, K.H.Y.; Lau, W.T.L.; Lau, A.P.Y.; Wat, P.Y. Non-Coplanar VMAT Plans for Postoperative Primary Brain Tumour to Reduce Dose to Hippocampus, Temporal Lobe and Cochlea: A Planning Study. BJR|Open 2021, 3, 20210009. [Google Scholar] [CrossRef] [PubMed]
- Cheung, E.Y.W.; Ng, S.S.H.; Yung, S.H.Y.; Cheng, D.Y.T.; Chan, F.Y.C.; Cheng, J.K.Y. Multi-Planar VMAT Plans for High-Grade Glioma and Glioblastoma Targeting the Hypothalamic-Pituitary Axis Sparing. Life 2022, 12, 195. [Google Scholar] [CrossRef] [PubMed]
- Baba, A.; Câtoi, C. Tumor Cell Morphology. In Comparative Oncology; The Publishing House of the Romanian Academy: Bucharest, Romania, 2007; Volume 3. [Google Scholar]
- Hao, Y.; Zhang, L.; Qiao, S.; Bai, Y.; Cheng, R.; Xue, H.; Hou, Y.; Zhang, W.; Zhang, G. Breast Cancer Histopathological Images Classification Based on Deep Semantic Features and Gray Level Co-Occurrence Matrix. PLoS ONE 2022, 17, e0267955. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Cooper, L.A.D.; Wang, F.; Gao, J.; Teodoro, G.; Scarpace, L.; Mikkelsen, T.; Schniederjan, M.J.; Moreno, C.S.; Saltz, J.H.; et al. Machine-Based Morphologic Analysis of Glioblastoma Using Whole-Slide Pathology Images Uncovers Clinically Relevant Molecular Correlates. PLoS ONE 2013, 8, e81049. [Google Scholar] [CrossRef] [PubMed]
- Madabhushi, A.; Lee, G. Image Analysis and Machine Learning in Digital Pathology: Challenges and Opportunities. Med. Image Anal. 2016, 33, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Janowczyk, A.; Chandran, S.; Madabhushi, A. A High-Throughput Active Contour Scheme for Segmentation of Histopathological Imagery. Med. Image Anal. 2011, 15, 851–862. [Google Scholar] [CrossRef]
- Davies, E.R. Texture Analysis. In Computer Vision; Elsevier: Amsterdam, The Netherlands, 2018; pp. 185–200. ISBN 978-0-12-809284-2. [Google Scholar]
- Galloway, M.M. Texture Analysis Using Gray Level Run Lengths. Comput. Graph. Image Process. 1975, 4, 172–179. [Google Scholar] [CrossRef]
- Hoffman, R.M.; Atallah, R.P.; Struble, R.D.; Badgett, R.G. Lung Cancer Screening with Low-Dose CT: A Meta-Analysis. J. Gen. Intern. Med. 2020, 35, 3015–3025. [Google Scholar] [CrossRef]
- Akazawa, M.; Hashimoto, K. Artificial Intelligence in Ovarian Cancer Diagnosis. Anticancer. Res. 2020, 40, 4795–4800. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, H.; Shaban, M.; Indave, B.I.; Santos-Silva, A.R.; Rajpoot, N.; Khurram, S.A. Use of Artificial Intelligence in Diagnosis of Head and Neck Precancerous and Cancerous Lesions: A Systematic Review. Oral. Oncol. 2020, 110, 104885. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Oikonomou, A.; Wong, A.; Haider, M.A.; Khalvati, F. Radiomics-Based Prognosis Analysis for Non-Small Cell Lung Cancer. Sci. Rep. 2017, 7, 46349. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Chu, Y.-W.; Cheung, E.Y.W. Radiomics AI Prediction for Head and Neck Squamous Cell Carcinoma (HNSCC) Prognosis and Recurrence with Target Volume Approach. BJR|Open 2021, 3, 20200073. [Google Scholar] [CrossRef]
- Cheung, E.Y.W.; Chau, A.C.M.; Tang, F.H.; on behalf of the Alzheimer’s Disease Neuroimaging Initiative. Radiomics-Based Artificial Intelligence Differentiation of Neurodegenerative Diseases with Reference to the Volumetry. Life 2022, 12, 514. [Google Scholar] [CrossRef] [PubMed]
- Men, K.; Dai, J.; Li, Y. Automatic Segmentation of the Clinical Target Volume and Organs at Risk in the Planning CT for Rectal Cancer Using Deep Dilated Convolutional Neural Networks. Med. Phys. 2017, 44, 6377–6389. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X. Automatic Segmentation of Multiple Organs on 3D CT Images by Using Deep Learning Approaches. Adv. Exp. Med. Biol. 2020, 1213, 135–147. [Google Scholar] [CrossRef]
- Reisert, M.; Sajonz, B.E.A.; Brugger, T.S.; Reinacher, P.C.; Russe, M.F.; Kellner, E.; Skibbe, H.; Coenen, V.A. Where Position Matters-Deep-Learning-Driven Normalization and Coregistration of Computed Tomography in the Postoperative Analysis of Deep Brain Stimulation. Neuromodulation 2023, 26, 302–309. [Google Scholar] [CrossRef]
- Pan, Z.; Zhang, R.; Shen, S.; Lin, Y.; Zhang, L.; Wang, X.; Ye, Q.; Wang, X.; Chen, J.; Zhao, Y.; et al. OWL: An Optimized and Independently Validated Machine Learning Prediction Model for Lung Cancer Screening Based on the UK Biobank, PLCO, and NLST Populations. eBioMedicine 2023, 88, 104443. [Google Scholar] [CrossRef]
- Mahmood, H.; Shaban, M.; Rajpoot, N.; Khurram, S.A. Artificial Intelligence-Based Methods in Head and Neck Cancer Diagnosis: An Overview. Br. J. Cancer 2021, 124, 1934–1940. [Google Scholar] [CrossRef]
- Hao, Y.; Qiao, S.; Zhang, L.; Xu, T.; Bai, Y.; Hu, H.; Zhang, W.; Zhang, G. Breast Cancer Histopathological Images Recognition Based on Low Dimensional Three-Channel Features. Front. Oncol. 2021, 11, 657560. [Google Scholar] [CrossRef] [PubMed]
- Belsare, A.D.; Mushrif, M.M.; Pangarkar, M.A.; Meshram, N. Classification of Breast Cancer Histopathology Images Using Texture Feature Analysis. In Proceedings of the TENCON 2015-2015 IEEE Region 10 Conference, Macao, China, 1–4 November 2015; pp. 1–5. [Google Scholar]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. A Dataset for Breast Cancer Histopathological Image Classification. IEEE Trans. Biomed. Eng. 2016, 63, 1455–1462. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Xiao, B.; Wei, H.; Huang, S.; Chen, T. SVM Classifier of Cervical Histopathology Images Based on Texture and Morphological Features. Technol. Health Care 2022, 31, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Scarpace, L.; Mikkelsen, T.; Cha, S.; Rao, S.; Tekchandani, S.; Gutman, D.; Saltz, J.H.; Erickson, B.J.; Pedano, N.; Flanders, A.E.; et al. The Cancer Genome Atlas Glioblastoma Multiforme Collection (TCGA-GBM). Cancer Imaging Arch. Publ. Online 2016. [Google Scholar]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol 2007, 114, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Wei, X. Gray Level Run Length Matrix Toolbox. Available online: https://www.mathworks.com/matlabcentral/fileexchange/17482-gray-level-run-length-matrix-toolbox (accessed on 24 October 2022).
- Tang, X. Texture Information in Run-Length Matrices. IEEE Trans. Image Process. 1998, 7, 1602–1609. [Google Scholar] [CrossRef] [PubMed]
- Soh, L.-K.; Tsatsoulis, C. Texture Analysis of SAR Sea Ice Imagery Using Gray Level Co-Occurrence Matrices. IEEE Trans. Geosci. Remote Sens. 1999, 37, 780–795. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- GLCM_Features4.m: Vectorized Version of GLCM_Features1.m [With Code Changes]. Available online: https://www.mathworks.com/matlabcentral/fileexchange/22354-glcm_features4-m-vectorized-version-of-glcm_features1-m-with-code-changes (accessed on 24 October 2022).
- Clausi, D.A. An Analysis of Co-Occurrence Texture Statistics as a Function of Grey Level Quantization. Can. J. Remote Sens. 2002, 28, 45–62. [Google Scholar] [CrossRef]
- Antoniou, G.; Deremer, D. (Eds.) Computing and Information Technologies: Exploring Emerging Technologies: Montclair State University, NJ, USA; World Scientific Pub. Co.: River Edge, NJ, USA, 2001; ISBN 978-981-02-4759-1. [Google Scholar]
- 3.1. Cross-Validation: Evaluating Estimator Performance. Available online: https://scikit-learn/stable/modules/cross_validation.html (accessed on 8 October 2022).
- Laws, K.I. Rapid Texture Identification; Wiener, T.F., Ed.; Optics & Photonics. Spie: San Diego, CA, USA, 1980; pp. 376–381. [Google Scholar]
- Willis, L.M. (Ed.) Professional Guide to Pathophysiology, 4th ed.; Wolters Kluwer: Philadelphia, PA, USA, 2019; ISBN 978-1-975107-69-7. [Google Scholar]
- Bhagat, P.K.; Choudhary, P.; Singh, K.M. A Comparative Study for Brain Tumor Detection in MRI Images Using Texture Features. In Sensors for Health Monitoring; Elsevier: Amsterdam, The Netherlands, 2019; pp. 259–287. ISBN 978-0-12-819361-7. [Google Scholar]
- Li, X.; Guindani, M.; Ng, C.S.; Hobbs, B.P. Spatial Bayesian Modeling of GLCM with Application to Malignant Lesion Characterization. J. Appl. Stat. 2018, 46, 230–246. [Google Scholar] [CrossRef]
- Shan, F.Y.; Zhao, D.; Tirado, C.A.; Fonkem, E.; Zhang, Y.; Feng, D.H.; Huang, J. Glioblastomas: Molecular Diagnosis and Pathology. In Glioblastoma-Current Evidence; Agrawal, A., Singh Kunwar, D., Eds.; IntechOpen: London, UK, 2023; ISBN 978-1-80356-128-8. [Google Scholar]
- Varuna Shree, N.; Kumar, T.N.R. Identification and Classification of Brain Tumor MRI Images with Feature Extraction Using DWT and Probabilistic Neural Network. Brain Inf. 2018, 5, 23–30. [Google Scholar] [CrossRef]
- Vapnik, V.N. Statistical Learning Theory. In Adaptive and Learning Systems for Signal Processing, Communications, and Control; Wiley: New York, NY, USA, 1998; ISBN 978-0-471-03003-4. [Google Scholar]
- Huang, M.-W.; Chen, C.-W.; Lin, W.-C.; Ke, S.-W.; Tsai, C.-F. SVM and SVM Ensembles in Breast Cancer Prediction. PLoS ONE 2017, 12, e0161501. [Google Scholar] [CrossRef]
- Galal, A.; Talal, M.; Moustafa, A. Applications of Machine Learning in Metabolomics: Disease Modeling and Classification. Front. Genet. 2022, 13, 1017340. [Google Scholar] [CrossRef]











| Cohort 1: for Model Building (TCGA–GBM) | Cohort 2: for Model Deployment (Local Hospital) | |||
|---|---|---|---|---|
| No of Participants | No of Images | No of Participants | No of Images | |
| GBM | 262 | 1500 | 60 | 702 |
| Normal | 40 | 1500 | 20 | 670 |
| Gray Level Co-Occurrence Matrix (GLCM) | Gray Level Run Length Matrix (GLRLM) [30,31] | |
|---|---|---|
| Autocorrelation [32] | Maximum Probability [32] | Short Run Emphasis |
| Contrast [32,33] | Sum of square [33] | Long Run Emphasis |
| Correlation 1 [34] | Sum average [33] | Gray Level Non-uniformity |
| Correlation 2 [32,33] | Sum variance [33] | Run Length Non-uniformity |
| Cluster prominence [32] | Sum entropy [33] | Run Percentage |
| Cluster Shade [32] | Difference variance [33] | Low Gray-level Run Emphasis |
| Dissimilarity [32] | Difference entropy [33] | High Gray-level Run Emphasis |
| Energy [32,33] | Information measure of correlation 1 [33] | Short Run Low Gray-level Run Emphasis |
| Entropy [32] | Information measure of correlation 2 [33] | Short Run High Gray-level Run Emphasis |
| Homogeneity 1 [34] | Inverse Difference normalized [35] | Long Run Low Gray-level Run Emphasis |
| Homogeneity 2 [32] | Inverse Difference moment normalized [35] | Long Rg equations to obtain each features were listed in the . es were y. Rrun High Gray-level Run Emphasis |
| Part 1 Model development | GBM group (n = 1500) | Normal Group (n = 1500) | |||||
| Group | Training | Validation | Testing | Training | Validation | Testing | |
| Percentage | 70% | 15% | 15% | 70% | 15% | 15% | |
| Sample size | 1050 | 225 | 225 | 1050 | 225 | 225 | |
| Part 2 Model deployment | GBM group (n = 702) | Normal Group (n = 670) | |||||
| Group | Training | Validation | Testing | Training | Validation | Testing | |
| Percentage | 70% | 15% | 15% | 70% | 15% | 15% | |
| Sample size | 492 | 105 | 105 | 470 | 100 | 100 | |
| Part 1 For Model Development | Part 2 For Model Deployment | |||
|---|---|---|---|---|
| GBM | Normal | GBM | Normal | |
| No of Participants | 262 | 40 | 60 | 20 |
| Age Range Mean ±SD Sex (M:F) | 14–86 58.96 ± 13.95 159:101 | NIL | 33–75 61 ± 11.06 42:18 | NIL |
| Algorithm | Overall Accuracy | Sensitivity | Specificity | Area under the ROC Curve | Precision | Recall | F1 Score |
|---|---|---|---|---|---|---|---|
| Decision Tree (DT) | 100% | 100% | 100% | 1 | 100% | 100% | 1 |
| Extreme Boost (EB) | 100% | 100% | 100% | 1 | 100% | 100% | 1 |
| Random Forest (RF) | 100% | 100% | 100% | 1 | 100% | 100% | 1 |
| Support Vector Machine (SVM) | 100% | 100% | 100% | 1 | 100% | 100% | 1 |
| Linear Model (LM) | 100% | 100% | 100% | 1 | 100% | 100% | 1 |
| Algorithm | Overall Accuracy | Sensitivity | Specificity | Area under the ROC Curve | Precision | Recall | F1 Score |
|---|---|---|---|---|---|---|---|
| DT-Model | 59.4% | 18.82% | 1 | 0.5357 | 17.2% | 100% | 29.3% |
| EB-Model | 59.4% | 18.82 | 1 | 0.5936 | 17.2% | 100% | 29.3% |
| RF-Model | 60.3% | 21.06% | 1 | 0.6283 | 20.8% | 100% | 34.4% |
| SVM-Model | 93.5% | 86.95% | 99.73% | 0.9908 | 82.9% | 99.7% | 90.5% |
| LM-Model | 55.0% | 11.80% | 1 | 0.5398 | 0% | 100% | 0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheung, E.Y.W.; Wu, R.W.K.; Li, A.S.M.; Chu, E.S.M. AI Deployment on GBM Diagnosis: A Novel Approach to Analyze Histopathological Images Using Image Feature-Based Analysis. Cancers 2023, 15, 5063. https://doi.org/10.3390/cancers15205063
Cheung EYW, Wu RWK, Li ASM, Chu ESM. AI Deployment on GBM Diagnosis: A Novel Approach to Analyze Histopathological Images Using Image Feature-Based Analysis. Cancers. 2023; 15(20):5063. https://doi.org/10.3390/cancers15205063
Chicago/Turabian StyleCheung, Eva Y. W., Ricky W. K. Wu, Albert S. M. Li, and Ellie S. M. Chu. 2023. "AI Deployment on GBM Diagnosis: A Novel Approach to Analyze Histopathological Images Using Image Feature-Based Analysis" Cancers 15, no. 20: 5063. https://doi.org/10.3390/cancers15205063
APA StyleCheung, E. Y. W., Wu, R. W. K., Li, A. S. M., & Chu, E. S. M. (2023). AI Deployment on GBM Diagnosis: A Novel Approach to Analyze Histopathological Images Using Image Feature-Based Analysis. Cancers, 15(20), 5063. https://doi.org/10.3390/cancers15205063






