The T Cell Immunoscore as a Reference for Biomarker Development Utilizing Real-World Data from Patients with Advanced Malignancies Treated with Immune Checkpoint Inhibitors †
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods and Materials
2.1. Patients and Datasets
2.2. RNA-Sequencing and Data Processing
2.3. Immunoscore Imputation
2.4. Construction of the Immunoscore
2.5. Statistical Analysis
3. Results
3.1. Baseline Characteristics
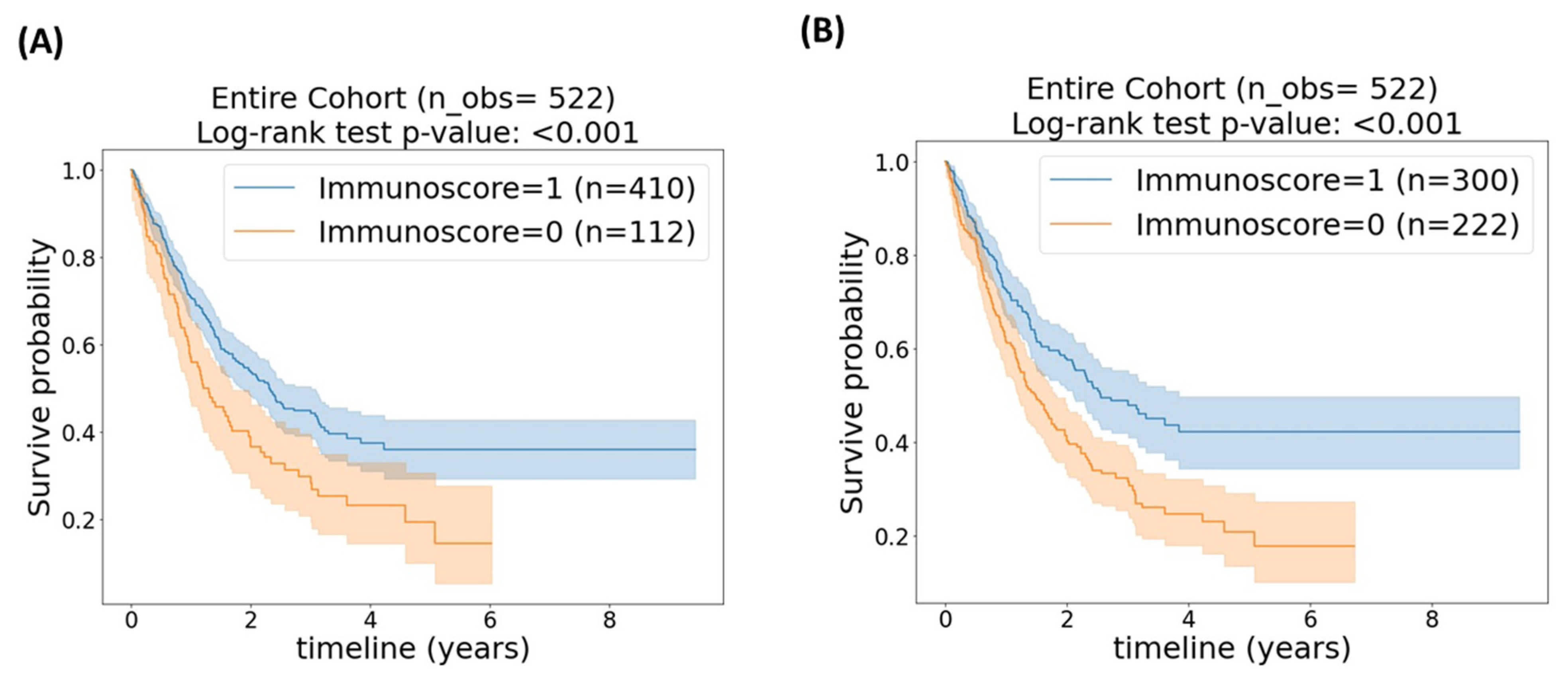
3.2. Entire Cohort
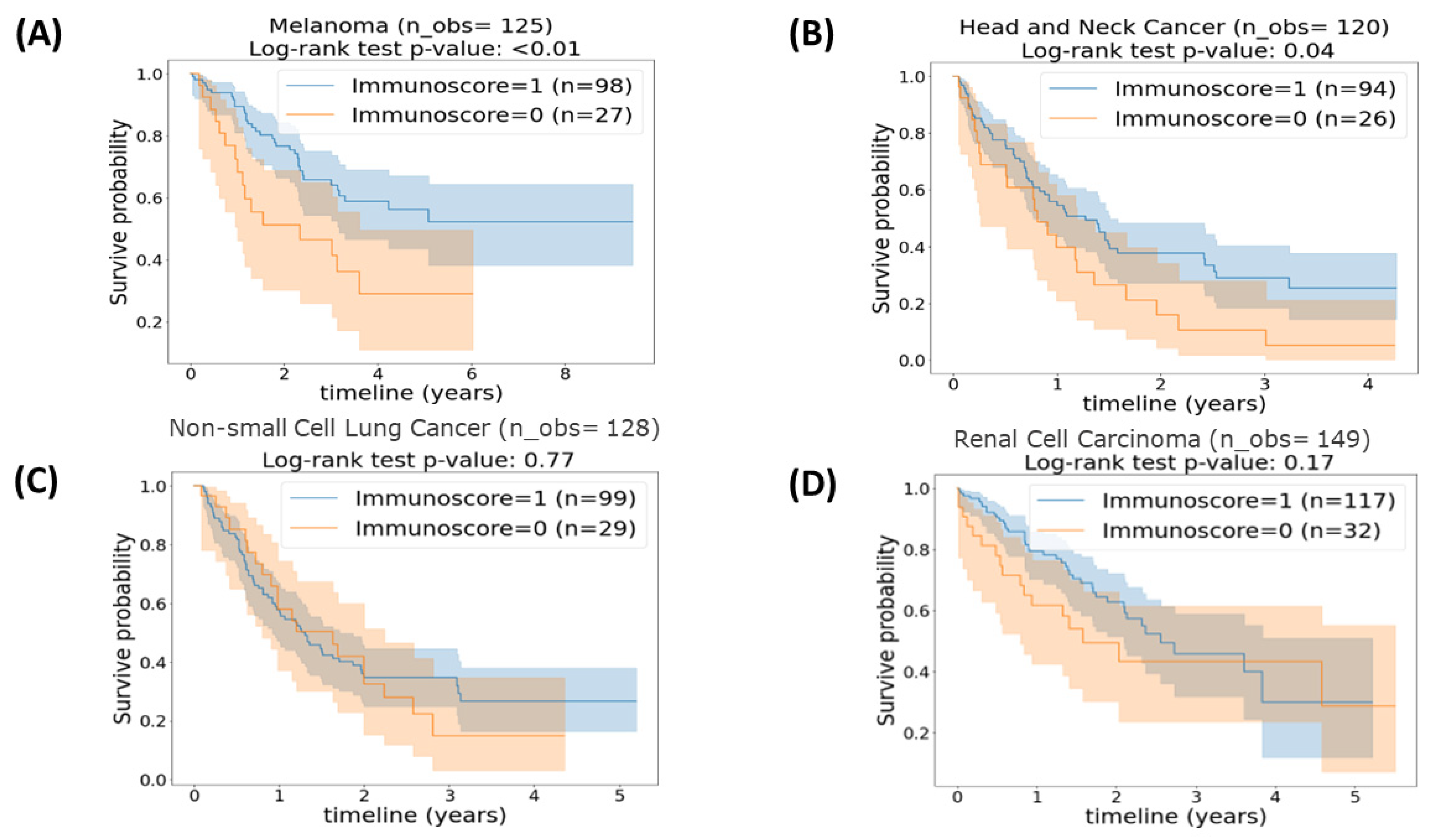
3.3. Cancer Categories
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Galon, J.; Lanzi, A. Immunoscore and its introduction in clinical practice. Q. J. Nucl. Med. Mol. Imaging 2020, 64, 152–161. [Google Scholar] [CrossRef]
- Yadav, D.; Kwak, M.; Chauhan, P.S.; Puranik, N.; Lee, P.C.W.; Jin, J.O. Cancer immunotherapy by immune checkpoint blockade and its advanced application using bio-nanomaterials. Semin. Cancer Biol. 2022, 86 Pt 2, 909–922. [Google Scholar] [CrossRef] [PubMed]
- Vale, C.L. Neoadjuvant chemotherapy in invasive bladder cancer: Update of a systematic review and meta-analysis of individual patient data advanced bladder cancer (ABC) meta-analysis collaboration. Eur. Urol. 2005, 48, 202–205; discussion 5-6. [Google Scholar] [CrossRef]
- Bondhopadhyay, B.; Sisodiya, S.; Chikara, A.; Khan, A.; Tanwar, P.; Afroze, D.; Singh, N.; Agrawal, U.; Mehrotra, R.; Hussain, S. Cancer immunotherapy: A promising dawn in cancer research. Am. J. Blood Res. 2020, 10, 375–385. [Google Scholar] [PubMed]
- Fares, C.M.; Van Allen, E.M.; Drake, C.G.; Allison, J.P.; Hu-Lieskovan, S. Mechanisms of Resistance to Immune Checkpoint Blockade: Why Does Checkpoint Inhibitor Immunotherapy Not Work for All Patients? Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 147–164. [Google Scholar] [CrossRef] [PubMed]
- Haslam, A.; Prasad, V. Estimation of the Percentage of US Patients With Cancer Who Are Eligible for and Respond to Checkpoint Inhibitor Immunotherapy Drugs. JAMA Netw. Open 2019, 2, e192535. [Google Scholar] [CrossRef]
- Kalbasi, A.; Ribas, A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 2020, 20, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Rodriguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csoszi, T.; Fulop, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Horn, L.; Spigel, D.R.; Vokes, E.E.; Holgado, E.; Ready, N.; Steins, M.; Poddubskaya, E.; Borghaei, H.; Felip, E.; Paz-Ares, L.; et al. Nivolumab Versus Docetaxel in Previously Treated Patients With Advanced Non-Small-Cell Lung Cancer: Two-Year Outcomes From Two Randomized, Open-Label, Phase III Trials (CheckMate 017 and CheckMate 057). J. Clin. Oncol. 2017, 35, 3924–3933. [Google Scholar] [CrossRef]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef] [PubMed]
- Bruni, D.; Angell, H.K.; Galon, J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer 2020, 20, 662–680. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Bruni, D. Tumor Immunology and Tumor Evolution: Intertwined Histories. Immunity 2020, 52, 55–81. [Google Scholar] [CrossRef] [PubMed]
- Ascierto, P.A.; Capone, M.; Urba, W.J.; Bifulco, C.B.; Botti, G.; Lugli, A.; Marincola, F.M.; Ciliberto, G.; Galon, J.; Fox, B.A. The additional facet of immunoscore: Immunoprofiling as a possible predictive tool for cancer treatment. J. Transl. Med. 2013, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef]
- Galon, J.; Fox, B.A.; Bifulco, C.B.; Masucci, G.; Rau, T.; Botti, G.; Marincola, F.M.; Ciliberto, G.; Pages, F.; Ascierto, P.A.; et al. Immunoscore and Immunoprofiling in cancer: An update from the melanoma and immunotherapy bridge 2015. J. Transl. Med. 2016, 14, 273. [Google Scholar] [CrossRef]
- Kirilovsky, A.; Marliot, F.; El Sissy, C.; Haicheur, N.; Galon, J.; Pages, F. Rational bases for the use of the Immunoscore in routine clinical settings as a prognostic and predictive biomarker in cancer patients. Int. Immunol. 2016, 28, 373–382. [Google Scholar] [CrossRef]
- Pages, F.; Galon, J.; Fridman, W.H. The essential role of the in situ immune reaction in human colorectal cancer. J. Leukoc. Biol. 2008, 84, 981–987. [Google Scholar] [CrossRef]
- Fridman, W.H.; Dieu-Nosjean, M.C.; Pages, F.; Cremer, I.; Damotte, D.; Sautes-Fridman, C.; Galon, J. The immune microenvironment of human tumors: General significance and clinical impact. Cancer Microenviron. 2013, 6, 117–122. [Google Scholar] [CrossRef]
- Marliot, F.; Pages, F.; Galon, J. Usefulness and robustness of Immunoscore for personalized management of cancer patients. Oncoimmunology 2020, 9, 1832324. [Google Scholar] [CrossRef]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pages, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P.; et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef]
- Galon, J.; Pages, F.; Marincola, F.M.; Thurin, M.; Trinchieri, G.; Fox, B.A.; Gajewski, T.F.; Ascierto, P.A. The immune score as a new possible approach for the classification of cancer. J. Transl. Med. 2012, 10, 1. [Google Scholar] [CrossRef]
- Anitei, M.G.; Zeitoun, G.; Mlecnik, B.; Marliot, F.; Haicheur, N.; Todosi, A.M.; Kirilovsky, A.; Lagorce, C.; Bindea, G.; Ferariu, D.; et al. Prognostic and predictive values of the immunoscore in patients with rectal cancer. Clin. Cancer Res. 2014, 20, 1891–1899. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.Y.; Shen, W.; Yue, J.Q.; Yao, W.Y.; Liu, S.L.; Jin, Y.P.; Dong, P.; Ma, F.; Wu, X.S.; Gong, W. Combining Immunoscore with Clinicopathologic Features in Cholangiocarcinoma: An Influential Prognostic Nomogram. Onco Targets Ther. 2020, 13, 11359–11376. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Mlecnik, B.; Bindea, G.; Angell, H.K.; Berger, A.; Lagorce, C.; Lugli, A.; Zlobec, I.; Hartmann, A.; Bifulco, C.; et al. Towards the introduction of the ‘Immunoscore’ in the classification of malignant tumours. J. Pathol. 2014, 232, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Pages, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018, 391, 2128–2139. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, Q.; Hu, Y.; Li, T.; Yu, J.; Zhao, L.; Ye, G.; Deng, H.; Mou, T.; Cai, S.; et al. ImmunoScore Signature: A Prognostic and Predictive Tool in Gastric Cancer. Ann. Surg. 2018, 267, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Li, X.D.; Huang, C.W.; Liu, Z.F.; Jiang, L.J.; Chen, J.W.; Xie, D.; Zhou, F.J.; Lu, H.M.; Liu, Z.W. Prognostic Role of the Immunoscore for Patients with Urothelial Carcinoma of the Bladder Who Underwent Radical Cystectomy. Ann. Surg. Oncol. 2019, 26, 4148–4156. [Google Scholar] [CrossRef] [PubMed]
- Angell, H.K.; Bruni, D.; Barrett, J.C.; Herbst, R.; Galon, J. The Immunoscore: Colon Cancer and Beyond. Clin. Cancer Res. 2020, 26, 332–339. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, J.; Du, L.; Zhou, Y.; Li, K. The prognostic value of Immunoscore in patients with cancer: A pooled analysis of 10,328 patients. Int. J. Biol. Markers 2020, 35, 3–13. [Google Scholar] [CrossRef]
- Nie, R.C.; Yuan, S.Q.; Wang, Y.; Chen, Y.B.; Cai, Y.Y.; Chen, S.; Li, S.M.; Zhou, J.; Chen, G.M.; Luo, T.Q.; et al. Robust immunoscore model to predict the response to anti-PD1 therapy in melanoma. Aging 2019, 11, 11576–11590. [Google Scholar] [CrossRef]
- Stoll, G.; Enot, D.; Mlecnik, B.; Galon, J.; Zitvogel, L.; Kroemer, G. Immune-related gene signatures predict the outcome of neoadjuvant chemotherapy. Oncoimmunology 2014, 3, e27884. [Google Scholar] [CrossRef] [PubMed]
- Vacchelli, E.; Galluzzi, L.; Fridman, W.H.; Galon, J.; Sautes-Fridman, C.; Tartour, E.; Kroemer, G. Trial watch: Chemotherapy with immunogenic cell death inducers. Oncoimmunology 2012, 1, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Dong, X.; Tang, X.; Qu, H.; Zhang, H.; Zhao, E. The prognostic value of immunoscore in patients with colorectal cancer: A systematic review and meta-analysis. Cancer Med. 2019, 8, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Noma, T.; Makino, T.; Ohshima, K.; Sugimura, K.; Miyata, H.; Honma, K.; Yamashita, K.; Saito, T.; Tanaka, K.; Yamamoto, K.; et al. Immunoscore Signatures in Surgical Specimens and Tumor-Infiltrating Lymphocytes in Pretreatment Biopsy Predict Treatment Efficacy and Survival in Esophageal Cancer. Ann. Surg. 2023, 277, e528–e537. [Google Scholar] [CrossRef]
- Nassif, E.F.; Thibault, C.; Oudard, S.; Galon, J. Precision immunity: Immunoscore and neoadjuvant treatment in bladder cancer. Oncoimmunology 2021, 10, 1888488. [Google Scholar] [CrossRef]
- Li, X.T.; Zhang, J.T.; Yan, H.H.; Su, J.; Cheng, M.L.; Sun, Q.H.; Zhong, W.Z.; Wu, Y.L.; Zhang, D.X.; Hou, D.J. Gene co-expression modules integrated with immunoscore predicts survival of non-small cell lung cancer. Cancer Treat. Res. Commun. 2021, 26, 100297. [Google Scholar] [CrossRef]
- Shen-Orr, S.S.; Gaujoux, R. Computational deconvolution: Extracting cell type-specific information from heterogeneous samples. Curr. Opin. Immunol. 2013, 25, 571–578. [Google Scholar] [CrossRef]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar]
- Tung, J.W.; Heydari, K.; Tirouvanziam, R.; Sahaf, B.; Parks, D.R.; Herzenberg, L.A.; Herzenberg, L.A. Modern flow cytometry: A practical approach. Clin. Lab. Med. 2007, 27, 453–468. [Google Scholar] [CrossRef]
- Abbas, A.R.; Wolslegel, K.; Seshasayee, D.; Modrusan, Z.; Clark, H.F. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS ONE. 2009, 4, e6098. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Weeks, J.C.; Cook, E.F.; O’Day, S.J.; Peterson, L.M.; Wenger, N.; Reding, D.; Harrell, F.E.; Kussin, P.; Dawson, N.V.; Connors, A.F., Jr.; et al. Relationship between cancer patients’ predictions of prognosis and their treatment preferences. JAMA 1998, 279, 1709–1714. [Google Scholar] [CrossRef] [PubMed]
- Erdag, G.; Schaefer, J.T.; Smolkin, M.E.; Deacon, D.H.; Shea, S.M.; Dengel, L.T.; Patterson, J.W.; Slingluff, C.L., Jr. Immunotype and immunohistologic characteristics of tumor-infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer Res. 2012, 72, 1070–1080. [Google Scholar] [CrossRef] [PubMed]
- Furgiuele, S.; Descamps, G.; Lechien, J.R.; Dequanter, D.; Journe, F.; Saussez, S. Immunoscore Combining CD8, FoxP3, and CD68-Positive Cells Density and Distribution Predicts the Prognosis of Head and Neck Cancer Patients. Cells 2022, 11, 2050. [Google Scholar] [CrossRef] [PubMed]
- Selvi, I.; Demirci, U.; Bozdogan, N.; Basar, H. The prognostic effect of immunoscore in patients with clear cell renal cell carcinoma: Preliminary results. Int. Urol. Nephrol. 2020, 52, 21–34. [Google Scholar] [CrossRef]


| Variable | Patients (N = 522) |
|---|---|
| Age (in years) | |
| Median (range) | 63 (19–90) |
| Sex, n (%) | 360 (69) |
| Male | 162 (31) |
| Female | |
| Race, n (%) | 496 (95) |
| White | 16 (3) |
| Black | 10 (2) |
| Other | |
| ECOG performance status at diagnosis, n (%) | |
| 0 | 99 (19) |
| 1 | 84 (16) |
| 2 | 10 (2) |
| Unknown | 329 (63) |
| Cancer type, n (%) | |
| Renal cell carcinoma | 149 (28.5%) |
| Non-small cell lung cancer | 128 (24.5%) |
| Melanoma | 125 (23.9%) |
| Head and neck cancer | 120 (23.0%) |
| Prior systemic therapy, n (%) | |
| 1 Prior line | 198 (38) |
| 2+ Prior line | 324 (62) |
| First Immune checkpoint inhibitors, n (%) | |
| Nivolumab | 219 (42.0%) |
| Pembrolizumab | 202 (38.7%) |
| Ipilimumab + nivolumab | 69 (13.2%) |
| Ipilimumab | 30 (5.6%) |
| Avelumab | 1 (0.2%) |
| Cemiplimab | 1 (0.2%) |
| Percentile Cut-Off | Avg. C-Index (95% CI) | Log-Rank Test p-Value * |
|---|---|---|
| Cut-off = 25th percentile | 0.5402 (0.5345, 0.5459) | <0.001 |
| Cut-off = 43.5th percentile | 0.5528 (0.5466, 0.5591) | <0.001 |
| Cancer Category | Percentile Cut-Off | Avg. C-Index (95% CI) | Log-Rank Test p-Value * |
|---|---|---|---|
| Head and neck | 43.7th | 0.55 (0.54, 0.56) | 0.04 |
| Renal Cell Carcinoma | 36.9th | 0.56 (0.55, 0.58) | 0.17 |
| Non-small cell lung | 57.75th | 0.47 (0.46, 0.48) | 0.77 |
| Melanoma | 49.13th | 0.58 (0.57, 0.59) | 0.009 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eljilany, I.; Saghand, P.G.; Chen, J.; Ratan, A.; McCarter, M.; Carpten, J.; Colman, H.; Ikeguchi, A.P.; Puzanov, I.; Arnold, S.; et al. The T Cell Immunoscore as a Reference for Biomarker Development Utilizing Real-World Data from Patients with Advanced Malignancies Treated with Immune Checkpoint Inhibitors. Cancers 2023, 15, 4913. https://doi.org/10.3390/cancers15204913
Eljilany I, Saghand PG, Chen J, Ratan A, McCarter M, Carpten J, Colman H, Ikeguchi AP, Puzanov I, Arnold S, et al. The T Cell Immunoscore as a Reference for Biomarker Development Utilizing Real-World Data from Patients with Advanced Malignancies Treated with Immune Checkpoint Inhibitors. Cancers. 2023; 15(20):4913. https://doi.org/10.3390/cancers15204913
Chicago/Turabian StyleEljilany, Islam, Payman Ghasemi Saghand, James Chen, Aakrosh Ratan, Martin McCarter, John Carpten, Howard Colman, Alexandra P. Ikeguchi, Igor Puzanov, Susanne Arnold, and et al. 2023. "The T Cell Immunoscore as a Reference for Biomarker Development Utilizing Real-World Data from Patients with Advanced Malignancies Treated with Immune Checkpoint Inhibitors" Cancers 15, no. 20: 4913. https://doi.org/10.3390/cancers15204913
APA StyleEljilany, I., Saghand, P. G., Chen, J., Ratan, A., McCarter, M., Carpten, J., Colman, H., Ikeguchi, A. P., Puzanov, I., Arnold, S., Churchman, M., Hwu, P., Conejo-Garcia, J., Dalton, W. S., Weiner, G. J., El Naqa, I. M., & Tarhini, A. A. (2023). The T Cell Immunoscore as a Reference for Biomarker Development Utilizing Real-World Data from Patients with Advanced Malignancies Treated with Immune Checkpoint Inhibitors. Cancers, 15(20), 4913. https://doi.org/10.3390/cancers15204913







