The Importance of Selected Dysregulated microRNAs in Diagnosis and Prognosis of Childhood B-Cell Precursor Acute Lymphoblastic Leukemia
Abstract
Simple Summary
Abstract
1. Introduction
2. Background
2.1. Non-Coding RNAs
2.2. MicroRNAs
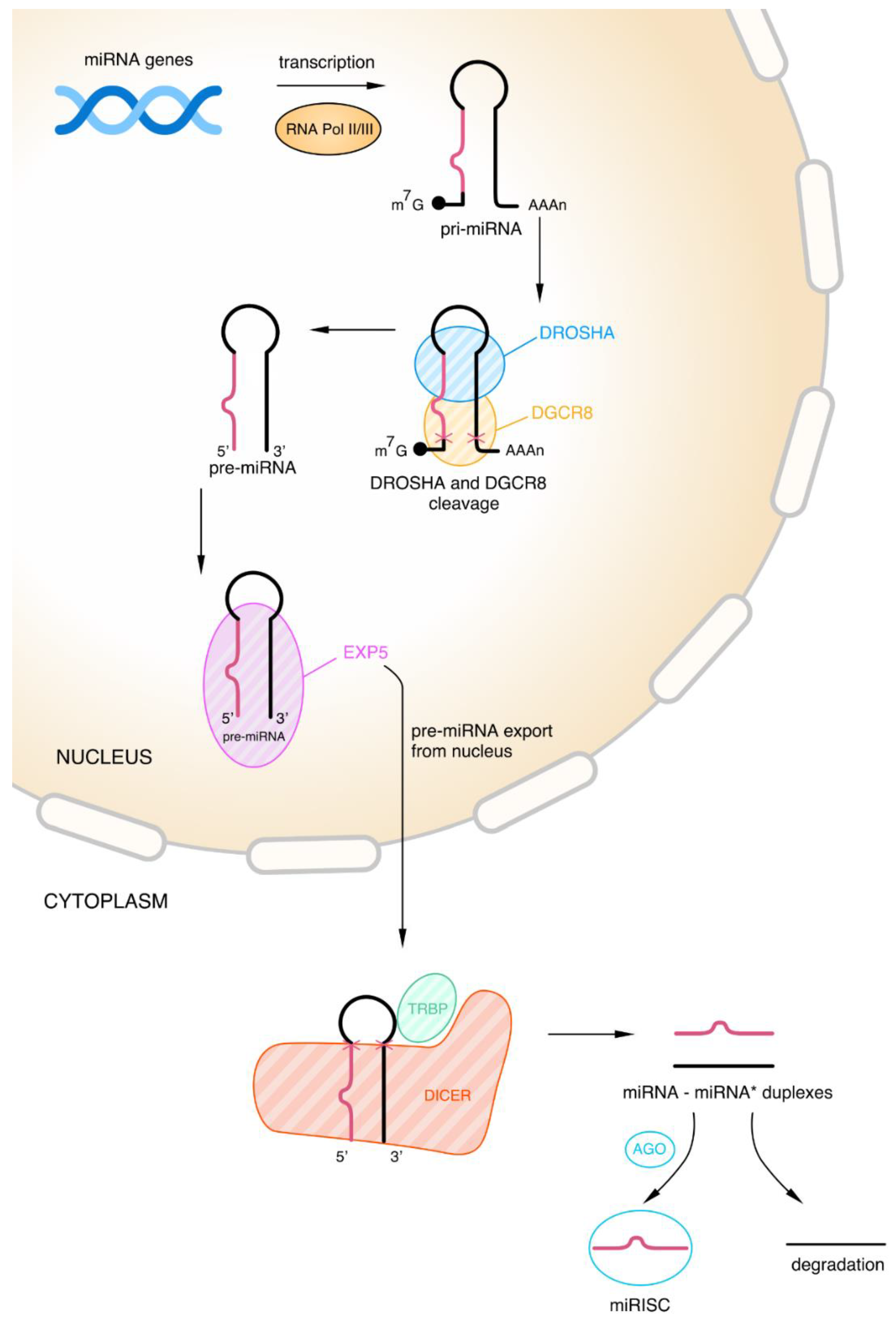
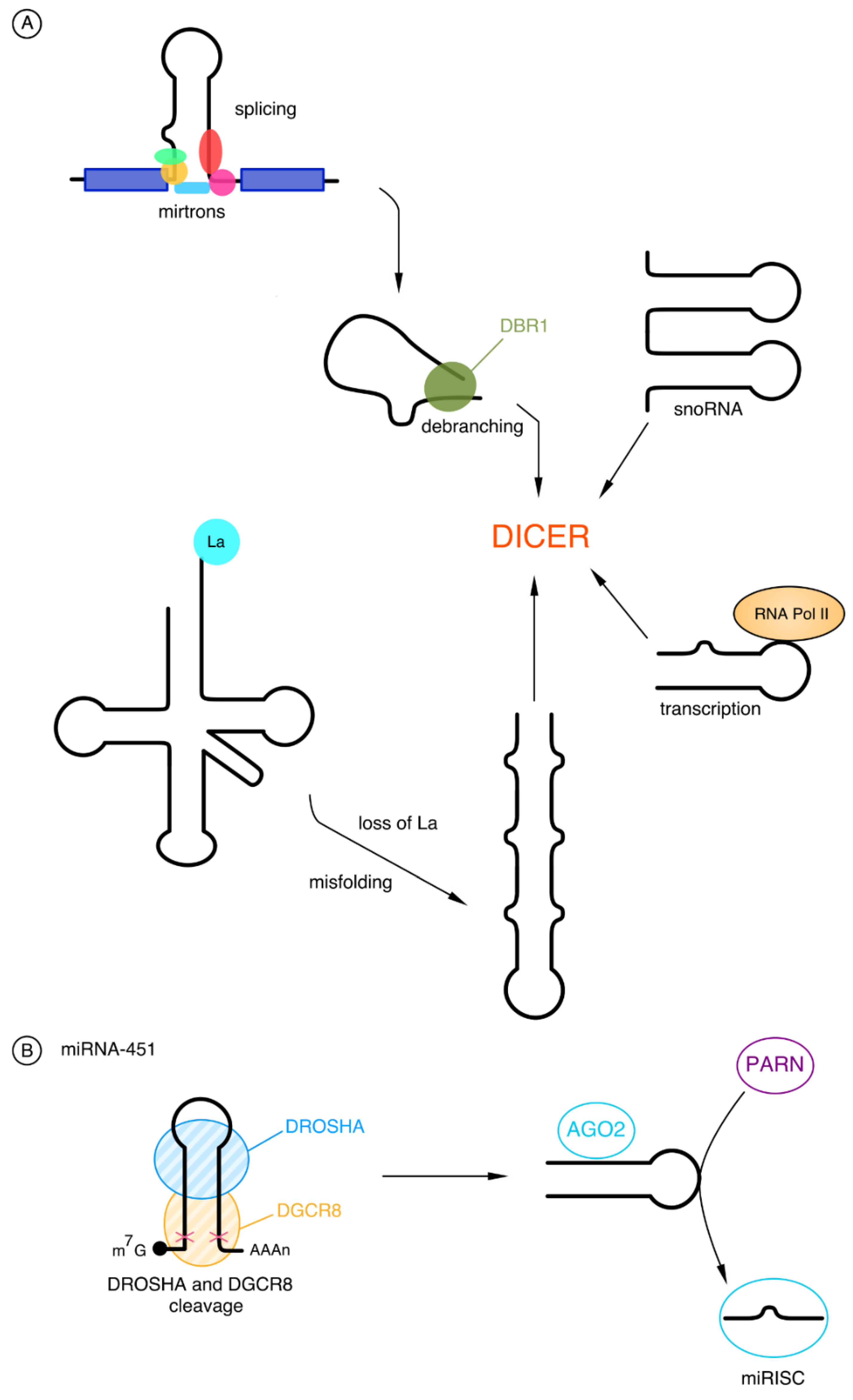
2.2.1. Biogenesis of microRNAs
2.2.2. Gene Regulation by microRNAs
3. miRNAs in Childhood BCP-ALL
3.1. Downregulated miRNAs
miRNA-200c and miRNA-326
3.2. Down- and Upregulated miRNAs
3.2.1. miRNA-100
3.2.2. miRNA-181a
3.3. Upregulated miRNAs
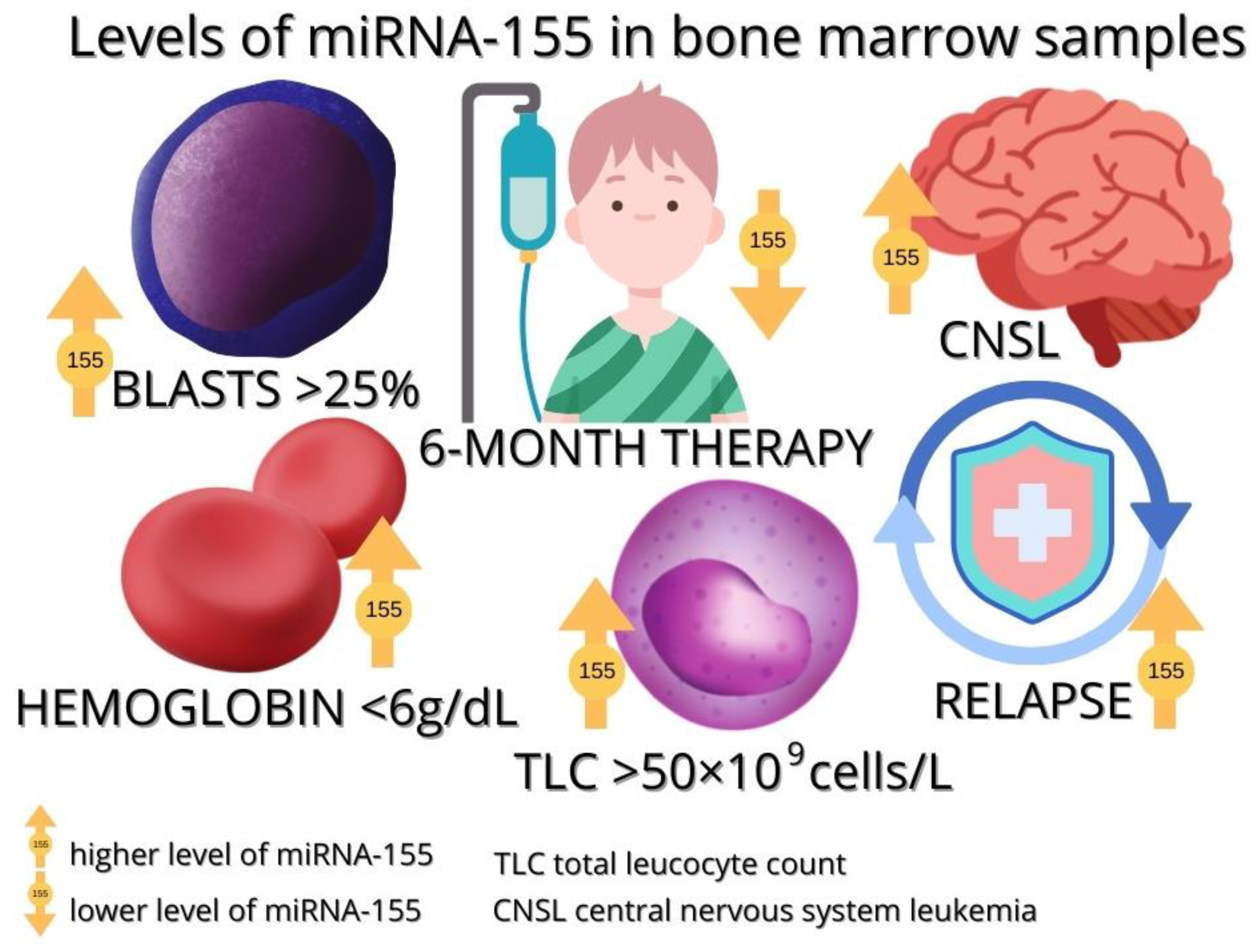
3.3.1. miRNA-155
3.3.2. miRNA-125b
3.3.3. miRNA-146a
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Schwab, C.; Harrison, C.J. Advances in B-cell Precursor Acute Lymphoblastic Leukemia Genomics. HemaSphere 2018, 2, e53. [Google Scholar] [CrossRef] [PubMed]
- Starý, J.; Zuna, J.; Zaliova, M. New biological and genetic classification and therapeutically relevant categories in childhood B-cell precursor acute lymphoblastic leukemia. F1000Research 2018, 7, 1569. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Zhu, S.; Peng, H.; Cheng, Z.; Zhang, G.; Wang, Z. B-cell acute lymphoblastic leukemia-related microRNAs: Uncovering their diverse and special roles. Am. J. Cancer Res. 2021, 11, 1104–1120. [Google Scholar]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs, and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef] [PubMed]
- Ning, S.; Li, X. Non-coding RNA Resources. Adv. Exp. Med. Biol. 2018, 1094, 1–7. [Google Scholar]
- He, Q.; Liu, Y.; Sun, W. Statistical analysis of non-coding RNA data. Cancer Lett. 2018, 417, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Ouimet, M.; Drouin, S.; Lajoie, M.; Caron, M.; St-Onge, P.; Gioia, R.; Richer, C.; Sinnett, D. A childhood acute lymphoblastic leukemia-specific lncRNA implicated in prednisolone resistance, cell proliferation, and migration. Oncotarget 2017, 8, 7477–7488. [Google Scholar] [CrossRef]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: IncRNA localization and function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef]
- Balatti, V.; Nigita, G.; Veneziano, D.; Drusco, A.; Stein, G.S.; Messier, T.L.; Farina, N.H.; Lian, J.B.; Tomasello, L.; Liu, C.G.; et al. tsRNA signatures in cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 8071–8076. [Google Scholar] [CrossRef]
- Park, J.; Ahn, S.H.; Shin, M.G.; Kim, H.K.; Chang, S. tRNA-Derived Small RNAs: Novel Epigenetic Regulators. Cancers 2020, 12, 2773. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dou, M.; Song, X.; Dong, Y.; Liu, S.; Liu, H.; Tao, J.; Li, W.; Yin, X.; Xu, W. The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 2019, 18, 123. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Munafò, M.; Ciabrelli, F.; Eastwood, E.L.; Fabry, M.H.; Kneuss, E.; Hannon, G.J. piRNA-Guided Genome Defense: From Biogenesis to Silencing. Annu. Rev. Genet. 2019, 52, 131–157. [Google Scholar] [CrossRef] [PubMed]
- Vishnoi, A.; Rani, S. miRNA Biogenesis and Regulation of Diseases: An Overview. Methods Mol. Biol. 2017, 1509, 1–10. [Google Scholar]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Yazarlou, F.; Kadkhoda, S.; Ghafouri-Fard, S. Emerging role of let-7 family in the pathogenesis of hematological malignancies. Biomed. Pharmacother. 2021, 144, 112334. [Google Scholar] [CrossRef]
- Olejniczak, M.; Kotowska-Zimmer, A.; Krzyzosiak, W. Stress-induced changes in miRNA biogenesis and functioning. Cell. Mol. Life Sci. CMLS 2018, 75, 177–191. [Google Scholar] [CrossRef]
- Kotipalli, A.; Gutti, R.; Mitra, C.K. Dynamics of miRNA biogenesis and nuclear transport. J. Integr. Bioinform. 2017, 13, 22–34. [Google Scholar] [CrossRef]
- Yu, T.; Xu, N.; Haque, N.; Gao, C.; Huang, W.; Huang, Z. Popular Computational Tools Used for miRNA Prediction and Their Future Development Prospects. Interdiscip. Sci. Comput. Life Sci. 2020, 12, 395–413. [Google Scholar] [CrossRef]
- Salim, U.; Kumar, A.; Kulshreshtha, R.; Vivekanandan, P. Biogenesis, characterization, and functions of mirtrons. Wiley Interdiscip. Rev. RNA 2022, 13, e1680. [Google Scholar] [CrossRef]
- Titov, I.I.; Vorozheykin, P.S. Comparing miRNA structure of mirtrons and non-mirtrons. BMC Genom. 2018, 19 (Suppl. S3), 114. [Google Scholar] [CrossRef] [PubMed]
- Michlewski, G.; Cáceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2019, 25, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Pong, S.K.; Gullerova, M. Noncanonical functions of microRNA pathway enzymes—Drosha, DGCR8, Dicer and Ago proteins. FEBS Lett. 2018, 592, 2973–2986. [Google Scholar] [CrossRef] [PubMed]
- Chukwurah, E.; Farabaugh, K.T.; Guan, B.J.; Ramakrishnan, P.; Hatzoglou, M. A tale of two proteins: PACT and PKR and their roles in inflammation. FEBS J. 2021, 288, 6365–6391. [Google Scholar] [CrossRef] [PubMed]
- Vergani-Junior, C.A.; Tonon-da-Silva, G.; Inan, M.D.; Mori, M.A. DICER: Structure, function, and regulation. Biophys. Rev. 2021, 13, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Song, M.S.; Rossi, J.J. Molecular Mechanisms of Dicer: Endonuclease and Enzymatic Activity. Biochem. J. 2017, 474, 1603–1618. [Google Scholar] [CrossRef]
- Medley, J.C.; Panzade, G.; Zinovyeva, A.Y. microRNA strand selection: Unwinding the rules. Wiley Interdiscip. Rev. RNA 2021, 12, e1627. [Google Scholar] [CrossRef]
- Sheu-Gruttadauria, J.; MacRae, I.J. Phase Transitions in the Assembly and Function of Human miRISC. Cell 2018, 173, 946–957.e16. [Google Scholar] [CrossRef]
- Nawalpuri, B.; Ravindran, S.; Muddashetty, R.S. The Role of Dynamic miRISC During Neuronal Development. Front. Mol. Biosci. 2020, 7, 8. [Google Scholar] [CrossRef]
- Park, S.; Kang, I.; Shin, C. MicroRNA clustering on the biogenesis of suboptimal microRNAs. Appl. Biol. Chem. 2021, 64, 51. [Google Scholar] [CrossRef]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Carrillo, E.; Berkhout, B. Dicer-independent processing of small RNA duplexes: Mechanistic insights and applications. Nucleic Acids Res. 2017, 45, 10369–10379. [Google Scholar] [CrossRef] [PubMed]
- Kretov, D.A.; Walawalkar, I.A.; Mora-Martin, A.; Shafik, A.M.; Moxon, S.; Cifuentes, D. Ago2-Dependent Processing Allows miR-451 to Evade the Global MicroRNA Turnover Elicited during Erythropoiesis. Mol. cell 2020, 78, 317–328.e6. [Google Scholar] [CrossRef] [PubMed]
- Pu, M.; Chen, J.; Tao, Z.; Miao, L.; Qi, X.; Wang, Y.; Ren, J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell. Mol. Life Sci. CMLS 2019, 76, 441–451. [Google Scholar] [CrossRef]
- Liu, H.; Lei, C.; He, Q.; Pan, Z.; Xiao, D.; Tao, Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol. Cancer 2018, 17, 64. [Google Scholar] [CrossRef]
- Szczepanek, J. Role of microRNA dysregulation in childhood acute leukemias: Diagnostics, monitoring and therapeutics: A comprehensive review. World J. Clin. Oncol. 2020, 11, 348–369. [Google Scholar] [CrossRef]
- Huang, G.L.; Sun, J.; Lu, Y.; Liu, Y.; Cao, H.; Zhang, H.; Calin, G.A. MiR-200 family and cancer: From a meta-analysis view. Mol. Asp. Med. 2019, 70, 57–71. [Google Scholar] [CrossRef]
- Choi, P.W.; Bahrampour, A.; Ng, S.K.; Liu, S.K.; Qiu, W.; Xie, F.; Kuo, W.P.; Kwong, J.; Hales, K.H.; Hales, D.B.; et al. Characterization of miR-200 family members as blood biomarkers for human and laying hen ovarian cancer. Sci. Rep. 2020, 10, 20071. [Google Scholar] [CrossRef]
- Namordizadeh, V. Genistein elicits its anticancer effects through up-regulation of E-Cadherin in Acute Lymphoblastic Leukemia (ALL) cells: An in vitro experimental study. Electron. Physician 2021, 11, 7391–7399. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, J.; Xie, B.; Li, H.; Shen, J.; Chen, J. MicroRNA-200 Family Profile: A Promising Ancillary Tool for Accurate Cancer Diagnosis. Am. J. Ther. 2016, 23, e388–e397. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.N.; Zhang, Q.M.; Ge, Y.Y.; Luo, B.; Xie, X.X. A Review of miR-326 and Female Related Diseases. Acta Histochem. Cytochem. 2021, 54, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.J.; Wan, J.; Wang, C.B. MiR-326: Promising Biomarker for Cancer. Cancer Manag. Res. 2019, 11, 10411–10418. [Google Scholar] [CrossRef] [PubMed]
- Ghodousi, E.S.; Rahgozar, S. MicroRNA-326 and microRNA-200c: Two novel biomarkers for diagnosis and prognosis of pediatric acute lymphoblastic leukemia. J. Cell. Biochem. 2018, 119, 6024–6032. [Google Scholar] [CrossRef]
- Hassan, N.M.; Refaat, L.A.; Ismail, G.N.; Abdellateif, M.; Fadel, S.A.; AbdelAziz, R.S. Diagnostic, prognostic and predictive values of miR-100 and miR-210 in pediatric acute lymphoblastic Leukemia. Hematology 2020, 25, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Swellam, M.; El-Khazragy, N. Clinical impact of circulating microRNAs as blood-based marker in childhood acute lymphoblastic leukemia. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2016, 37, 10571–10576. [Google Scholar] [CrossRef]
- Schotte, D.; Chau, J.C.; Sylvester, G.; Liu, G.; Chen, C.; van der Velden, V.H.; Broekhuis, M.J.; Peters, T.C.; Pieters, R.; den Boer, M.L. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia 2009, 23, 313–322. [Google Scholar] [CrossRef]
- Xue, Y.; Yang, X.; Hu, S.; Kang, M.; Chen, J.; Fang, Y. A genetic variant in miR-100 is a protective factor of childhood acute lymphoblastic leukemia. Cancer Med. 2019, 8, 2553–2560. [Google Scholar] [CrossRef]
- Li, X.J.; Luo, X.Q.; Han, B.W.; Duan, F.T.; Wei, P.P.; Chen, Y.Q. MicroRNA-100/99a, deregulated in acute lymphoblastic leukaemia, suppress proliferation and promote apoptosis by regulating the FKBP51 and IGF1R/mTOR signalling pathways. Br. J. Cancer 2013, 109, 2189–2198. [Google Scholar] [CrossRef]
- De Oliveira, J.C.; Scrideli, C.A.; Brassesco, M.S.; Morales, A.G.; Pezuk, J.A.; Queiroz, R.D.P.; Yunes, J.A.; Brandalise, S.R.; Tone, L.G. Differential miRNA expression in childhood acute lymphoblastic leukemia and association with clinical and biological features. Leuk. Res. 2012, 36, 293–298. [Google Scholar] [CrossRef]
- Zhang, H.; Luo, X.-Q.; Zhang, P.; Huang, L.-B.; Zheng, Y.-S.; Wu, J.; Zhou, H.; Qu, L.-H.; Xu, L.; Chen, Y.-Q. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS ONE 2009, 4, e7826. [Google Scholar] [CrossRef] [PubMed]
- Schotte, D.; De Menezes, R.X.; Moqadam, F.A.; Khankahdani, L.M.; Lange-Turenhout, E.; Chen, C.; Pieters, R.; Boer, M.L.D. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica 2011, 96, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Kyriakidis, I.; Kyriakidis, K.; Tsezou, A. MicroRNAs and the Diagnosis of Childhood Acute Lymphoblastic Leukemia: Systematic Review, Meta-Analysis and Re-Analysis with Novel Small RNA-Seq Tools. Cancers 2022, 14, 3976. [Google Scholar] [CrossRef]
- Shafik, R.E.; El Wahab, N.A.; Mokhtar, M.M.; A El Taweel, M.; Ebeid, E. Expression of microRNA-181a and microRNA-196b in Egyptian Pediatric acute Lymphoblastic Leukemia. Asian Pac. J. Cancer Prev. 2020, 21, 3429–3434. [Google Scholar] [CrossRef]
- Rzepiel, A.; Kutszegi, N.; Gézsi, A.; Sági, J.C.; Egyed, B.; Péter, G.; Butz, H.; Nyírő, G.; Müller, J.; Kovács, G.T.; et al. Circulating microRNAs as minimal residual disease biomarkers in childhood acute lymphoblastic leukemia. J. Transl. 2019, 17, 372. [Google Scholar] [CrossRef]
- Luna-Aguirre, C.M.; Martinez-Fierro, M.D.L.L.; Mar-Aguilar, F.; Garza-Veloz, I.; Treviño-Alvarado, V.; Rojas-Martinez, A.; Jaime-Perez, J.C.; Malagon-Santiago, G.I.; Gutierrez-Aguirre, C.H.; Gonzalez-Llano, O.; et al. Circulating microRNA expression profile in B-cell acute lymphoblastic leukemia. Cancer Biomark. 2015, 15, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Verduci, L.; Azzalin, G.; Gioiosa, S.; Carissimi, C.; Laudadio, I.; Fulci, V.; Macino, G. microRNA-181a enhances cell proliferation in acute lymphoblastic leukemia by targeting EGR1. Leuk. Res. 2015, 39, 479–485. [Google Scholar] [CrossRef]
- Weng, H.; Lal, K.; Yang, F.F.; Chen, J. The pathological role and prognostic impact of miR-181 in acute myeloid leukemia. Cancergenetics 2015, 208, 225–229. [Google Scholar] [CrossRef]
- Indrieri, A.; Carrella, S.; Carotenuto, P.; Banfi, S.; Franco, B. The Pervasive Role of the miR-181 Family in Development, Neurodegeneration, and Cancer. Int. J. Mol. 2020, 21, 2092. [Google Scholar] [CrossRef]
- Chen, C.Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef]
- Szczepański, T.; Harrison, C.J.; van Dongen, J.J. Genetic aberrations in paediatric acute leukaemias and implications for management of patients. Lancet Oncol. 2010, 11, 880–889. [Google Scholar] [CrossRef] [PubMed]
- Hickey, C.; Schwind, S.; Becker, H.; Alachkar, H.; Garzon, R.; Wu, Y.Z.; Liu, S.; Perrotti, D.; Marcucci, G. MicroRNA-181a Targets TEL/AML1 Expression and Impairs Cell Proliferation in t (12; 21) Acute Lymphocytic Leukemia (ALL) Cells. Blood 2009, 114, 766. [Google Scholar] [CrossRef]
- Yang, Y.L.; Yen, C.T.; Pai, C.H.; Chen, H.Y.; Yu, S.L.; Lin, C.Y.; Hu, C.Y.; Jou, S.T.; Lin, D.T.; Lin, S.R.; et al. A double negative loop comprising ETV6/RUNX1 and MIR181A1 contributes to differentiation block in t (12; 21)-positive acute lymphoblastic leukemia. PloS ONE 2015, 10, e0142863. [Google Scholar] [CrossRef]
- Nabhan, M.; Louka, M.L.; Khairy, E.; Tash, F.; Ali-Labib, R.; El-Habashy, S. MicroRNA-181a and its target Smad 7 as potential biomarkers for tracking child acute lymphoblastic leukemia. Gene 2017, 628, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Bedewy, A.; Elmaghraby, S.M.; Shehata, A.A.; Kandil, N.S. Prognostic Value of miRNA-155 Expression in B-Cell Non-Hodgkin Lymphoma. Turk. J. Hematol. 2017, 34, 207–212. [Google Scholar]
- Mashima, R. Physiological roles of miR-155. Immunology 2015, 145, 323–333. [Google Scholar] [CrossRef]
- Zhao, Y.; Jaber, V.; Percy, M.E.; Lukiw, W.J. A microRNA cluster (let-7c, miRNA-99a, miRNA-125b, miRNA-155 and miRNA-802) encoded at chr21q21.1-chr21q21.3 and the phenotypic diversity of Down’s syndrome (DS; trisomy 21). J. Nat. Sci. 2017, 3, e446. [Google Scholar]
- Zhang, X.; Wang, Y.; Guo, Q.; Diao, Y.; Liu, H.; Song, G.; Wang, W.; Zhang, Z.; Yin, H.; Li, L. Prognostic role of microRNA-155 in patients with leukemia: A meta-analysis. Clin. Chim. Acta 2018, 483, 6–13. [Google Scholar] [CrossRef]
- Bansal, S.; Itabashi, Y.; Perincheri, S.; Poulson, C.; Bharat, A.; Smith, M.A.; Bremner, R.M.; Mohanakumar, T. The role of miRNA-155 in the immunopathogenesis of obliterative airway disease in mice induced by circulating exosomes from human lung transplant recipients with chronic lung allograft dysfunction. Cell. Immunol. 2020, 355, 104172. [Google Scholar] [CrossRef]
- Liang, C.; Li, Y.; Wang, L.N.; Zhang, X.L.; Luo, J.S.; Peng, C.J.; Tang, W.Y.; Huang, L.B.; Tang, Y.L.; Luo, X.Q. Up-regulated miR-155 is associated with poor prognosis in childhood acute lymphoblastic leukemia and promotes cell proliferation targeting ZNF238. Hematology 2021, 26, 16–25. [Google Scholar] [CrossRef]
- Pedicone, C.; Meyer, S.T.; Chisholm, J.D.; Kerr, W.G. Targeting SHIP1 and SHIP2 in Cancer. Cancers 2021, 13, 890. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Wang, C. miR-155 Regulates the Proliferation of Glioma Cells Through PI3K/AKT Signaling. Front. Neurol. 2020, 11, 297. [Google Scholar] [CrossRef] [PubMed]
- Bogoch, Y.; Friedlander-Malik, G.; Lupu, L.; Bondar, E.; Zohar, N.; Langier, S.; Ram, Z.; Nachmany, I.; Klausner, J.M.; Pencovich, N. Augmented expression of RUNX1 deregulates the global gene expression of U87 glioblastoma multiforme cells and inhibits tumor growth in mice. Tumour Biol. 2017, 39, 1010428317698357. [Google Scholar] [CrossRef] [PubMed]
- Akpinar, F.; Polat, A.; Balci, Y.I.; Akça, H.; Şenol, H.; Tokgun, O. Microrna Expression Profiles and Changes with Treatment on Childhood Leukemias. 2020. Available online: http://hdl.handle.net/11499/37538 (accessed on 5 January 2023).
- El-Khazragy, N.; Noshi, M.A.; Abdel-Malak, C.; Zahran, R.F.; Swellam, M. miRNA-155 and miRNA-181a as prognostic biomarkers for pediatric acute lymphoblastic leukemia. J. Cell. 2019, 120, 6315–6321. [Google Scholar] [CrossRef]
- Duyu, M.; Durmaz, B.; Gunduz, C.; Vergin, C.; YilmazKarapinar, D.; Aksoylar, S.; Kavakli, K.; Cetingul, N.; Irken, G.; Yaman, Y.; et al. Prospective evaluation of whole genome microRNA expression profiling in childhood acute lymphoblastic leukemia. BioMed Res. Int. 2014, 2014, 967585. [Google Scholar] [CrossRef]
- Hassan, S.S.; El-Khazragy, N.; Elshimy, A.A.; Aboelhussein, M.M.; Saleh, S.A.; Fadel, S.; Atia, H.A.; Matbouly, S.; Tamer, N. In vitro knock-out of miR-155 suppresses leukemic and HCV virus loads in pediatric HCV-4-associated acute lymphoid leukemia: A promising target therapy. J. Cell. 2020, 121, 2811–2817. [Google Scholar] [CrossRef]
- Vendramini, E.; Giordan, M.; Giarin, E.; Michielotto, B.; Fazio, G.; Cazzaniga, G.; Biondi, A.; Silvestri, D.; Valsecchi, M.G.; Muckenthaler, M.U.; et al. High expression of miR-125b-2 and SNORD116 noncoding RNA clusters characterize ERG-related B cell precursor acute lymphoblastic leukemia. Oncotarget 2017, 8, 42398–42413. [Google Scholar] [CrossRef]
- Peng, B.; Theng, P.Y.; Le, M.T.N. Essential functions of miR-125b in cancer. Cell Prolif. 2021, 54, e12913. [Google Scholar] [CrossRef]
- Sonoki, T.; Iwanaga, E.; Mitsuya, H.; Asou, N. Insertion of microRNA-125b-1, a human homologue of lin-4, into a rearranged immunoglobulin heavy chain gene locus in a patient with precursor B-cell acute lymphoblastic leukemia. Leukemia 2005, 19, 2009–2010. [Google Scholar] [CrossRef]
- Li, G.; So, A.Y.; Sookram, R.; Wong, S.; Wang, J.K.; Ouyang, Y.; He, P.; Su, Y.; Casellas, R.; Baltimore, D. Epigenetic silencing of miR-125b is required for normal B-cell development. Blood 2018, 131, 1920–1930. [Google Scholar] [CrossRef]
- Chapiro, E.; Russell, L.J.; Struski, S.; Cavé, H.; Radford-Weiss, I.; Valle, V.D.; Lachenaud, J.; Brousset, P.; Bernard, O.A.; Harrison, C.J.; et al. A new recurrent translocation t(11;14)(q24;q32) involving IGH@ and miR-125b-1 in B-cell progenitor acute lymphoblastic leukemia. Leukemia 2010, 24, 1362–1364. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, M.; Harris, M.H.; Zhou, B.; Lodish, H.F. MicroRNA miR-125b causes leukemia. Proc. Natl. Acad. Sci. USA 2010, 107, 21558–21563. [Google Scholar] [CrossRef] [PubMed]
- Puissegur, M.P.; Eichner, R.; Quelen, C.; Coyaud, E.; Mari, B.; Lebrigand, K.; Broccardo, C.; Nguyen-Khac, F.; Bousquet, M.; Brousset, P. B-cell regulator of immunoglobulin heavy-chain transcription (Bright)/ARID3a is a direct target of the oncomir microRNA-125b in progenitor B-cells. Leukemia 2012, 26, 2224–2232. [Google Scholar] [CrossRef] [PubMed]
- Gefen, N.; Binder, V.; Zaliova, M.; Linka, Y.; Morrow, M.; Novosel, A.; Edry, L.; Hertzberg, L.; Shomron, N.; Williams, O.; et al. Hsa-mir-125b-2 is highly expressed in childhood ETV6/RUNX1 (TEL/AML1) leukemias and confers survival advantage to growth inhibitory signals independent of p53. Leukemia 2010, 24, 89–96. [Google Scholar] [CrossRef]
- Sanddhya, N.S.; Sachdanandam, P.; Thilagavathy, S.; Shanthi, P. Role of miR-125b and miR-203 expressions in the pathogenesis of BCR-ABL+ acute lymphoblastic leukemia (ALL). Gene Rep. 2016, 4, 253–257. [Google Scholar] [CrossRef]
- Piatopoulou, D.; Avgeris, M.; Marmarinos, A.; Xagorari, M.; Baka, M.; Doganis, D.; Kossiva, L.; Scorilas, A.; Gourgiotis, D. miR-125b predicts childhood acute lymphoblastic leukaemia poor response to BFM chemotherapy treatment. Br. J. Cancer 2017, 117, 801–812. [Google Scholar] [CrossRef]
- Shahid, S.; Shahid, W.; Shaheen, J.; Akhtar, M.W.; Sadaf, S. Circulating miR-146a expression as a non-invasive predictive biomarker for acute lymphoblastic leukemia. Sci. Rep. 2021, 11, 22783. [Google Scholar] [CrossRef]
- MICRO RNA 146A; MIR146A. Available online: https://omim.org/entry/610566 (accessed on 6 December 2022).
- Cameron, J.E.; Yin, Q.; Fewell, C.; Lacey, M.; McBride, J.; Wang, X.; Lin, Z.; Schaefer, B.C.; Flemington, E.K. Epstein-Barr virus latent membrane protein 1 induces cellular MicroRNA miR-146a, a modulator of lymphocyte signaling pathways. J. Virol. 2008, 82, 1946–1958. [Google Scholar] [CrossRef]
- Monk, C.E.; Hutvagner, G.; Arthur, J.S. Regulation of miRNA transcription in macrophages in response to Candida albicans. PloS ONE 2010, 5, e13669. [Google Scholar] [CrossRef]
- Jurkin, J.; Schichl, Y.M.; Koeffel, R.; Bauer, T.; Richter, S.; Konradi, S.; Gesslbauer, B.; Strobl, H. miR-146a is differentially expressed by myeloid dendritic cell subsets and desensitizes cells to TLR2-dependent activation. J. Immunol. 2010, 184, 4955–4965. [Google Scholar] [CrossRef]
- Etzrodt, M.; Cortez-Retamozo, V.; Newton, A.; Zhao, J.; Ng, A.; Wildgruber, M.; Romero, P.; Wurdinger, T.; Xavier, R.; Geissmann, F.; et al. Regulation of monocyte functional heterogeneity by miR-146a and Relb. Cell Rep. 2012, 1, 317–324. [Google Scholar] [CrossRef] [PubMed]
- So, A.Y.; Zhao, J.L.; Baltimore, D. The Yin and Yang of microRNAs: Leukemia and immunity. Immunol. Rev. 2013, 253, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, H.; Lei, D. microRNA-146a Promotes Growth of Acute Leukemia Cells by Downregulating Ciliary Neurotrophic Factor Receptor and Activating JAK2/STAT3 Signaling. Yonsei Med. J. 2019, 60, 924–934. [Google Scholar] [CrossRef]
- King, J.K.; Ung, N.M.; Paing, M.H.; Contreras, J.R.; Alberti, M.O.; Fernando, T.R.; Zhang, K.; Pellegrini, M.; Rao, D.S. Regulation of marginal zone B-cell differentiation by microRNA-146a. Front. Immunol. 2017, 7, 670. [Google Scholar] [CrossRef] [PubMed]
- Akbari Moqadam, F.; Lange-Turenhout, E.A.; van der Veer, A.; Marchante, J.R.; Boer, J.M.; Pieters, R.; den Boer, M. MicroRNA signature in BCR-ABL1-like and BCR-ABL1-positive childhood acute lymphoblastic leukemia: Similarities and dissimilarities. Leuk. Lymphoma 2014, 55, 1942–1945. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.; He, C.; Wang, D.; Yuan, X.; Chen, J.; Jin, J. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol. Dis. 2010, 44, 191–197. [Google Scholar] [CrossRef]
- Durmaz, B.; Bagca, B.G.; Cogulu, O.; Susluer, S.Y.; Alpay, A.; Aksoylar, S.; Gunduz, C. Antileukemic Effects of Anti-miR-146a, Anti-miR-155, Anti-miR-181a, and Prednisolone on Childhood Acute Lymphoblastic Leukemia. BioMed Res. Int. 2021, 2021, 3207328. [Google Scholar] [CrossRef]
- Kalfert, D.; Pesta, M.; Kulda, Y.; Topolcan, O.; Ryska, A.; Celakovsky, P.; Laco, J.; Ludvikova, M. MicroRNA Profile in Site-specific Head and Neck Squamous Cell Cancer. Anticancer Res. 2015, 35, 2455–2463. [Google Scholar]
- Lundgren, K.; Tobin, N.P.; Lehn, S.; Stål, O.; Rydén, L.; Jirström, K.; Landberg, G. Stromal expression of β-arrestin-1 predicts clinical outcome and tamoxifen response in breast cancer. J. Mol. Diagn. 2011, 13, 340–351. [Google Scholar] [CrossRef]
- Wang, W.; Liu, Y.; Guo, J.; He, H.; Mi, X.; Chen, C.; Xie, J.; Wang, S.; Wu, P.; Cao, F.; et al. miR-100 maintains phenotype of tumor-associated macrophages by targeting mTOR to promote tumor metastasis via Stat5a/IL-1ra pathway in mouse breast cancer. Oncogenesis 2018, 7, 97. [Google Scholar] [CrossRef]
- Tang, L.; Peng, Y.-Z.; Li, C.-G.; Jiang, H.-W.; Mei, H.; Hu, Y. Prognostic and Clinicopathological Significance of MiR-155 in Hematologic Malignancies: A Systematic Review and Meta-analysis. J. Cancer 2019, 10, 654–664. [Google Scholar] [CrossRef] [PubMed]
| Locus on Chromosome | microRNA | Expression in BCP-ALL | Target of miRNA | Function mRNA of miRNA/Target | Ref. |
|---|---|---|---|---|---|
| 12p13 | miRNA-200c | downregulation | ABCA2, ABCA3 EMT/MET | inhibition of tumor metastases, involved in leukemogenesis (?) multidrug resistance | [3,38,42,44,100] |
| 11q1 | miRNA-326 | downregulation | ABCA2, ABCA3 | inhibition of invasion and metastasis, multidrug resistance chemoresistance | [3,43,44,101] |
| 1q32.1 * 9q33.3 ** | miRNA-181a | downregulation | Smad7 ↑ ETV6::RUNX1 | increase in proliferation and decrease in apoptosis | [59,60] |
| upregulation | TLR4, TLR8 IRF8, IL6R | inhibition of innate immunity and inflammation | [3] | ||
| 11q24.1 | miRNA-100 | downregulation | FKBP51 ↑ the antiapoptotic gene MCL1 ↑ | regulation of cell proliferation and dexamethasone-induced apoptosis | [45,46,47,102] |
| upregulation | IGF1R/mTOR (MCL1 ↓) | cell proliferation, differentiation, and survival stimulation; rapamycin-mediated enhancement of dexamethasone-induced apoptosis | |||
| 21q21.3 | miRNA-155 | upregulation | SHIP1 Mxd1/Mad1 (BCL6↓) HDAC4 SMAD5 | promotes B cell proliferation, carcinogen, inhibits the proliferation of human hematopoietic progenitor cells by signaling TGF-β pathway, chemoresistance | [3,66,67,103] |
| 21q21.1 | miRNA-125b | upregulation | PPP1CA, BTG2, PTEN ↓ P53, Bak1, Bmf transcript ↓ | increase in proliferation and decrease in apoptosis, oncogene | [3] |
| 5q33.3 | miRNA-146a | upregulation | N-RAS, RAS, AMPK-alpha, PBX2, ErbB4, TRAF6, LIN28, NUMB, CNTFR | increased proliferation, influenced maturation of B lymphocytes, inhibited apoptosis | [85,88,89] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ziętara, K.J.; Lejman, J.; Wojciechowska, K.; Lejman, M. The Importance of Selected Dysregulated microRNAs in Diagnosis and Prognosis of Childhood B-Cell Precursor Acute Lymphoblastic Leukemia. Cancers 2023, 15, 428. https://doi.org/10.3390/cancers15020428
Ziętara KJ, Lejman J, Wojciechowska K, Lejman M. The Importance of Selected Dysregulated microRNAs in Diagnosis and Prognosis of Childhood B-Cell Precursor Acute Lymphoblastic Leukemia. Cancers. 2023; 15(2):428. https://doi.org/10.3390/cancers15020428
Chicago/Turabian StyleZiętara, Karolina Joanna, Jan Lejman, Katarzyna Wojciechowska, and Monika Lejman. 2023. "The Importance of Selected Dysregulated microRNAs in Diagnosis and Prognosis of Childhood B-Cell Precursor Acute Lymphoblastic Leukemia" Cancers 15, no. 2: 428. https://doi.org/10.3390/cancers15020428
APA StyleZiętara, K. J., Lejman, J., Wojciechowska, K., & Lejman, M. (2023). The Importance of Selected Dysregulated microRNAs in Diagnosis and Prognosis of Childhood B-Cell Precursor Acute Lymphoblastic Leukemia. Cancers, 15(2), 428. https://doi.org/10.3390/cancers15020428






