Simple Summary
Aquaporins (AQPs) are transmembrane channel proteins that primarily transport water across the cellular membranes. AQPs have been found to be overexpressed in various human cancers, including prostate cancer. Clinical data suggest ideal prospects for AQPs as biomarkers. This review article mainly focuses on the opportunities for the development of AQPs as prognostic markers in prostate cancer.
Abstract
Prostate cancer is a complex heterogeneous disease that affects millions of males worldwide. Despite rapid advances in molecular biology and innovation in technology, few biomarkers have been forthcoming in prostate cancer. The currently available biomarkers for the prognosis of prostate cancer are inadequate and face challenges, thus having limited clinical utility. To date, there are a number of prognostic and predictive biomarkers identified for prostate cancer but lack specificity and sensitivity to guide clinical decision making. There is still tremendous scope for specific biomarkers to understand the natural history and complex biology of this heterogeneous disease, and to identify early treatment responses. Accumulative studies indicate that aquaporins (AQPs) a family of membrane water channels may serve as a prognostic biomarker for prostate cancer in monitoring disease advancement. In the present review, we discuss the existing prostate cancer biomarkers, their limitations, and aquaporins as a prospective biomarker of prognostic significance in prostate cancer.
1. Introduction
Prostate cancer (PC) is a biologically heterogeneous disease and the most common cancer among males. According to the American Cancer Society, in the year 2022, approximately 268,490 new cases and 34,500 deaths occurred in the United States alone [1]. Inflammation, genetic modifications, and increased cellular proliferation are major critical factors for the initiation of prostate cancer [2]. Prostate cancer in humans exhibits a unique spectrum of features: multifocality, heterogeneity, variable clinical progression, propensity to metastasize to bone, and the emergence of androgen-independent disease forms [2]. Long-term clinical outcomes for men can vary greatly, even when they are diagnosed with organ-confined disease [3]. There is substantial variability among patients and within tumors in terms of histologic and molecular characteristics [4]. Progress in the treatment of prostate cancer has also been hindered by the fact that histologically identical cancers in different patients may exhibit widely variant biologic behavior [4]. These challenges pose major implications in the clinical management of prostate cancer.
The current prognosis of prostate cancer is highly variable and depends on the degree of cancer and its stage at the time of diagnosis. Acceptance of screening based upon the measurement of prostate-specific antigen (PSA) has led to earlier detection of prostate cancer however lack of specificity to detect a significant number of PSA-negative tumors limits its detection efficacy [5]. In fact, PSA has been shown to test positive for common confounders such as benign prostatic hyperplasia [6]. As a result, over 90% of the low-risk Gleason score (GS) (6 or less) prostate cancer patients receive aggressive treatment to avoid potential cancer-related deaths [7]. This has led to concerns regarding overdiagnosis and the overtreatment of prostate cancer [8]. Therefore, there is an urgent need to identify novel prognostic biomarkers to determine those patients that are at higher risk for progression and might benefit from more aggressive treatment, and patients that might be spared from unnecessary and potentially harmful interventions.
Recent advancements in genomic and proteomic techniques combined with progress in bioinformatics demonstrate great promise in the identification of several new biomarkers of diagnostic and prognostic value in prostate cancer [9]. In the present review, we discuss the existing prognostic biomarkers of prostate cancer, their limitations, and aquaporins as a prospective biomarker of prognostic significance in prostate cancer.
2. Prognostic Biomarkers and Their Limitations in Prostate Cancer
Prognostic biomarkers aim to evaluate objectively patient’s overall outcome and are essential to provide important clinical decisions for prostate cancer patients. Besides predicting clinical progression, it is usually considered that such prognostic biomarkers also provide valuable information about disease mechanisms and the underlying molecular processes. Although, prognostic biomarkers, even after a long history of research, are still not recommended for use in clinical settings although quite a few of them are considered potential candidates. However, with no data currently available about their clinical relevance in the long term, also prospective validation is lacking. A list of commercially available prognostic biomarkers and their limitations are listed in Table 1.

Table 1.
Commercially available prognostic biomarkers in prostate cancer.
There are a number of prognostic tests available for commercial gene expression signatures. These subsets of biomarkers assist clinicians in discriminating against aggressive vs. indolent prostate tumors. The ConfirmMDx, Prostate Core Mitomic Test (PCMT), phosphatase and tensin homolog (PTEN) gene, TMPRSS2-ERG gene fusion, and ProMark are few commercially available tests [24]. Epigenetic-based ConfirmMDx detects epigenetic field effect associated with cancerization in the DNA [25]. It is useful for identifying patients with true negative biopsy results from those with occult cancers. Prostate Core Mitomic Test detects malignant cells in normal-appearing prostate tissue across a wide area by identifying a large-scale depletion in mitochondrial DNA associated with undiagnosed prostate cancer [26]. Prostate cancers with the TMPRSS2-ERG gene fusion account for 40 to 80% of total cases [27]. Clinically significant prostate cancer is associated with high urine TMPRSS2-ERG levels based on Epstein criteria that stratify disease aggressiveness using PSA density and biopsy characteristics including the percentage of normal and tumor prostate tissue detected, Gleason score, and the number of tumor cores [27]. A defective tumor suppressor PTEN gene, involved in the regulation of the cell cycle, steadily correlates with a poor prognosis in prostate cancer [28]. Accumulative evidence showed a positive association between deletion of the PTEN gene and progression risk, higher Gleason grades, and recurrence after therapy [28]. Aside from this, it has been linked to advanced localized or metastatic disease as well as death [28]. ProMark is another biopsy-based prostate cancer test that quantifies biomarker expression and classifies patients’ tumors based on analysis of immunofluorescent imaging [29]. On the basis of formalin-fixed, paraffin-embedded tissue data from a clinical validation study, ProMark is capable of differentiating indolent from aggressive disease [29].
Several genetic tests of biopsy specimens are now commercially available for risk stratification of patients with prostate cancer such as Prolaris, Oncotype DX and Decipher [30]. The Prolaris test evaluates the expression of 31 cell-cycle–related genes and 15 housekeeping genes [31]. The results are represented as a cell-cycle progression (CCP) score. Combining standard clinicopathologic parameters with CCP score provides predictive genomic data regarding prostate cancer-specific progression and disease-specific mortality. This assay has been validated in multiple cohorts. Based on validation studies, Prolaris can identify patients with low risk who can be treated conservatively and those with a high-risk disease that may take advantage from earlier definitive treatment [32]. The Oncotype Dx test evaluates the expression of 12 cancer-related genes and five housekeeping genes [33]. The results are represented as a Genomic Prostate Score (GPS). In men harboring very low, low, and low-intermediate risk prostate cancer, in a prospective study, Oncotype DX has been demonstrated to predict adverse pathology based on biopsy [33]. Beyond clinical and pathological measures, the GPS offers independent predictive information. Using biopsy tumor volumes from very small biopsy specimens, GPS assesses underlying biology to determine disease aggressiveness with greater accuracy, considering tumor heterogeneity and undersampling [34]. The Decipher test evaluates 22 cancer-related genes. The results are represented as a Genomic Classifier score intended to help predict the risk of metastasis after radical prostatectomy [35]. A high-risk surgical cohort has shown that this assay is independently prognostic of prostate cancer death. These genomic-based tests demonstrate rigorous quality criteria including reproducibility, linearity, analytical accuracy, precision, and are reliable prognostic tools for the prediction of biochemical recurrence or prostate cancer-specific survival albeit their systematic use in prostate cancer is currently not recommended due to insufficient evidence.
3. Aquaporin Family and Their Function in Normal and Cancer Pathophysiology
Aquaporin families are small-size (24–30 kDa) pore-forming integral membrane proteins. The Aquaporin gene encodes six bilayer spanning domains integral membrane protein that forms water channels and permits osmotic gradient-mediated (passive) transport (Figure 1).
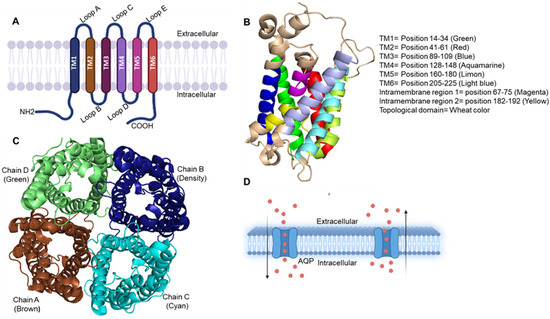
Figure 1.
(A) Representation of three-dimensional structure of aquaporin and its topology. The illustration shows AQP protein structure, a membrane-bound protein composed of six transmembrane helices and has both the carboxyl and amino terminus regions positioned in the cytoplasm. All six transmembrane helices show connectivity with five loops named as A–E. (B) Monomer structure (side view) of AQP visualized by PyMOL software. It shows six transmembrane, two intramembrane and all topological domains present in the AQP with defined position and color. (C) Tetramer structure (top view and active form) of AQP, which is made up of four AQP monomer chains A–D (represented with different color) which in altogether forms pore in the middle for solute transfer passively. PyMOL software was used to visualize the figure. (D) Passive transport of water molecules through aquaporins across the membrane.
To date, thirteen isoforms (AQP0–AQP12) of the AQP family have been recognized in humans and categorized into three subfamilies [36]. The first subfamily of aquaporin is water-selective pore-forming proteins, which are known as classical aquaporins including AQP0, AQP1, AQP2, AQP4, AQP5, AQP6 and AQP8 [37]. Numerous studies have been conducted on this subfamily of AQPs, which have been useful for better understanding the role they might play in physiological and pathophysiological conditions [38]. Recent studies revealed that AQP6 and AQP8 are unorthodox aquaporins since AQP6 is highly water permeable and AQP8 has unique phylogenetics setting it apart from other aquaporins [39]. In mice lacking AQP1, urine concentration is greatly impaired because it cannot concentrate in the descending vasa recta, the descending limb of the loop of Henle, and the epithelium of the renal proximal tubule [40]. AQP4 is the primary water channel expressed in astrocytes throughout the central nervous system [40,41,42]. This protein is known to participate in water transport between the brain and spinal cord, in neuroexcitation and in astrocyte migration following injury [43]. Studies reported the abundance of AQP2 and AQP4 expression mainly in kidney-collecting duct epithelial cells [44]. The second subfamily of aquaporins is denoted by aquaglyceroporins that are permeable to small uncharged molecules, water, and other molecules such as urea, ammonia, and glycerol [45]. Aquaglyceroporins play an important role in metalloid homeostasis and facilitate arsenite and antimonite diffusion [46]. A comparison of amino acid sequence alignments can distinguish the aquaporins from aquaglyceroporins (AQP3, AQP7, AQP9, and AQP10) [47]. First cloned mammalian aquaglyceroporin was AQP3 which was able to facilitate water and glycerol transportation [48], however in xenopus oocytes, AQP7, AQP9, and AQP10 transport water, glycerol, and urea [49]. Oocytes also allow the flow of a wide range of solutes through AQP9 [50]. Glycerol and urea are transported by aquaglyceroporins, which are still poorly understood. Unlike the first two subfamilies, the third aquaporin subfamily is characterized by low conservation of amino acid sequences around the asparagine-proline-alanine (NPA) boxes [51]. In mammals, there are only two super aquaporins, named AQP11 and AQP12 [51]. With a homology of less than 20%, these two AQPs appear to belong to a supergene family of AQPs, as their NPA boxes are highly different from those of other classical AQPs [52]. At present, little is known about the structure and function of AQP11 and AQP12.
Upregulation of AQPs has been demonstrated in many tumor types, such as breast, prostate, lungs, brain, liver, cervical, ovarian, skin, renal, stomach, esophageal, thyroid, and colorectal cancer [53]. There is evidence to suggest that AQPs play a crucial role in cancer metastasis and progression. Silencing of AQP1 in mice has been shown to reduce tumor growth and angiogenesis [54]. Studies reported that increased angiogenesis induced by AQP1 through endothelial cell stimulation is via estrogen receptors [55]. Additionally, AQP3 may facilitate glycerol transport into the mammary gland, fueling growth demands by increasing intracellular ATP [56]. Furthermore, AQP4 knockdown inhibits cell invasion in human glioma cells, while AQP8 overexpression promotes cervical cancer cell invasion [57,58]. Researchers have also demonstrated that co-expression of AQP3 and AQP5 is associated with aggressive tumor progression as well as poor outcomes in esophageal squamous cell carcinoma [59]. Moreover, it has also been shown that silencing AQP3 improves the effectiveness of cryotherapy for prostate cancer [60]. AQP5 and AQP9 are associated with drug resistance in colorectal chemotherapy [61]. Several studies reported the prognostic potential of AQPs in breast cancer [62], renal cell carcinoma [63], ovarian cancer [64], and lung adenocarcinoma [65]. In this sequence, AQPs could be proposed as prognostic biomarker(s) for prostate cancer.
4. Aquaporin Expression in Prostate Cancer
Aquaporin 1 (AQP1) regulates the permeability of epithelial and endothelial barriers by assisting water movement across cell membranes. Different levels of AQP1 expression has been shown to correlate with tumor stage in cancer patients [66]. A study reported that AQP1 facilitates interstitial fluid pressure and high vascular permeability in the carcinoma of the colon, brain, pancreas, and breast [67]. In addition, AQP1 is involved in the effusion or edema fluid development that stimulates tumor angiogenesis [67]. A study was conducted for comparative abundance and distribution analysis of AQP1 in tumor and normal tissue of the prostate. The outcome of the study demonstrated that AQP1 was expressed in capillary endothelia of all normal tissues and slightly higher in microvascular structures. This suggests that overexpression of AQP1 may be a consequence of angiogenesis and perform a significant role in tumor edema formation or clearance [67]. Another study examined specimens from benign prostate hyperplasia and prostate cancer and demonstrated that AQP1 is majorly expressed in venules and capillaries of the prostate [68]. A clinical trial (NCT00851994) was conducted to evaluate the specificity and sensitivity of AQP1 concentration to diagnose the clear cell or papillary renal cell carcinoma (RCC) by comparing urine AQP1 concentrations in RCC, bladder cancer, non-cancer renal masses, and prostate cancer patients [69]. The results demonstrate that AQP1 could be a suitable biomarker having excellent specificity and sensitivity in the urine sample of RCC patients. Another study reported that cell density-induced pericellular hypoxia and cobalt (II) chloride (CoCl(2))-induced hypoxia phosphorylates p38 mitogen-activated protein kinase (MAPK) which enhance AQP1 expression [66]. Furthermore, protein kinase C (PKC) and intracellular calcium ion (Ca2+) activates p38 MAPK pathway enhancing AQP1 expression. Induction of AQP1 expression is also dependent on the lower oxygen (O2) levels [66]. One of the study highlighted that certain secretory proteins indirectly induce AQP1 expression [70]. Other studies reported that malnutrition increases the prevalence of non-communicable chronic diseases such as cancer [71,72]. In another study, researchers found increased expression of AQP1 and oxidative stress levels in malnourished rat model together contribute to prostate carcinogenesis in offspring [73]. A study assessed the AQP1 expression and their clinico-pathological significance in prostate adenocarcinoma. Tissue microarray analysis of paired malignant and benign prostatic tissues revealed higher expression of AQP1 in 17.2% specimens with both low and high Gleason scores. Positive association of AQP1 overexpression and higher Gleason score was associated with higher pathologic stages, and biochemical recurrence [74]. A transcriptomic analysis revealed that increased transcript levels of AQP1 was significantly associated with poor survival of prostate cancer patients [58].
Aquaporin 3 (AQP3) channel proteins transport water, nonionic small solutes such as urea and glycerol and other small solutes across the cell membrane [75]. Aberrant AQP3 expression has been reported in colon cancer [76], lung cancer [77,78], and esophageal and oral squamous cell carcinoma [79]. A cDNA microarray-based study demonstrates that prostate cancer cells exhibit overexpression of AQP3 protein [80]. Chen et al. (2015) showed that AQP3 silencing by small interfering RNA (siRNA) inhibited motility and invasiveness in prostate cancer cells through a reduction in extracellular signal-regulated kinase (ERK) ½ activation [80]. The study also showed that AQP3 upregulates the matrix metalloproteinase 3 expression and its secretion in prostate cancer through activation of the ERK signal pathway [80]. Subcellular localization of AQP3 in normal human prostate cells is restricted to the cell membrane whereas in prostate cancer, AQP3 is frequently located in the cytoplasm [81]. Furthermore, localization of AQP3 was limited to the basolateral cell membranes in the normal epithelia of the prostate. However, in cancer, AQP3 expression was not observed on the cell membranes. Nejsum and Nelson, (2007) confirmed that AQP3 co-localizes with E-cadherin during the early stages of cell–cell contact formation [82]. AQP3-E-cadherin co-localization depends upon the level of E-cadherin and increased E-cadherin levels simultaneously increase AQP3 expression in the plasma membranes of prostate epithelial cells [83]. Another study reported that the knockdown of RAS such as proto-oncogene A (RalA) facilitates increased AQP3 expression onto the plasma membrane [83]. The absence of RalA suppressed cell motility and invasion in prostate cancer cells. This mechanistic study revealed that redistribution of AQP3 in prostate cancer occurs through RalA/PKA/cAMP signaling pathways [83]. The role of hypoxia in AQP3 expression and its cellular localization reveals that both chronic and acute hypoxia plays a crucial role in the adaptation of AQP3 in the plasma membrane [84]. Khan et al. (2021) demonstrated that regulation of the AQP3 gene occurs by estrogen response elements (ERE) in prostate cancer [85].
Aquaporin 5 (AQP5) is an androgen-regulated member of a family of small hydrophobic integral transmembrane water channel proteins regulating cellular water homeostasis and growth signaling. AQP5 is overexpressed in colon cancer [86,87], lung cancer [88], cervical cancer [89], leukemia [90], esophageal cancer [68], ovarian cancer [91], and hepatic cancer [92]. Research revealed that AQP5 expression is associated with PTEN deletion and ERG positivity. AQP5 positivity was observed in ERG-positive (15.5%), ERG-negative (5.8%), with PTEN deletion (14.7%) and without PTEN deletion (9.4%) prostate cancers. Notably, both AQP5 positivity and AQP5 negativity were associated with disease aggressiveness [93]. The clinical significance of AQP5 and its correlation with key genomic alterations in prostate cancer was investigated by Pust et al. (2016) on a tissue microarray containing 12,427 prostate tumors [93]. The study revealed lower expression of AQP5 in normal prostate epithelium, whereas in prostate cancer, its expression showed a dichotomous pattern. Immunostaining showed 25.0% negative, 32.5% weak, 32.5% moderate, and 10.0% strong AQP5 staining in 10,239 interpretable tumors. Furthermore, another research group attempted to propose AQP5 as a prognostic biomarker by evaluating AQP5 expression in 60 prostate cancer specimens and prostate cancer cell lines. The result showed that 31.7% (19) patients exhibited high levels of AQP5 expression, 50.0% (30) showed intermediate, and 18.3% (11) showed absence of AQP5. Increased AQP5 expression frequently accompanies gene amplification associated with TNM stage and lymph node metastasis. However, the association between tumor size and age with AQP5 expression was not noteworthy. A positive correlation between circulating tumor cells and negative cumulative survival rate was observed with AQP5 expression in prostate cancer patients [94]. On the contrary, the transcriptomic analysis revealed that increased AQP5 mRNA levels were highly associated with poor survival [58]. A study performed by Park and Yoon (2017) showed that AQP5 expression negatively correlate with neoplastic and non-neoplastic tissue and established no correlation with clinicopathological parameters [74].
Aquaporin 9 (AQP9) overexpression has been linked with carcinoma of the kidneys, liver, lungs, colorectum, brain, ovarian, prostate, and liver [95]. AQP9 is expressed in the cytoplasm of prostate epithelial cells both in the benign and malignant stages [48]. Transcriptomic analysis revealed that elevated AQP9 mRNA levels were positively associated with poor survival [58]. AQP9 gene regulation occurs by estrogen response elements (ERE) in prostate cancer [85]. A research group evaluated and confirmed the androgen-dependent upregulation of AQP9 in prostate cancer [96]. A study explored the androgen-independent expression of AQP9 in prostate cancer PC3 cells, and in prostate cancer samples and adjacent cancer tissues [97]. AQP9 gene silencing in PC3 cells inhibits proliferation. Furthermore, the absence of AQP9 decreases anti-apoptotic protein Bcl-2 expression and increases apoptotic protein expression (cleaved caspase 3 and Bax) suggesting that AQP9 regulates apoptosis in prostate cancer. AQP9 expression also affects the motility and invasiveness of prostate cancer cells. In-depth analysis revealed that the absence of AQP9 led to reduced ERK1/2 phosphorylation which suggests that activation of the ERK pathway requires AQP9 channel proteins [97].
Several others AQPs such as AQP4, AQP7, AQP8, AQP10 and AQP11 are also overexpressed in prostate cancer cells and in both benign and malignant human prostate tissue at the transcript level. Immunofluorescence microscopy confirmed AQP4 and AQP7 protein expression in human prostate tissue [98]. A list of AQPs that have been expressed in prostate cancer is shown in Table 2.

Table 2.
Studies conducted on AQPs in prostate cancer.
5. Do Aquaporins Serve as Prognostic Biomarkers for Prostate Cancer?
The present scenario of prostate cancer prognosis involves various biomarkers which can serve in the primary detection of prostate cancer however their role in discrimination against malignant progression is still untrustworthy. In this direction, research studies indicating that aquaporins have capability to compete as a successful biomarker for prognosis of prostate cancer. Aquaporins play important roles in the maintenance of water balance, including cellular migration, cellular expansion, and cellular adhesion facilitation together with a significant association between aquaporins expression and tumor grade [53]. Several research findings indicate that AQPs such as AQP1, AQP2, AQP3, and AQP5 have been confirmed as useful biomarkers for hepatocellular carcinoma, breast cancer, lung adenocarcinoma and colorectal cancer [53,61,85]. However, AQPs as prognostic biomarker-related research studies are limited in prostate cancer. For the first time, Park and Yoon (2017) reported that AQP1 can perform as a prognostic factor for biochemical recurrence in prostate adenocarcinoma [74]. In another study, researchers measured AQP5 expression in prostate cancer tissues and cell lines and established its expression is highly correlative with tumor (T), nodes (N), and metastases (M) (TNM) stage [94]. Even though AQPs including AQP1, AQP3, AQP5 and AQP9 have exhibited significant roles that contribute to prostate cancer prognosis, very little progress has been made in identifying AQPs as biomarker(s) for their use in prostate cancer. OncoPrint data analysis of prostate cancer patients further uncovers the possibilities of AQP1, AQP3, AQP5 and AQP9 being developed as prognostic biomarkers for prostate cancer (Figure 2).
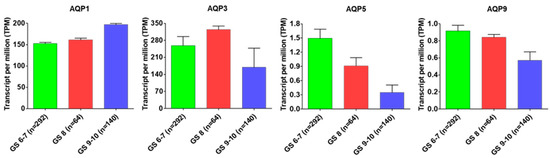
Figure 2.
AQPs expression status with Gleason score. AQP1, AQP3, AQP5 and AQP9 expression status with Gleason score in prostate adenocarcinoma (PRAD) based on the cancer genome atlas (TCGA) dataset.
Results revealed that AQP1 exhibited a positive correlation with the Gleason score; and with a progressive increase in the Gleason score AQP1 expression increases in prostate tumor. AQP3 expression also positively correlate with tumor progression, but in advance-stage prostate tumors with high Gleason score (GS, 9–10), a decrease was observed in its expression. Furthermore, a progressive decrease in the expression of AQP5 and AQP9 was observed during prostate cancer progression that negatively correlates with their expression.
Studies on protein–protein interaction between AQP1, AQP3, AQP5, and AQP9 have identified some key molecules that play a critical role in driving cellular processes including cell–cell adhesion, proliferation, migration, angiogenesis, and stemness. Protein–protein interaction search revealed that major aquaporins such as AQP1, AQP3, AQP5 and AQP9 interact with other proteins including androgen receptor (AR), signal transducer and activator of transcription (STATs) (STAT5A, STAT3), sma- and mad- related protein (SMADs) (SMAD2, SMAD3 SMAD4), stemness-related transcription factors such as SOX2, SOX9, SOX11, NANOG, MYC, epigenetic modifiers such as enhancer of zeste 2 polycomb repressive complex 2 (EZH2), suppressor of zeste 12 protein homolog (SUZ12), kruppel-like factors (KLF1, KLF4) and transcriptomic regulators including E2F1, cyclic AMP-responsive element-binding protein 1 (CREB1), zinc finger protein 281 (ZNF281), Yes-associated protein 1 (YAP1). Some of these proteins including AR, SMAD, NANOG, EZH2, and KLF have demonstrated prognostic significance in advance-stage prostate cancer. Rebello et al. (2021) reviewed the role of AR in prostate cancer progression [2]. Androgen deprivation therapy suppresses hormone-naïve prostate cancer, but after a certain period of time prostate tumor adapts to survive under low levels of androgens. KLF functions as an AR activator and positively correlates with prostate cancer progression [100]. Liu et al. (2020) established the role of NANOG in prostate cancer stem cell proliferation and its regulation through the SMAD signaling pathway [101]. NANOG silencing decreases phosphorylation events in TGF-β/SMAD signaling components which leads to the inhibition of prostate cancer stem cells proliferation, cell cycle arrest and induction of apoptosis [102]. Overexpression of EZH2 in prostate cancer promotes its progression whereas downregulation of EZH2 inhibits cell proliferation, cell cycle, and invasion in vitro, and tumor reduction in vivo [103]. It is likely that a subset of these molecules together with AQPs might have a better prognostic ability for prostate cancer than a single molecule. Additional studies are required to confirm this hypothesis (Figure 3).
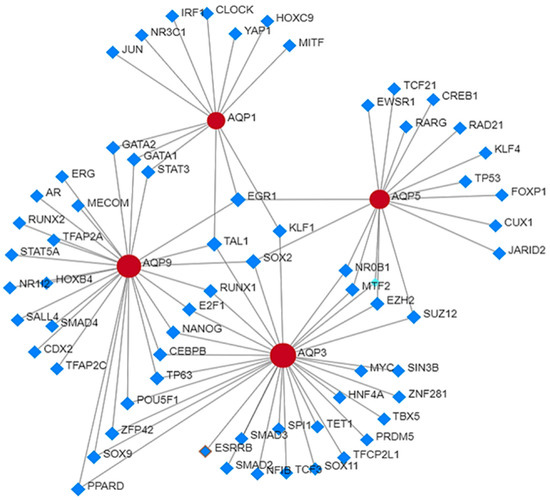
Figure 3.
Protein–protein interaction analysis of AQPs with other proteins in prostate cancer.
To the best of our knowledge, the prognostic significance of other AQPs including AQP2, AQP4, AQP7, and AQP8 is still enigmatic. Preliminary studies indicate that these AQPs are positively associated with prostate cancer initiation, progression, and recurrence development after therapy. Additional studies are required to establish AQPs as a prognostic biomarker for prostate cancer.
6. Conclusions and Future Directions
In conclusion, research over the last decade has shown that AQPs play essential biological roles in cancer, contributing to critical cellular processes such as cell proliferation, migration, and tumor growth. Studies on structural and functional assessment highlight a strong biological relationship between AQPs protein expression, localization, and key biological functions in normal and prostate cancer tissues, where aberrant AQP1, AQP3 and AQP5 expression correlate with tumorigenesis and metastasis. To overcome the prognostic challenges of prostate cancer including stage, grade, and lymph node status additional biomarkers are urgently needed. The research finding from the previous studies indicates having potential that AQPs may serve as prognostic biomarker(s) for prostate cancer. Research investigations found that AQP1 and AQP5 can serve as prognostic biomarker(s) in prostate cancer. Our analysis revealed that AQP1 and AQP3 have prognostic value and may be developed as prognostic biomarkers either alone or together with other identified network proteins. However, additional investigation is still needed. Understanding the unidentified AQP pathophysiology is also essential in order to recognize other AQPs as prognostic biomarkers in prostate cancer.
Author Contributions
S.G., conceived and designed the study; P.P.K., wrote the original draft of the manuscript; P.P.K. and S.V., prepared figures and table; S.G., revised and edited the manuscript; All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Department of Defense Grants W81XWH-18-1-0618, W81XWH-19-1-0720 to S.G.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- American Cancer Society. Cancer Facts & Figures; American Cancer Society: Atlanta, GA, USA, 2022. [Google Scholar]
- Rebello, R.J.; Christoph, O.; Knudsen, K.E.; Loeb, S.; Johnson, D.C.; Reiter, R.E.; Silke Gillessen, S.; Kwast, T.V.D.; Bristow, R.G. Prostate cancer. Nat. Rev. Dis. Prim. 2021, 7, 9. [Google Scholar] [CrossRef]
- Milonas, D.; Venclovas, Z.; Gudinaviciene, I.; Zviniene, K.; Matjosaitis, A.J. Long-term oncological outcomes for young men undergoing radical prostatectomy for localized prostate cancer. Biomed. Res. Int. 2017, 2017, 9858923. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, P.A. Histopathology of prostate cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a030411. [Google Scholar] [CrossRef]
- Bickers, B.; Aukim-Hastie, C. New molecular biomarkers for the prognosis and management of prostate cancer-the post PSA era. Anticancer Res. 2009, 29, 3289–3298. [Google Scholar] [PubMed]
- Lilja, H.; Ulmert, D.; Vickers, A.J. Prostate-specific antigen and prostate cancer: Prediction, detection and monitoring. Nat. Rev. Cancer 2008, 8, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Eggener, S.E.; Badani, K.; Barocas, D.A.; Barrisford, G.W.; Cheng, J.S.; Chin, A.I.; Corcoran, A.; Epstein, J.I.; George, A.K.; Gupta, G.N.; et al. Gleason 6 prostate cancer: Translating biology into population health. J. Urol. 2015, 194, 626–634. [Google Scholar] [CrossRef]
- Ferro, M.; Crocetto, F.; Bruzzese, D.; Imbriaco, M.; Fusco, F.; Longo, N.; Napolitano, L.; Civita, E.L.; Cennamo, M.; Liotti, A.; et al. Prostate health index and multiparametric MRI: Partners in crime fighting overdiagnosis and overtreatment in prostate cancer. Cancers 2021, 13, 4723. [Google Scholar] [CrossRef]
- Crocetto, F.; Russo, G.; Di Zazzo, E.; Pisapia, P.; Mirto, B.F.; Palmieri, A.; Pepe, F.; Bellevicine, C.; Russo, A.; Civita, E.L.; et al. Liquid biopsy in prostate cancer management—Current challenges and future perspectives. Cancers 2022, 14, 3272. [Google Scholar] [CrossRef]
- Kretschmer, A.; Tilki, D. Biomarkers in prostate cancer–current clinical utility and future perspectives. Crit. Rev. Oncol. Hematol. 2017, 120, 180–193. [Google Scholar] [CrossRef]
- Matuszczak, M.; Schalken, J.A.; Salagierski, M. Prostate cancer liquid biopsy biomarkers’ clinical utility in diagnosis and prognosis. Cancers 2021, 13, 3373. [Google Scholar] [CrossRef]
- Legisi, L.; DeSa, E.; Qureshi, M.N. Use of the prostate core mitomic test in repeated biopsy decision-making: Real-world assessment of clinical utility in a multicenter patient population. Am. Health Drug Benefits 2016, 9, 497–502. [Google Scholar] [PubMed]
- Falzarano, S.M.; Ferro, M.; Bollito, E.; Klein, E.A.; Carrieri, G.; Magi-Galluzzi, C. Novel biomarkers and genomic tests in prostate cancer: A critical analysis. Ital. J. Urol. Nephrol. 2015, 67, 211–231. [Google Scholar]
- Bazzichetto, C.; Conciatori, F.; Pallocca, M.; Falcone, I.; Fanciulli, M.; Cognetti, F.; Milella, M.; Ciuffreda, L. PTEN as a prognostic/predictive biomarker in cancer: An unfulfilled promise? Cancers 2019, 11, 435. [Google Scholar] [CrossRef] [PubMed]
- Lotan, T.L.; Wei, W.; Ludkovski, O.; Morais, C.L.; Guedes, L.B.; Jamaspishvili, T.; Lopez, K.; Hawley, S.T.; Feng, Z.; Fazli, L.; et al. Analytic validation of a clinical-grade PTEN immunohistochemistry assay in prostate cancer by comparison with PTEN FISH. Mod. Pathol. 2016, 29, 904–914. [Google Scholar] [CrossRef]
- Kish, E.K.; Choudhry, M.; Gamallat, Y.; Buharideen, S.M.; Bismar, T.A. The expression of proto-oncogene ETS-related gene (ERG) plays a central role in the oncogenic mechanism involved in the development and progression of prostate cancer. Int. J. Mol. Sci. 2022, 23, 4772. [Google Scholar] [CrossRef]
- Krumbholz, M.; Agaimy, A.; Stoehr, R.; Burger, M.; Wach, S.; Taubert, H.; Wullich, B.; Hartmann, A.; Metzler, M. Molecular composition of genomic TMPRSS2-ERG rearrangements in prostate cancer. Dis. Markers 2019, 2019, 5085373. [Google Scholar] [CrossRef]
- Olleik, G.; Kassouf, W.; Aprikian, A.; Hu, J.; Vanhuyse, M.; Cury, F.; Peacock, S.; Bonnevier, E.; Dragomir, A. Evaluation of new tests and interventions for prostate cancer management: A systematic review. J. Natl. Compr. Cancer Netw. 2018, 16, 1340–1351. [Google Scholar] [CrossRef]
- Zhuang, L.; Johnson, M.T. How precisely can prostate cancer be managed? Int. Neurourol. J. 2016, 20 (Suppl. S2), S120–S130. [Google Scholar] [CrossRef]
- Visser, W.C.; de Jong, H.; Melchers, W.J.; Mulders, P.F.; Schalken, J.A. Commercialized blood-, urinary-and tissue-based biomarker tests for prostate cancer diagnosis and prognosis. Cancers 2020, 12, 3790. [Google Scholar] [CrossRef]
- Toribio-Vázquez, C.; Rivas, J.G.; Yebes, Á.; Carrión, D.M.; Barrado, M.Y.; Álvarez-Maestro, M.; Martinez-Piñeiro, L. New strategies for decision making in prostate cancer. The role of oncotypedx. Actas Urol. Esp. 2022, S2173–S5786. [Google Scholar] [CrossRef]
- Lone, Z.; Benidir, T.; Rainey, M.; Nair, M.; Davicioni, E.; Gibb, E.A.; Williamson, S.; Gupta, S.; Ornstein, M.C.; Tendulkar, R.; et al. Transcriptomic features of cribriform and intraductal carcinoma of the prostate. Eur. Urol. Focus. 2022, 8, 1575–1582. [Google Scholar] [CrossRef] [PubMed]
- Boström, P.J.; Bjartell, A.S.; Catto, J.W.; Eggener, S.E.; Lilja, H.; Loeb, S.; Schalken, J.; Schlomm, T.; Cooperberg, M.R. Genomic predictors of outcome in prostate cancer. Eur. Urol. 2015, 68, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Khatami, F.; Aghamir, S.M.K.; Salmaninejad, A.; Shivarani, S.; Khorrami, M.H. Biomarkers for prostate cancer diagnosis from genetic perspectives. Transl. Res. Urol. 2020, 2, 51–58. [Google Scholar] [CrossRef]
- Carlsson, S.V.; Roobol, M.J. Improving the evaluation and diagnosis of clinically significant prostate cancer in 2017. Curr. Opin. Urol. 2017, 27, 198–204. [Google Scholar] [CrossRef] [PubMed]
- McGrowder, D.; Anderson-Jackson, L.; Dilworth, L.; Mohansingh, S.; Cross, M.A.; Bryan, S.; Miller, F.; Wilson-Clarke, C.; Nwokocha, C.; Alexander-Lindo, R.; et al. The clinical usefulness of prostate cancer biomarkers: Current and future directions. In Cancer Bioinformatics, 1st ed.; Kais, G., Hamdi, Y., Eds.; IntechOpen: London, UK, 2022; Volume 79, pp. 1–36. [Google Scholar] [CrossRef]
- Sun, Q.P.; Li, L.Y.; Chen, Z.; Pang, J.; Yang, W.J.; Zhou, X.F.; Qiu, J.G.; Su, Z.L.; He, D.; Gao, X. Detection of TMPRSS2-ETS fusions by a multiprobe fluorescence in situ hybridization assay for the early diagnosis of prostate cancer: A pilot study. J. Mol. Diagn. 2010, 12, 718–724. [Google Scholar] [CrossRef]
- Geybels, M.S.; Fang, M.; Wright, J.L.; Qu, X.; Bibikova, M.; Klotzle, B.; Fan, J.B.; Feng, Z.; Ostrander, E.A.; Nelson, P.S.; et al. PTEN loss is associated with prostate cancer recurrence and alterations in tumor DNA methylation profiles. Oncotarget 2017, 8, 84338–84348. [Google Scholar] [CrossRef]
- Alarcón-Zendejas, A.P.; Scavuzzo, A.; Jiménez-Ríos, M.A.; Álvarez-Gómez, R.M.; Montiel-Manríquez, R.; Castro-Hernández, C.; Jiménez-Dávila, M.A.; Pérez-Montiel, D.; González-Barrios, R.; Jiménez-Trejo, F.; et al. The promising role of new molecular biomarkers in prostate cancer: From coding and non-coding genes to artificial intelligence approaches. Prostate Cancer Prostatic Dis. 2022, 25, 431–443. [Google Scholar] [CrossRef]
- Alam, S.; Tortora, J.; Staff, I.; McLaughlin, T.; Wagner, J. Prostate cancer genomics: Comparing results from three molecular assays. Can. J. Urol. 2019, 26, 9758–9762. [Google Scholar]
- Health Quality Ontario. Prolaris Cell Cycle Progression Test for Localized Prostate Cancer: A Health Technology Assessment. Ont. Health Technol. Assess. Ser. 2017, 17, 1–75. [Google Scholar]
- Brawer, M.K.; Cuzick, J.M.; Cooperberg, M.R.; Swanson, G.P.; Freedland, S.J.; Reid, J.E.; Fisher, G.; Lanchbury, J.S.; Gutin, A.; Stone, S.; et al. Prolaris: A novel genetic test for prostate cancer prognosis. J. Clin. Oncol. 2013, 15, 5005. [Google Scholar] [CrossRef]
- Murphy, A.B.; Carbunaru, S.; Nettey, O.S.; Gornbein, C.; Dixon, M.A.; Macias, V.; Sharifi, R.; Kittles, R.A.; Yang, X.; Kajdacsy-Balla, A.; et al. A 17-gene panel genomic prostate score has similar predictive accuracy for adverse pathology at radical prostatectomy in African American and European American Men. Urology 2020, 142, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Basourakos, S.P.; Tzeng, M.; Lewicki, P.J.; Patel, K.; Awamlh, B.A.H.A.; Venkat, S.; Shoag, J.E.; Gorin, M.A.; Barbieri, C.E.; Hu, J.C. Tissue-based biomarkers for the risk stratification of men with clinically localized prostate cancer. Front. Oncol. 2021, 11, 676716. [Google Scholar] [CrossRef] [PubMed]
- Marrone, M.; Potosky, A.L.; Penson, D.; Freedman, A.N. A 22 gene-expression assay, Decipher® (GenomeDx Biosciences) to predict five-year risk of metastatic prostate cancer in men treated with radical prostatectomy. PLoS Curr. 2015, 7. [Google Scholar] [CrossRef] [PubMed]
- Kruse, E.; Uehlein, N.; Kaldenhoff, R. The aquaporins. Genome Biol. 2006, 7, 206. [Google Scholar] [CrossRef] [PubMed]
- Zannetti, A.; Benga, G.; Brunetti, A.; Napolitano, F.; Avallone, L.; Pelagalli, A. Role of aquaporins in the physiological functions of mesenchymal stem cells. Cells 2020, 9, 2678. [Google Scholar] [CrossRef] [PubMed]
- Pelagalli, A.; Squillacioti, C.; Mirabella, N.; Meli, R. Aquaporins in health and disease: An overview focusing on the gut of different species. Int. J. Mol. Sci. 2016, 17, 1213. [Google Scholar] [CrossRef]
- King, L.S.; Kozono, D.; Agre, P. From structure to disease: The evolving tale of aquaporin biology. Nat. Rev. Mol. Cell Biol. 2004, 5, 687–698. [Google Scholar] [CrossRef]
- Ma, T.; Yang, B.; Gillespie, A.; Carlson, E.J.; Epstein, C.J.; Verkman, A.S. Severely impaired urinary concentrating ability in transgenic mice lacking aquaporin-1 water channels. J. Biol. Chem. 1998, 273, 4296–4299. [Google Scholar] [CrossRef]
- Rash, J.E.; Yasumura, T.; Hudson, C.S.; Agre, P.; Nielsen, S. Direct immunogold labeling of aquaporin-4 in square arrays of astrocyte and ependymocyte plasma membranes in rat brain and spinal cord. Proc. Natl. Acad. Sci. USA 1998, 95, 11981–11986. [Google Scholar] [CrossRef]
- Smith, A.J.; Jin, B.J.; Ratelade, J.; Verkman, A.S. Aggregation state determines the localization and function of M1-and M23-aquaporin-4 in astrocytes. J. Cell. Biol. 2014, 204, 559–573. [Google Scholar] [CrossRef]
- Verkman, A.S.; Ratelade, J.; Rossi, A.; Zhang, H.; Tradtrantip, L. Aquaporin-4: Orthogonal array assembly, CNS functions, and role in neuromyelitis optica. Acta Pharmacol. Sin. 2011, 32, 702–710. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Yang, B. Aquaporins in renal diseases. Int. J. Mol. Sci. 2019, 20, 366. [Google Scholar] [CrossRef] [PubMed]
- Calamita, G.; Perret, J.; Delporte, C. Aquaglyceroporins: Drug targets for metabolic diseases? Front. Physiol. 2018, 9, 851. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, R.; Bhattacharjee, H.; Rosen, B.P. Aquaglyceroporins: Generalized metalloid channels. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 1583–1591. [Google Scholar] [CrossRef] [PubMed]
- Borgnia, M.; Nielsen, S.; Engel, A.; Agre, P. Cellular and molecular biology of the aquaporin water channels. Annu. Rev. Biochem. 1999, 68, 425–458. [Google Scholar] [CrossRef]
- Yang, B.; Verkman, A.S. Water and glycerol permeabilities of aquaporins 1–5 and MIP determined quantitatively by expression of epitope-tagged constructs in Xenopus oocytes. J. Biol. Chem. 1997, 272, 16140–16146. [Google Scholar] [CrossRef]
- Li, S.; Li, C.; Wang, W. Molecular aspects of aquaporins. Vitam. Horm. 2020, 113, 129–181. [Google Scholar] [CrossRef]
- Tsukaguchi, H.; Weremowicz, S.; Morton, C.C.; Hediger, M.A. Functional and molecular characterization of the human neutral solute channel aquaporin-9. Am. J. Physiol. Ren. Physiol. 1999, 277, F685–F696. [Google Scholar] [CrossRef]
- Ishibashi, K. Aquaporin subfamily with unusual NPA boxes. Biochim. Biophys. Acta Biomembr. 2006, 1758, 989–993. [Google Scholar] [CrossRef]
- Yang, B. Aquaporins, 1st ed.; Springer: Dordrecht, The Netherlands, 2017; pp. 1–277. [Google Scholar] [CrossRef]
- Dajani, S.; Saripalli, A.; Sharma-Walia, N. Water transport proteins–aquaporins (AQPs) in cancer biology. Oncotarget 2018, 9, 36392–36405. [Google Scholar] [CrossRef]
- Simone, L.; Gargano, C.D.; Pisani, F.; Cibelli, A.; Mola, M.G.; Frigeri, A.; Svelto, M.; Nicchia, G.P. Aquaporin-1 inhibition reduces metastatic formation in a mouse model of melanoma. J. Cell. Mol. Med. 2018, 22, 904–912. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.B.; Shi, S.; Zhang, R.J.; Wang, T.T.; Tan, Y.J.; Zhang, D.; Fei, X.Y.; Ding, G.L.; Gao, Q.; Chen, C.; et al. Aquaporin-1 plays a crucial role in estrogen-induced tubulogenesis of vascular endothelial cells. J. Clin. Endocr. Metab. 2013, 98, E672–E682. [Google Scholar] [CrossRef] [PubMed]
- Milković, L.; Čipak Gašparović, A. AQP3 and AQP5-potential regulators of redox status in breast cancer. Molecules 2021, 26, 2613. [Google Scholar] [CrossRef] [PubMed]
- De Ieso, M.L.; Yool, A.J. Mechanisms of aquaporin-facilitated cancer invasion and metastasis. Front. Chem. 2018, 6, 135. [Google Scholar] [CrossRef] [PubMed]
- Chow, P.H.; Bowen, J.; Yool, A.J. Combined systematic review and transcriptomic analyses of mammalian aquaporin classes 1 to 10 as biomarkers and prognostic indicators in diverse cancers. Cancers 2020, 12, 1911. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, S.; Jiang, H.; Yang, Y.; Jiang, Y. Co-expression of AQP3 and AQP5 in esophageal squamous cell carcinoma correlates with aggressive tumor progression and poor prognosis. Med. Oncol. 2013, 30, 636. [Google Scholar] [CrossRef]
- Ismail, M.; Bokaee, S.; Davies, J.; Harrington, K.J.; Pandha, H. Inhibition of the aquaporin 3 water channel increases the sensitivity of prostate cancer cells to cryotherapy. Br. J. Cancer 2009, 100, 1889–1895. [Google Scholar] [CrossRef]
- Wang, J.; Feng, L.; Zhu, Z.; Zheng, M.; Wang, D.; Chen, Z.; Sun, H. Aquaporins as diagnostic and therapeutic targets in cancer: How far we are? J. Transl. Med. 2015, 13, 96. [Google Scholar] [CrossRef]
- Zhu, L.; Ma, N.; Wang, B.; Wang, L.; Zhou, C.; Yan, Y.; He, J.; Ren, Y. Significant prognostic values of aquaporin mRNA expression in breast cancer. Cancer Manag. Res. 2019, 11, 1503–1515. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, W.; Ding, Z.; Xu, T.; Zhang, X.; Xu, K. Comprehensive exploration of the expression and prognostic value of AQPs in clear cell renal cell carcinoma. Medicine 2022, 101, e29344. [Google Scholar] [CrossRef]
- Abdelrahman, A.E.; Fathy, A.; Elsebai, E.A.; Nawar, N.; Etman, W.M. Prognostic impact of Apaf-1, Cyclin D1, and AQP-5 in serous ovarian carcinoma treated with the first-line chemotherapy. Ann. Diagn. Pathol. 2018, 35, 27–37. [Google Scholar] [CrossRef]
- Bellezza, G.; Vannucci, J.; Bianconi, F.; Metro, G.; Del Sordo, R.; Andolfi, M.; Ferri, I.; Siccu, P.; Ludovini, V.; Puma, F.; et al. Prognostic implication of aquaporin 1 overexpression in resected lung adenocarcinoma. Interact. Cardiovasc. Thorac. Surg. 2017, 25, 856–861. [Google Scholar] [CrossRef]
- Tie, L.; Lu, N.; Pan, X.Y.; Pan, Y.; An, Y.; Gao, J.W.; Lin, Y.H.; Yu, H.M.; Li, X.J. Hypoxia-induced up-regulation of aquaporin-1 protein in prostate cancer cells in a p38-dependent manner. Cell. Physiol. Biochem. 2012, 29, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Mobasheri, A.; Airley, R.; Hewitt, S.M.; Marples, D. Heterogeneous expression of the aquaporin 1 (AQP1) water channel in tumors of the prostate, breast, ovary, colon and lung: A study using high density multiple human tumor tissue microarrays. Int. J. Oncol. 2005, 26, 1149–1158. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Jung, S.I.; Hwang, E.C.; Song, S.H.; Lee, H.S.; Kim, S.O.; Kang, T.W.; Kwon, D.; Park, K. Expression and localization of aquaporins in benign prostate hyperplasia and prostate cancer. Chonnam Med. J. 2012, 48, 174–178. [Google Scholar] [CrossRef]
- Morrissey, J.J.; London, A.N.; Lambert, M.C.; Kharasch, E.D. Sensitivity and specificity of urinary neutrophil gelatinase-associated lipocalin and kidney injury molecule-1 for the diagnosis of renal cell carcinoma. Am. J. Nephrol. 2011, 34, 391–398. [Google Scholar] [CrossRef]
- Arnaoutova, I.; Cawley, N.X.; Patel, N.; Kim, T.; Rathod, T.; Loh, Y.P. Aquaporin 1 is important for maintaining secretory granule biogenesis in endocrine cells. Mol. Endocrinol. 2008, 22, 1924–1934. [Google Scholar] [CrossRef][Green Version]
- Bullock, A.F.; Greenley, S.L.; McKenzie, G.A.; Paton, L.W.; Johnson, M.J. Relationship between markers of malnutrition and clinical outcomes in older adults with cancer: Systematic review, narrative synthesis and meta-analysis. Eur. J. Clin. Nutr. 2020, 74, 1519–1535. [Google Scholar] [CrossRef] [PubMed]
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef] [PubMed]
- Portela, L.M.; Santos, S.A.; Constantino, F.B.; Camargo, A.C.; Colombelli, K.T.; Fioretto, M.N.; Barquilha, C.N.; Périco, L.L.; Hiruma-Lima, C.A.; Scarano, W.R.; et al. Increased oxidative stress and cancer biomarkers in the ventral prostate of older rats submitted to maternal malnutrition. Mol. Cell. Endocrinol. 2021, 523, 111148. [Google Scholar] [CrossRef]
- Park, J.Y.; Yoon, G. Overexpression of aquaporin-1 is a prognostic factor for biochemical recurrence in prostate adenocarcinoma. Pathol. Oncol. Res. 2017, 23, 189–196. [Google Scholar] [CrossRef]
- Ribatti, D.; Ranieri, G.; Annese, T.; Nico, B. Aquaporins in cancer. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 1550–1553. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Lu, D.; Zhang, Y.; Li, J.; Fang, Y.; Li, F.; Sun, J. Critical role of aquaporin-3 in epidermal growth factor-induced migration of colorectal carcinoma cells and its clinical significance. Oncol. Rep. 2013, 29, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.L.; Matsuzaki, T.; Nakazawa, T.; Murata, S.; Nakamura, N.; Kondo, T.; Iwashina, M.; Mochizuki, K.; Yamane, T.; Takata, K.; et al. Expression of aquaporin 3 (AQP3) in normal and neoplastic lung tissues. Hum. Pathol. 2007, 38, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Machida, Y.; Ueda, Y.; Shimasaki, M.; Sato, K.; Sagawa, M.; Katsuda, S.; Sakuma, T. Relationship of aquaporin 1, 3, and 5 expression in lung cancer cells to cellular differentiation, invasive growth, and metastasis potential. Hum. Pathol. 2011, 42, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Kusayama, M.; Wada, K.; Nagata, M.; Ishimoto, S.; Takahashi, H.; Yoneda, M.; Nakajima, A.; Okura, M.; Kogo, M.; Kamisaki, Y. Critical role of aquaporin 3 on growth of human esophageal and oral squamous cell carcinoma. Cancer Sci. 2011, 102, 1128–1136. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Z.; Xu, D.; Liu, Y.; Gao, Y. Aquaporin 3 promotes prostate cancer cell motility and invasion via extracellular signal-regulated kinase 1/2-mediated matrix metalloproteinase-3 secretion. Mol. Med. Rep. 2015, 11, 2882–2888. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tanji, N.; Kikugawa, T.; Shudou, M.; Song, X.; Yokoyama, M. Expression of aquaporin 3 in the human prostate. Int. J. Urol. 2007, 14, 1088–1092. [Google Scholar] [CrossRef] [PubMed]
- Nejsum, L.N.; Nelson, W.J. A molecular mechanism directly linking E-cadherin adhesion to initiation of epithelial cell surface polarity. J. Cell. Biol. 2007, 178, 323–335. [Google Scholar] [CrossRef]
- Chen, Q.; Zhu, L.; Zong, H.; Song, X.; Wang, L.; Wang, X.; Yanh, D.; Wang, J. Subcellular localization of aquaporin 3 in prostate cancer is regulated by RalA. Oncol. Rep. 2018, 39, 2171–2177. [Google Scholar] [CrossRef]
- de Almeida, A.; Parthimos, D.; Dew, H.; Smart, O.; Wiltshire, M.; Errington, R.J. Aquaglyceroporin-3’s expression and cellular localization is differentially modulated by hypoxia in prostate cancer cell lines. Cells 2021, 10, 838. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Ricciardelli, C.; Yool, A.J. Targeting aquaporins in novel therapies for male and female breast and reproductive cancers. Cells 2021, 10, 215. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.K.; Chae, Y.K.; Woo, J.; Kim, M.S.; Park, J.C.; Lee, J.; Soria, J.C.; Jang, S.J.; Sidransky, D.; Moon, C. Role of human aquaporin 5 in colorectal carcinogenesis. Am. J. Pathol. 2008, 173, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, Q.; Yang, T.; Bai, G.; Li, D.; Li, Q.; Sun, H. Expression of AQP5 and AQP8 in human colorectal carcinoma and their clinical significance. World J. Surg. Oncol. 2012, 10, 242. [Google Scholar] [CrossRef] [PubMed]
- Chae, Y.K.; Woo, J.; Kim, M.J.; Kang, S.K.; Kim, M.S.; Lee, J.; Lee, S.K.; Gong, G.; Kim, Y.H.; Soria, J.C.; et al. Expression of aquaporin 5 (AQP5) promotes tumor invasion in human non-small cell lung cancer. PLoS ONE 2008, 3, e2162. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, C.; Chen, D.; Zhou, Z. Overexpression of AQP5 in cervical cancer: Correlation with clinicopathological features and prognosis. Med. Oncol. 2012, 29, 1998–2004. [Google Scholar] [CrossRef]
- Chae, Y.K.; Kang, S.K.; Kim, M.S.; Woo, J.; Lee, J.; Chang, S.; Kim, D.W.; Kim, M.; Park, S.; Kim, I.; et al. Human AQP5 plays a role in the progression of chronic myelogenous leukemia (CML). PLoS ONE 2008, 3, e2594. [Google Scholar] [CrossRef]
- Yan, C.; Zhu, Y.; Zhang, X.; Chen, X.; Zheng, W.; Yang, J. Down-regulated aquaporin 5 inhibits proliferation and migration of human epithelial ovarian cancer 3AO cells. J. Ovarian Res. 2014, 7, 78. [Google Scholar] [CrossRef]
- Guo, X.; Sun, T.; Yang, M.; Li, Z.; Li, Z.; Gao, Y. Prognostic value of combined aquaporin 3 and aquaporin 5 overexpression in hepatocellular carcinoma. BioMed Res. Int. 2013, 2013, 206525. [Google Scholar] [CrossRef]
- Pust, A.; Kylies, D.; Hube-Magg, C.; Kluth, M.; Minner, S.; Koop, C.; Grob, T.; Graefen, M.; Salomon, G.; Tsourlakis, M.C.; et al. Aquaporin 5 expression is frequent in prostate cancer and shows a dichotomous correlation with tumor phenotype and PSA recurrence. Hum. Pathol. 2016, 48, 102–110. [Google Scholar] [CrossRef]
- Li, J.; Wang, Z.; Chong, T.; Chen, H.; Li, H.; Li, G.; Zhai, X.; Li, Y. Over-expression of a poor prognostic marker in prostate cancer: AQP5 promotes cells growth and local invasion. World J. Surg. Oncol. 2014, 12, 284. [Google Scholar] [CrossRef] [PubMed]
- Lindskog, C.; Asplund, A.; Catrina, A.; Nielsen, S.; Rützler, M. A systematic characterization of aquaporin-9 expression in human normal and pathological tissues. J. Histochem. Cytochem. 2016, 64, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tanji, N.; Sasaki, T.; Kikugawa, T.; Song, X.; Yokoyama, M. Androgens upregulate aquaporin 9 expression in the prostate. Int. J. Urol. 2008, 15, 936–941. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhu, L.; Zheng, B.; Wang, J.; Song, X.; Zheng, W.; Wang, L.; Yang, D.; Wang, J. Effect of AQP9 expression in androgen-independent prostate cancer cell PC3. Int. J. Mol. Sci. 2016, 17, 738. [Google Scholar] [CrossRef]
- Bründl, J.; Wallinger, S.; Breyer, J.; Weber, F.; Evert, M.; Georgopoulos, N.T.; Rosenhammer, B.; Burger, M.; Otto, W.; Rubenwolf, P. Expression, localisation and potential significance of aquaporins in benign and malignant human prostate tissue. BMC Urol. 2018, 18, 75. [Google Scholar] [CrossRef]
- Pan, X.Y.; Guo, H.; Han, J.; Hao, F.; An, Y.; Xu, Y.; Xiaokaiti, Y.; Pan, Y.; Li, X.J. Ginsenoside Rg3 attenuates cell migration via inhibition of aquaporin 1 expression in PC-3M prostate cancer cells. Eur. J. Pharmacol. 2012, 683, 27–34. [Google Scholar] [CrossRef]
- Siu, M.K.; Suau, F.; Chen, W.Y.; Tsai, Y.C.; Tsai, H.Y.; Yeh, H.L.; Liu, Y. KLF4 functions as an activator of the androgen receptor through reciprocal feedback. Oncogenesis 2016, 5, e282. [Google Scholar] [CrossRef]
- Liu, C.; Sheng, M.; Lin, L.; Li, H.; Guo, S.; Zhang, J.; Chen, G.; Chen, H. NANOG regulates the proliferation of PCSCs via the TGF-β1/SMAD pathway. Open Med. 2020, 15, 841–849. [Google Scholar] [CrossRef]
- Cackowski, F.C.; Heath, E.I. Prostate cancer dormancy and recurrence. Cancer Lett. 2022, 524, 103–108. [Google Scholar] [CrossRef]
- Park, S.H.; Fong, K.W.; Mong, E.; Martin, M.C.; Schiltz, G.E.; Yu, J. Going beyond polycomb: EZH2 functions in prostate cancer. Oncogene 2021, 40, 5788–5798. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).