GradWise: A Novel Application of a Rank-Based Weighted Hybrid Filter and Embedded Feature Selection Method for Glioma Grading with Clinical and Molecular Characteristics
Abstract
:Simple Summary
Abstract
1. Introduction
- We combine the advantages of various feature selection methods via a rank-based feature-weighting approach for glioma grading on two commonly used glioma datasets (TCGA and CGGA).
- We utilize feature-weighting to determine which features are significant, enabling validation of this method for glioma grading tasks.
- We conduct a comprehensive computational analysis comparing our feature selection methods, given that these are two commonly employed glioma datasets that share similarities but also exhibit differences.
- Our objective is to determine the optimal combination of feature subsets and learning models during the feature selection stage, aiming to achieve high accuracy with a minimal number of features while accounting for dataset variability in large-scale datasets. This approach seeks to provide accurate results that can be transferred and applied effectively across different scenarios.
- We introduce a TCGA- and CGGA-specific shared feature set and connect identified features for glioma grading with described mutations in glioma and identify potential mechanistic implications for progression to higher grade.
2. Methods
2.1. The Utilized Methodology for Glioma Grading
2.2. Feature Selection and Feature-Weighting
2.3. Classification
3. Experimental Work
3.1. Experimental Process
3.2. Dataset
3.3. Performance Metrics
3.4. Computational Results
3.4.1. The Effects of Using Feature Selection Methods
3.4.2. The Effects of Using LASSO and mRMR Feature Selection and Feature-Weighting Methods
3.4.3. Other Performance Results Based on Feature Selection and Weighting Process
3.4.4. Comparison with the Related Methods for Glioma Grading
4. Discussion
5. Conclusions and Future Work
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACC | Accuracy |
| AdaBoost | Adaptive Boosting |
| AUC | Area Under the ROC Curve |
| CGGA | Chinese Glioma Genome Atlas |
| CNS | Central Nervous System |
| F1 | F-Measure |
| GBM | Glioblastoma Multiforme |
| HGG | High-Grade Glioma |
| IDH | Isocitrate Dehydrogenase |
| IPA | Ingenuity Pathway Analysis |
| KNN | K Nearest Neighbors |
| LGG | Low-Grade Glioma |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| LR | Logistic Regression |
| mRMR | Minimum Redundancy—Maximum Relevance |
| NCI | National Cancer Institute |
| NIDAP | NIH Integrated Data Analysis Platform |
| NIH | National Institutes of Health |
| PRE | Precision |
| REC | Recall |
| RF | Random Forest |
| ROC | Receiver Operating Characteristics |
| RT | Radiation Therapy |
| SPEC | Specificity |
| SVM | Support Vector Machine |
| TCGA | The Cancer Genome Atlas |
| TMZ | Temozolomide |
| WHO | World Health Organization |
References
- Marquet, G.; Dameron, O.; Saikali, S.; Mosser, J.; Burgun, A. Grading glioma tumors using OWL-DL and NCI thesaurus. In Proceedings of the AMIA Annual Symposium Proceedings, Chicago, IL, USA, 10–14 November 2007; American Medical Informatics Association: Washington, DC, USA. [Google Scholar]
- Pereira, S.; Meier, R.; Alves, V.; Reyes, M.; Silva, C.A. Automatic brain tumor grading from MRI data using convolutional neural networks and quality assessment. In Understanding and Interpreting Machine Learning in Medical Image Computing Applications; Springer: Berlin/Heidelberg, Germany, 2018; pp. 106–114. [Google Scholar]
- Tasci, E.; Ugur, A.; Camphausen, K.; Zhuge, Y.; Zhao, R.; Krauze, A.V. 3D Multimodal Brain Tumor Segmentation and Grading Scheme based on Machine, Deep, and Transfer Learning Approaches. Int. J. Bioinfor. Intell. Comput. 2022, 1, 77–95. [Google Scholar]
- Tasci, E.; Zhuge, Y.; Kaur, H.; Camphausen, K.; Krauze, A.V. Hierarchical Voting-Based Feature Selection and Ensemble Learning Model Scheme for Glioma Grading with Clinical and Molecular Characteristics. Int. J. Mol. Sci. 2022, 23, 14155. [Google Scholar] [CrossRef]
- Krauze, A. Using Artificial Intelligence and Magnetic Resonance Imaging to Address Limitations in Response Assessment in Glioma. Oncol. Insights 2022, 2022, 616. [Google Scholar] [CrossRef]
- Gaillard, F. WHO Classification of CNS Tumors. Reference Article, Radiopaedia.org. Available online: https://radiopaedia.org/articles/who-classification-of-cns-tumours-1?lang=us (accessed on 2 September 2022).
- Mirchia, K.; Richardson, T.E. Beyond IDH-mutation: Emerging molecular diagnostic and prognostic features in adult diffuse gliomas. Cancers 2020, 12, 1817. [Google Scholar] [CrossRef] [PubMed]
- Vigneswaran, K.; Neill, S.; Hadjipanayis, C.G. Beyond the World Health Organization grading of infiltrating gliomas: Advances in the molecular genetics of glioma classification. Ann. Transl. Med. 2015, 3, 95. [Google Scholar] [PubMed]
- DeWitt, J.C.; Jordan, J.T.; Frosch, M.P.; Samore, W.R.; Iafrate, A.J.; Louis, D.N.; Lennerz, J.K. Cost-effectiveness of IDH testing in diffuse gliomas according to the 2016 WHO classification of tumors of the central nervous system recommendations. Neuro-Oncol. 2017, 19, 1640–1650. [Google Scholar] [CrossRef] [PubMed]
- Krauze, A.; Zhuge, Y.; Zhao, R.; Tasci, E.; Camphausen, K. AI-Driven Image Analysis in Central Nervous System Tumors-Traditional Machine Learning, Deep Learning and Hybrid Models. J. Biotechnol. Biomed. 2022, 5, 1–19. [Google Scholar]
- Diaz Rosario, M.; Kaur, H.; Tasci, E.; Shankavaram, U.; Sproull, M.; Zhuge, Y.; Camphausen, K.; Krauze, A. The Next Frontier in Health Disparities—A Closer Look at Exploring Sex Differences in Glioma Data and Omics Analysis, from Bench to Bedside and Back. Biomolecules 2022, 12, 1203. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Gokalp, O.; Tasci, E.; Ugur, A. A novel wrapper feature selection algorithm based on iterated greedy metaheuristic for sentiment classification. Expert Syst. Appl. 2020, 146, 113176. [Google Scholar] [CrossRef]
- Taşci, E.; Gökalp, O.; Uğur, A. Development of a novel feature weighting method using cma-es optimization. In Proceedings of the 2018 26th Signal Processing and Communications Applications Conference (SIU), Izmir, Turkey, 2–5 May 2018. [Google Scholar]
- Taşcı, E.; Uğur, A. Shape and texture based novel features for automated juxtapleural nodule detection in lung CTs. J. Med. Syst. 2015, 39, 1–13. [Google Scholar] [CrossRef]
- Zanella, L.; Facco, P.; Bezzo, F.; Cimetta, E. Feature Selection and Molecular Classification of Cancer Phenotypes: A Comparative Study. Int. J. Mol. Sci. 2022, 23, 9087. [Google Scholar] [CrossRef]
- Tasci, E.; Ugur, A. A novel pattern recognition framework based on ensemble of handcrafted features on images. Multimed. Tools Appl. 2022, 81, 30195–30218. [Google Scholar] [CrossRef]
- Tasci, E.; Zhuge, Y.; Camphausen, K.; Krauze, A.V. Bias and Class Imbalance in Oncologic Data—Towards Inclusive and Transferrable AI in Large Scale Oncology Data Sets. Cancers 2022, 14, 2897. [Google Scholar] [CrossRef] [PubMed]
- Tasci, E.; Jagasia, S.; Zhuge, Y.; Sproull, M.; Cooley Zgela, T.; Mackey, M.; Camphausen, K.; Krauze, A.V. RadWise: A Rank-Based Hybrid Feature Weighting and Selection Method for Proteomic Categorization of Chemoirradiation in Patients with Glioblastoma. Cancers 2023, 15, 2672. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, G.; Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Li, J.; Cheng, K.; Wang, S.; Morstatter, F.; Trevino, R.P.; Tang, J.; Liu, H. Feature selection: A data perspective. ACM Comput. Surv. 2017, 50, 1–45. [Google Scholar] [CrossRef]
- Tang, J.; Alelyani, S.; Liu, H. Feature selection for classification: A review. Data Classif. Algorithms Appl. 2014, 37, 65–92. [Google Scholar]
- Tahir, M.A.; Bouridane, A.; Kurugollu, F. Simultaneous feature selection and feature weighting using Hybrid Tabu Search/K-nearest neighbor classifier. Pattern Recognit. Lett. 2007, 28, 438–446. [Google Scholar] [CrossRef]
- Tasci, E.; Zhuge, Y.; Camphausen, K.; Krauze, A.V. Glioma Grading Clinical and Mutation Features Dataset; UCI Machine Learning Repository: Irvine, CA, USA, 2022. [Google Scholar]
- Zhao, Z.; Zhang, K.-N.; Wang, Q.; Li, G.; Zeng, F.; Zhang, Y.; Wu, F.; Chai, R.; Wang, Z.; Zhang, C. Chinese Glioma Genome Atlas (CGGA): A comprehensive resource with functional genomic data from Chinese glioma patients. Genom. Proteom. Bioinform. 2021, 19, 1–12. [Google Scholar] [CrossRef]
- Palantir Foundry—The NIH Integrated Data Analysis Platform (NIDAP); NCI Center for Biomedical Informatics & Information Technology (CBIIT); Software Provided by Palantir Technologies Inc. Available online: https://www.palantir.com (accessed on 5 June 2023).
- Yan, Y.; Takayasu, T.; Hines, G.; Dono, A.; Hsu, S.H.; Zhu, J.J.; Riascos-Castaneda, R.F.; Kamali, A.; Bhattacharjee, M.B.; Blanco, A.I.; et al. Landscape of Genomic Alterations in IDH Wild-Type Glioblastoma Identifies PI3K as a Favorable Prognostic Factor. JCO Precis. Oncol. 2020, 4, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Mu, Q.; Bao, Z.; Chen, Y.; Liu, Y.; Chen, J.; Wang, K.; Wang, Z.; Nam, Y.; Jiang, B.; et al. Mutational Landscape of Secondary Glioblastoma Guides MET-Targeted Trial in Brain Tumor. Cell 2018, 175, 1665–1678.e18. [Google Scholar] [CrossRef]
- Fawcett, T. An Introduction to ROC Analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Li, Y.; Mansmann, U.; Du, S.; Hornung, R. Benchmark study of feature selection strategies for multi-omics data. BMC Bioinform. 2022, 23, 412. [Google Scholar] [CrossRef] [PubMed]
- Bhadra, T.; Mallik, S.; Hasan, N.; Zhao, Z. Comparison of five supervised feature selection algorithms leading to top features and gene signatures from multi-omics data in cancer. BMC Bioinform. 2022, 23, 153. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.W.; Ou, G.Y.; Zhao, W.J. Mutational profiling of low-grade gliomas identifies prognosis and immunotherapy-related biomarkers and tumour immune microenvironment characteristics. J. Cell. Mol. Med. 2021, 25, 10111–10125. [Google Scholar] [CrossRef]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Meng, X.; Liu, L.; Li, S.; Li, Y.; Ali, S.; Li, S.; Xiong, J.; Liu, X.; Li, S.; et al. Identification of the Prognostic Signatures of Glioma With Different PTEN Status. Front. Oncol. 2021, 11, 633357. [Google Scholar] [CrossRef]
- Georgescu, M.M.; Olar, A. Genetic and histologic spatiotemporal evolution of recurrent, multifocal, multicentric and metastatic glioblastoma. Acta Neuropathol. Commun. 2020, 8, 10. [Google Scholar] [CrossRef]
- Lobbous, M.; Bernstock, J.D.; Coffee, E.; Friedman, G.K.; Metrock, L.K.; Chagoya, G.; Elsayed, G.; Nakano, I.; Hackney, J.R.; Korf, B.R.; et al. An Update on Neurofibromatosis Type 1-Associated Gliomas. Cancers 2020, 12, 114. [Google Scholar] [CrossRef]
- Sakthikumar, S.; Roy, A.; Haseeb, L.; Pettersson, M.E.; Sundström, E.; Marinescu, V.D.; Lindblad-Toh, K.; Forsberg-Nilsson, K. Whole-genome sequencing of glioblastoma reveals enrichment of non-coding constraint mutations in known and novel genes. Genome Biol. 2020, 21, 127. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, V.P. MUC16 mutation is associated with tumor grade, clinical features, and prognosis in glioma patients. Cancer Genet. 2023, 270–271, 22–30. [Google Scholar] [CrossRef]
- Hu, W.; Duan, H.; Zhong, S.; Zeng, J.; Mou, Y. High frequency of PDGFRA and MUC family gene mutations in diffuse hemispheric glioma, H3 G34-mutant: A glimmer of hope? J. Transl. Med. 2022, 20, 64. [Google Scholar] [CrossRef] [PubMed]
- Bunda, S.; Heir, P.; Metcalf, J.; Li, A.S.C.; Agnihotri, S.; Pusch, S.; Yasin, M.; Li, M.; Burrell, K.; Mansouri, S.; et al. CIC protein instability contributes to tumorigenesis in glioblastoma. Nat. Commun. 2019, 10, 661. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Zadeh Shirazi, A.; McDonnell, M.D.; Fornaciari, E.; Bagherian, N.S.; Scheer, K.G.; Samuel, M.S.; Yaghoobi, M.; Ormsby, R.J.; Poonnoose, S.; Tumes, D.J.; et al. A deep convolutional neural network for segmentation of whole-slide pathology images identifies novel tumour cell-perivascular niche interactions that are associated with poor survival in glioblastoma. Br. J. Cancer 2021, 125, 337–350. [Google Scholar] [CrossRef]
- Krigers, A.; Demetz, M.; Thomé, C.; Freyschlag, C.F. Age is associated with unfavorable neuropathological and radiological features and poor outcome in patients with WHO grade 2 and 3 gliomas. Sci. Rep. 2021, 11, 17380. [Google Scholar] [CrossRef]
- Han, S.; Liu, Y.; Cai, S.J.; Qian, M.; Ding, J.; Larion, M.; Gilbert, M.R.; Yang, C. IDH mutation in glioma: Molecular mechanisms and potential therapeutic targets. Br. J. Cancer 2020, 122, 1580–1589. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Oprita, A.; Baloi, S.C.; Staicu, G.A.; Alexandru, O.; Tache, D.E.; Danoiu, S.; Micu, E.S.; Sevastre, A.S. Updated Insights on EGFR Signaling Pathways in Glioma. Int. J. Mol. Sci. 2021, 22, 587. [Google Scholar] [CrossRef]
- Quayle, S.N.; Lee, J.Y.; Cheung, L.W.; Ding, L.; Wiedemeyer, R.; Dewan, R.W.; Huang-Hobbs, E.; Zhuang, L.; Wilson, R.K.; Ligon, K.L.; et al. Somatic mutations of PIK3R1 promote gliomagenesis. PLoS ONE 2012, 7, e49466. [Google Scholar] [CrossRef]
- Nandakumar, P.; Mansouri, A.; Das, S. The Role of ATRX in Glioma Biology. Front. Oncol. 2017, 7, 236. [Google Scholar] [CrossRef] [PubMed]
- Parmigiani, E.; Taylor, V.; Giachino, C. Oncogenic and Tumor-Suppressive Functions of NOTCH Signaling in Glioma. Cells 2020, 9, 2304. [Google Scholar] [CrossRef] [PubMed]
- Felder, M.; Kapur, A.; Gonzalez-Bosquet, J.; Horibata, S.; Heintz, J.; Albrecht, R.; Fass, L.; Kaur, J.; Hu, K.; Shojaei, H.; et al. MUC16 (CA125): Tumor biomarker to cancer therapy, a work in progress. Mol. Cancer 2014, 13, 129. [Google Scholar] [CrossRef] [PubMed]

| 1. Input: Clinical and molecular predictors with labels |
| 2. Feature Selection with cross-validation: For each fold:
4. Evaluation:
|
| # | Type | Name | # | Type | Name | # | Type | Name |
|---|---|---|---|---|---|---|---|---|
| 1 | Clinical | Gender | 9 | Molecular | CIC | 17 | Molecular | BCOR |
| 2 | Clinical | Age | 10 | Molecular | MUC16 | 18 | Molecular | CSMD3 |
| 3 | Clinical | Race | 11 | Molecular | PIK3CA | 19 | Molecular | SMARCA4 |
| 4 | Molecular | IDH1 | 12 | Molecular | NF1 | 20 | Molecular | GRIN2A |
| 5 | Molecular | TP53 | 13 | Molecular | PIK3R1 | 21 | Molecular | IDH2 |
| 6 | Molecular | ATRX | 14 | Molecular | FUBP1 | 22 | Molecular | FAT4 |
| 7 | Molecular | PTEN | 15 | Molecular | RB1 | 23 | Molecular | PDGFRA |
| 8 | Molecular | EGFR | 16 | Molecular | NOTCH1 | 24 | Class | Grade |
| ML-ACC | Without FS | LASSO | mRMR |
|---|---|---|---|
| SVM | 86.769 | 87.007 | 74.733 |
| LR | 86.414 | 86.414 | 85.935 |
| KNN | 82.837 | 83.313 | 82.839 |
| RF | 82.841 | 82.362 | 81.886 |
| AdaBoost | 85.339 | 85.101 | 84.621 |
| ML-ACC | Without FS | LASSO | mRMR |
|---|---|---|---|
| SVM | 76.564 | 76.915 | 73.085 |
| LR | 76.570 | 76.921 | 76.933 |
| KNN | 74.816 | 76.576 | 71.670 |
| RF | 74.840 | 72.741 | 73.442 |
| AdaBoost | 74.834 | 72.033 | 76.576 |
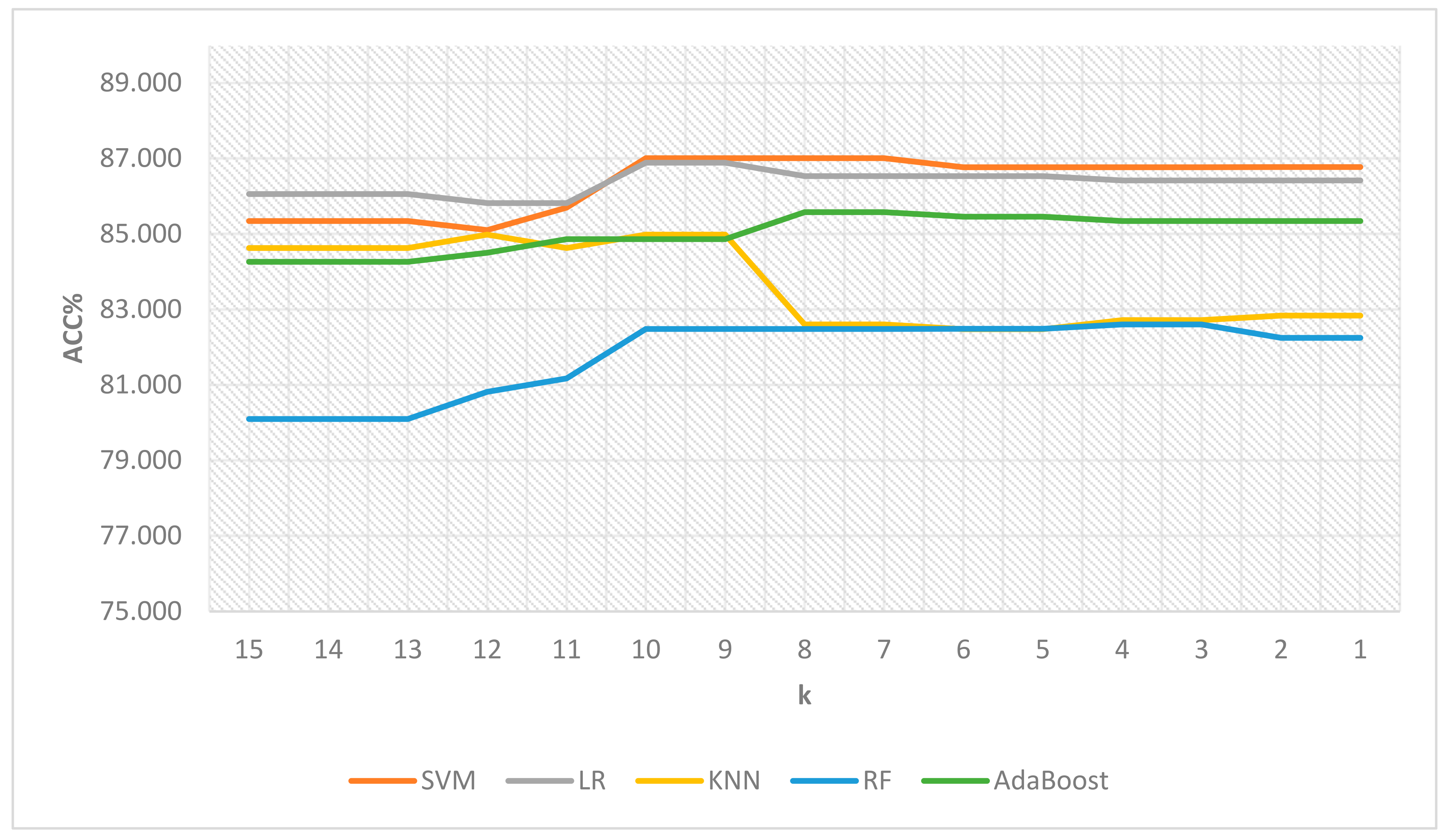
| k | # of Features | SVM | LR | KNN | RF | AdaBoost |
|---|---|---|---|---|---|---|
| 15 | 4 | 85.340 | 86.054 | 84.626 | 80.100 | 84.264 |
| 14 | 4 | 85.340 | 86.054 | 84.626 | 80.100 | 84.264 |
| 13 | 4 | 85.340 | 86.054 | 84.626 | 80.100 | 84.264 |
| 12 | 5 | 85.102 | 85.816 | 84.983 | 80.814 | 84.502 |
| 11 | 6 | 85.698 | 85.816 | 84.627 | 81.172 | 84.859 |
| 10 | 13 | 87.007 | 86.890 | 84.983 | 82.481 | 84.862 |
| 9 | 13 | 87.007 | 86.890 | 84.983 | 82.481 | 84.862 |
| 8 | 18 | 87.007 | 86.533 | 82.599 | 82.481 | 85.577 |
| 7 | 18 | 87.007 | 86.533 | 82.599 | 82.481 | 85.577 |
| 6 | 20 | 86.768 | 86.533 | 82.479 | 82.484 | 85.458 |
| 5 | 20 | 86.768 | 86.533 | 82.479 | 82.484 | 85.458 |
| 4 | 22 | 86.768 | 86.414 | 82.718 | 82.603 | 85.339 |
| 3 | 22 | 86.768 | 86.414 | 82.718 | 82.603 | 85.339 |
| 2 | 23 | 86.769 | 86.414 | 82.837 | 82.244 | 85.339 |
| 1 | 23 | 86.769 | 86.414 | 82.837 | 82.244 | 85.339 |
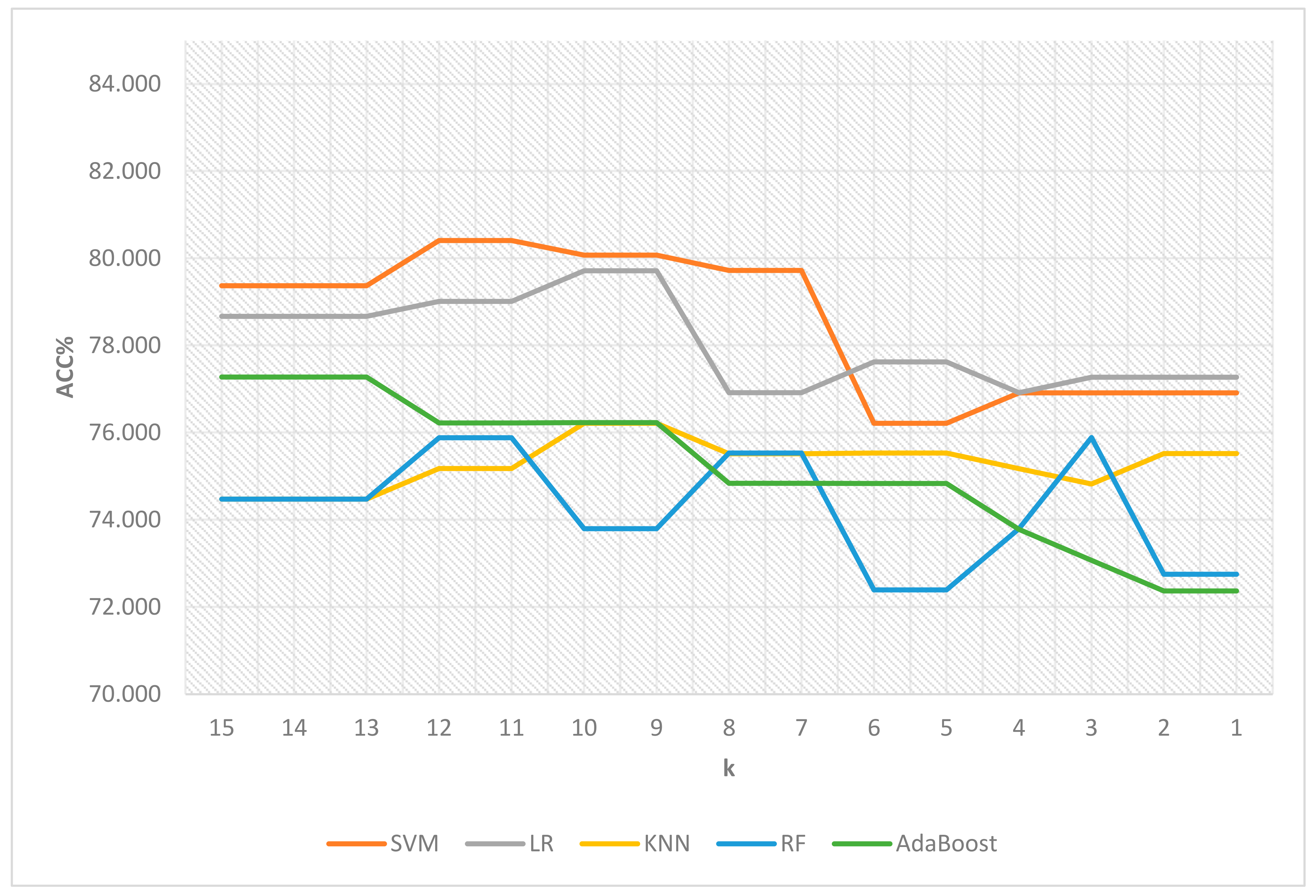
| k | # of Features | SVM | LR | KNN | RF | AdaBoost |
|---|---|---|---|---|---|---|
| 15 | 4 | 79.371 | 78.669 | 74.477 | 74.476 | 77.278 |
| 14 | 4 | 79.371 | 78.669 | 74.477 | 74.476 | 77.278 |
| 13 | 4 | 79.371 | 78.669 | 74.477 | 74.476 | 77.278 |
| 12 | 5 | 80.412 | 79.014 | 75.178 | 75.886 | 76.225 |
| 11 | 5 | 80.412 | 79.014 | 75.178 | 75.886 | 76.225 |
| 10 | 8 | 80.073 | 79.716 | 76.219 | 73.799 | 76.231 |
| 9 | 8 | 80.073 | 79.716 | 76.219 | 73.799 | 76.231 |
| 8 | 10 | 79.722 | 76.921 | 75.517 | 75.535 | 74.840 |
| 7 | 10 | 79.722 | 76.921 | 75.517 | 75.535 | 74.840 |
| 6 | 11 | 76.219 | 77.623 | 75.535 | 72.396 | 74.834 |
| 5 | 11 | 76.219 | 77.623 | 75.535 | 72.396 | 74.834 |
| 4 | 13 | 76.915 | 76.921 | 75.173 | 73.799 | 73.781 |
| 3 | 14 | 76.915 | 77.272 | 74.822 | 75.892 | 73.073 |
| 2 | 16 | 76.915 | 77.272 | 75.523 | 72.752 | 72.371 |
| 1 | 16 | 76.915 | 77.272 | 75.523 | 72.752 | 72.371 |
| Without FS | With FW and FS | Without FS | With FW and FS | |||
|---|---|---|---|---|---|---|
| ML | ACC% | AUC | ||||
| SVM | 86.769 | 87.007 | 0.904 | 0.911 | ||
| LR | 86.414 | 86.890 | 0.918 | 0.923 | ||
| KNN | 82.837 | 84.983 | 0.893 | 0.906 | ||
| RF | 82.841 | 82.481 | 0.897 | 0.900 | ||
| AdaBoost | 85.339 | 84.862 | 0.905 | 0.908 | ||
| Without FS | With FW and FS | Without FS | With FW and FS | |||
| ML | F1 | PRE | ||||
| SVM | 0.852 | 0.855 | 0.801 | 0.804 | ||
| LR | 0.847 | 0.852 | 0.805 | 0.808 | ||
| KNN | 0.802 | 0.826 | 0.782 | 0.802 | ||
| RF | 0.793 | 0.792 | 0.796 | 0.786 | ||
| AdaBoost | 0.832 | 0.829 | 0.803 | 0.789 | ||
| Without FS | With FW and FS | Without FS | With FW and FS | |||
| ML | REC | SPEC | ||||
| SVM | 0.912 | 0.915 | 0.837 | 0.839 | ||
| LR | 0.897 | 0.905 | 0.843 | 0.845 | ||
| KNN | 0.827 | 0.856 | 0.832 | 0.846 | ||
| RF | 0.796 | 0.802 | 0.853 | 0.842 | ||
| AdaBoost | 0.869 | 0.878 | 0.845 | 0.830 | ||
| Without FS | With FW and FS | Without FS | With FW and FS | ||
|---|---|---|---|---|---|
| ML | ACC% | AUC | |||
| SVM | 76.564 | 80.412 | 0.815 | 0.798 | |
| LR | 76.570 | 79.014 | 0.792 | 0.788 | |
| KNN | 74.816 | 75.178 | 0.772 | 0.753 | |
| RF | 74.840 | 75.886 | 0.758 | 0.767 | |
| AdaBoost | 74.834 | 76.225 | 0.759 | 0.749 | |
| Without FS | With FW and FS | Without FS | With FW and FS | ||
| ML | F1 | PRE | |||
| SVM | 0.609 | 0.679 | 0.759 | 0.807 | |
| LR | 0.633 | 0.656 | 0.706 | 0.788 | |
| KNN | 0.555 | 0.577 | 0.743 | 0.706 | |
| RF | 0.592 | 0.629 | 0.659 | 0.663 | |
| AdaBoost | 0.625 | 0.603 | 0.661 | 0.717 | |
| Without FS | With FW and FS | Without FS | With FW and FS | ||
| ML | REC | SPEC | |||
| SVM | 0.527 | 0.607 | 0.901 | 0.913 | |
| LR | 0.584 | 0.582 | 0.862 | 0.907 | |
| KNN | 0.454 | 0.516 | 0.908 | 0.880 | |
| RF | 0.549 | 0.610 | 0.855 | 0.835 | |
| AdaBoost | 0.605 | 0.536 | 0.829 | 0.888 | |
| Dataset | TCGA | CGGA | ||
|---|---|---|---|---|
| Total # of Features | 23 | 22 | ||
| Study | Our Method | [4] | Our Method | [4] |
| Selected # of Features | 13 | 14.9 | 5 | 17.6 |
| ACC % | 87.007 | 87.606 | 80.412 | 79.668 |
| Study | Our Method | [4] | ||
| Method | mRMR + LASSO | Hierarchical voting-based ensemble scheme | ||
| Advantages | Effective, more realistic, and consistent results, and identified feature names | The method employs an ensemble procedure | ||
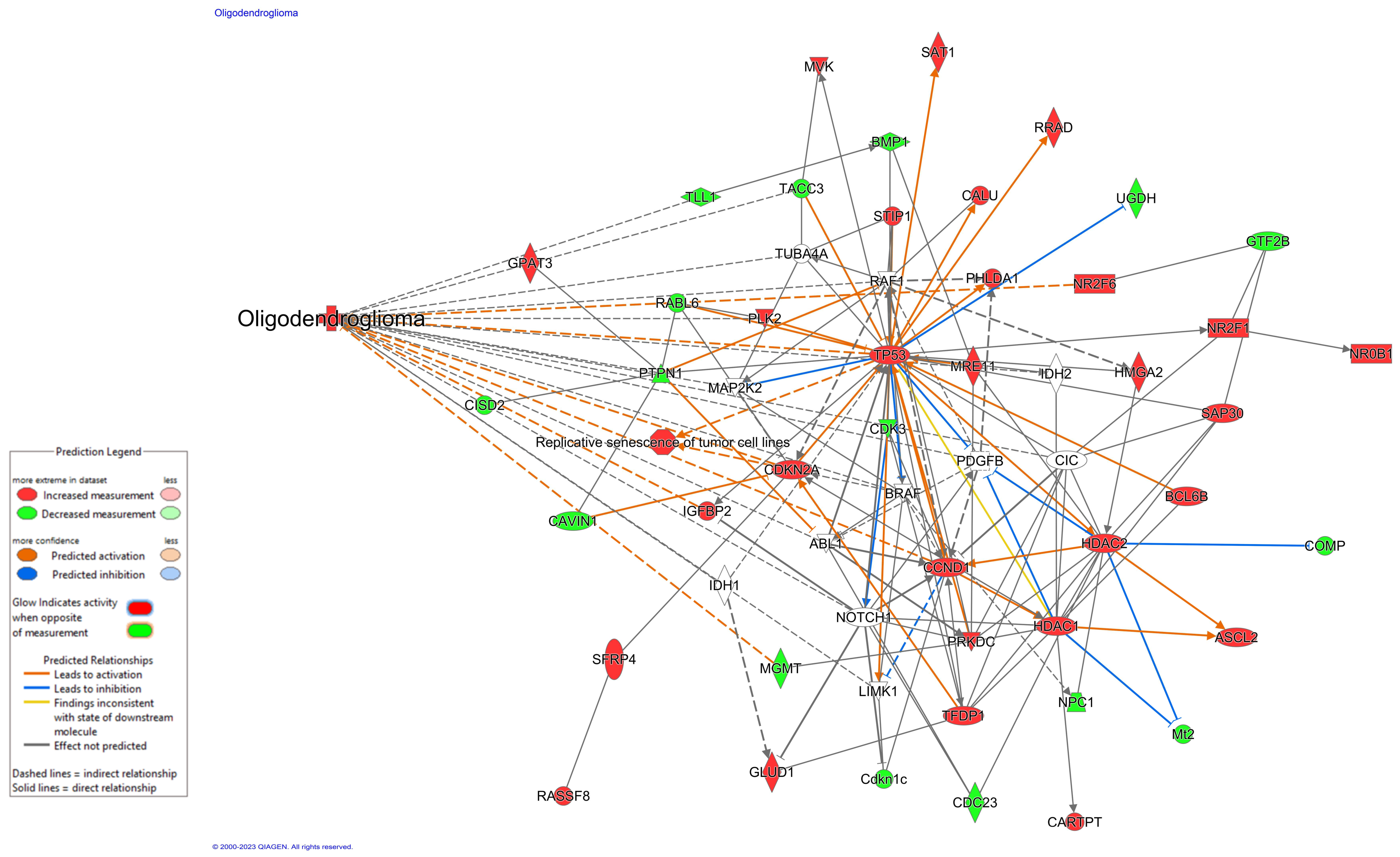
| Feature | Frequency of Mutated Genes in TCGA in GBM [37] | Somatic Genomic Alterations in GBM [33] | % GBM Patients Harboring Specific Oncogenic Mutations in TCGA [42] | Mutation Landscape of LGG [32] | Current Role in Oncology | Mechanistic Connections | |
|---|---|---|---|---|---|---|---|
| Literature Evidence | Use in Clinic | ||||||
| Age | n/a | n/a | n/a | n/a | Age-associated with unfavorable neuropathological and radiological features in gliomas [43] | Yes, for clinical decision-making via recursive partitioning criteria | Investigational |
| IDH1/IDH2 | n/a | n/a | 3% | 77% | IDH mutation in glioma: molecular mechanisms and therapeutic targets [44,45] | Yes, for tumor molecular characterization | HIF-1α |
| PTEN | 34% | 31% | 19% | n/a | Identification of the Prognostic Signatures of Glioma With Different PTEN Status [34] | Yes, for tumor molecular characterization | TP53, GRIN2A |
| NF1 | 11% | 11% | 9% | n/a | An Update on Neurofibromatosis Type 1-Associated Gliomas [36] | Yes, for clinical decision-making and management discussion | EGFR, PTEN |
| EGFR | 26% | 26% | 15% | 6% | Updated Insights on EGFR Signaling Pathways in Glioma [46] | Yes, for tumor molecular characterization | NOTCH1 |
| TP53 | 34% | 29% | 16% | 46% | Genetic and histologic spatiotemporal evolution of recurrent, multifocal, multicentric and metastatic glioblastoma [35] | Yes, for tumor molecular characterization | PTEN, GRIN2A |
| PIK3R1 | 18% | 11% | 6% | n/a | Somatic Mutations of PIK3R1 Promote Gliomagenesis [47] | Not currently used in the clinic | PI3K |
| ATRX | n/a | 6% | 5% | 33% | The Role of ATRX in Glioma Biology [48] | Yes, for tumor molecular characterization | ATM |
| PDGFRA | n/a | 4% | 5% | n/a | High frequency of PDGFRA and MUC family gene mutations in diffuse hemispheric glioma, H3 G34-mutant: a glimmer of hope? [39] | Investigational | MUC16 |
| NOTCH1 | n/a | n/a | n/a | n/a | Oncogenic and Tumor-Suppressive Functions of NOTCH Signaling in Glioma [49] | Investigational | EGFR |
| GRIN2A | n/a | n/a | 4% | n/a | Somatic mutation of GRIN2A in malignant melanoma results in loss of tumor suppressor activity via aberrant NMDAR complex formation [35] | Investigational | PTEN, TP53 |
| MUC16 (CA-125) | 11% | n/a | n/a | n/a | MUC16 mutation is associated with tumor grade, clinical features, and prognosis in glioma patients [38] | Used as a serum biomarker in ovarian cancer with implications for other cancers as well [50] | PDGFRA |
| CIC | n/a | n/a | n/a | 20% | CIC protein instability contributes to tumorigenesis in glioblastoma [40] | Not currently used in clinic | EGFR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tasci, E.; Jagasia, S.; Zhuge, Y.; Camphausen, K.; Krauze, A.V. GradWise: A Novel Application of a Rank-Based Weighted Hybrid Filter and Embedded Feature Selection Method for Glioma Grading with Clinical and Molecular Characteristics. Cancers 2023, 15, 4628. https://doi.org/10.3390/cancers15184628
Tasci E, Jagasia S, Zhuge Y, Camphausen K, Krauze AV. GradWise: A Novel Application of a Rank-Based Weighted Hybrid Filter and Embedded Feature Selection Method for Glioma Grading with Clinical and Molecular Characteristics. Cancers. 2023; 15(18):4628. https://doi.org/10.3390/cancers15184628
Chicago/Turabian StyleTasci, Erdal, Sarisha Jagasia, Ying Zhuge, Kevin Camphausen, and Andra Valentina Krauze. 2023. "GradWise: A Novel Application of a Rank-Based Weighted Hybrid Filter and Embedded Feature Selection Method for Glioma Grading with Clinical and Molecular Characteristics" Cancers 15, no. 18: 4628. https://doi.org/10.3390/cancers15184628
APA StyleTasci, E., Jagasia, S., Zhuge, Y., Camphausen, K., & Krauze, A. V. (2023). GradWise: A Novel Application of a Rank-Based Weighted Hybrid Filter and Embedded Feature Selection Method for Glioma Grading with Clinical and Molecular Characteristics. Cancers, 15(18), 4628. https://doi.org/10.3390/cancers15184628






