Simple Summary
Colonoscopies are effective in the prevention of colorectal cancer (CRC) but considered burdensome. An alternative for or addition to this procedure might be found in circulating tumor DNA (ctDNA) analysis. However, to date, it is not clear which analysis is most suitable for such a ctDNA-based blood test for CRC. Therefore, we assessed this in ten patients with colonoscopies for Lynch syndrome or in the context of the Dutch national screening program who were diagnosed with CRC or its precursor lesion (advanced adenoma). The results of this proof-of-principle study could form the foundation for subsequent studies on ctDNA-based blood test development for CRC screening and management, specifically in carriers of Lynch syndrome.
Abstract
Colorectal cancer (CRC) colonoscopic surveillance is effective but burdensome. Circulating tumor DNA (ctDNA) analysis has emerged as a promising, minimally invasive tool for disease detection and management. Here, we assessed which ctDNA assay might be most suitable for a ctDNA-based CRC screening/surveillance blood test. In this prospective, proof-of-concept study, patients with colonoscopies for Lynch surveillance or the National Colorectal Cancer screening program were included between 7 July 2019 and 3 June 2022. Blood was drawn, and if advanced neoplasia (adenoma with villous component, high-grade dysplasia, ≥10 mm, or CRC) was detected, it was analyzed for chromosomal copy number variations, single nucleotide variants, and genome-wide methylation (MeD-seq). Outcomes were compared with corresponding patients’ tissues and the MeD-seq results of healthy blood donors. Two Lynch carriers and eight screening program patients were included: five with CRC and five with advanced adenomas. cfDNA showed copy number variations and single nucleotide variants in one patient with CRC and liver metastases. Eight patients analyzed with MeD-seq showed clustering of Lynch-associated and sporadic microsatellite instable lesions separate from microsatellite stable lesions, as did healthy blood donors. In conclusion, whereas copy number changes and single nucleotide variants were only detected in one patient, cfDNA methylation profiles could discriminate all microsatellite instable advanced neoplasia, rendering this tool particularly promising for LS surveillance. Larger studies are warranted to validate these findings.
1. Introduction
Colorectal cancer (CRC) is the third-leading cause of cancer-related death worldwide [1]. For this purpose, an increasing number of developed countries implemented dedicated CRC screening programs. The Dutch National Colorectal Cancer screening program (SP) was initiated in 2014 and invites all Dutch inhabitants aged 55–75 biennially for a fecal immunohistochemical test (iFOBT), which is followed by colonoscopy in cases of positive results. This way of screening proved to be effective in detecting CRC and its precursor lesions, advanced adenoma (AA), and therefore, it contributes to a decrease in CRC incidence and mortality by detecting CRC earlier [2].
Besides this nationwide program, additional dedicated screening programs exist for individuals who are at high risk of developing CRC, such as carriers of a pathogenic variant in one of the DNA mismatch repair (MMR) genes, resulting in Lynch Syndrome (LS) [3,4,5,6,7]. LS carriers have up to a 55% chance of developing CRC during their lives, but exact risk estimates depend on the specific MMR gene involved [8,9,10]. A hallmark of LS-associated tumors is microsatellite instability (MSI-H), although this is also detected in a minority of sporadic CRCs [11,12]. To prevent CRC development, LS carriers are advised to undergo biennial colonoscopy [13]. This surveillance strategy proved to be effective in diminishing CRC-related deaths in LS carriers [14].
Despite its high efficiency, the iFOBT has a low sensitivity. Nearly half of the people who are advised to have a colonoscopy do not have colorectal anomalies after all. Similarly, the majority of LS carriers do not have CRC or AA during biennial colonoscopic surveillance. Additionally, colonoscopies are burdensome. Therefore, new, less invasive strategies for early CRC detection are warranted.
Cell-free DNA (cfDNA)-based blood tests seem to be particularly promising for this purpose [15]. cfDNA is shed into the circulation by dying cells and can thus also contain a fraction of circulating tumor DNA (ctDNA) [16]. For ctDNA detection in the total pool of cfDNA, a myriad of tests can be performed, including the assessment of single nucleotide variants (SNVs), chromosomal copy number variations (CNVs), microsatellite instability (MSI), and DNA methylation, or a combination of all the above, and ctDNA is often determined via a multitude of assays currently applied in various settings [17]. For example, tumor-specific SNV or methylation patterns detected in ctDNA correlate with prognosis [18,19] and can be used for disease monitoring [20,21,22], while genomic copy number instability (CNI) scores based on CNVs detected in the ctDNA are helpful in determining more tailored treatment strategies [23]. Theoretically, ctDNA analyses can also be used for cancer diagnosis; however, generally, more ctDNA is shed into the circulation in more advanced stages [24,25,26], indicating that a highly sensitive assay is required for this purpose. In addition, even though SNVs and short indels in cfDNA can be detected with high sensitivity and are predictive of CRC recurrence, these should ideally be known upfront to develop a patient-specific screening test and are thus less useful for primary cancer screening.
Ideally, cfDNA could even be used for the detection of precursor lesions to prevent cancer outgrowth and for locating the origin of this lesion. This is particularly of importance to LS carriers and other individuals with a hereditary predisposition to develop cancer. Specifically, cfDNA methylation profiles seem to be promising for this purpose. Previous research showed clustering of AA separate from healthy blood donors based on 11 methylation markers [27]. More insight into genome-wide methylation patterns may provide us with an alternative method for both colorectal and extra-colonic surveillance in hereditary predisposed patients. Conventional genome-wide methylation analyses are, however, extremely expensive at this moment and require high amounts of input DNA.
Recently, we presented a cheaper alternative to genome-wide methylation analysis (MeD-seq) [28] and showed that this method can also reliably be applied to lower amounts of cfDNA [29]. Therefore, we aimed to assess which assay or combination of assays would be most promising for the development of a cfDNA-based blood test (MeD-seq, CNV, and SNV analyses) to promptly detect AA and CRC in the context of the nationwide CRC screening program and LS surveillance.
2. Materials and Methods
For this proof-of-concept study, we approached participants of the National CRC screening program with a positive iFOBT, as well as molecularly proven LS carriers, with a planned colonoscopy in the Erasmus MC, Rotterdam (The Netherlands) between 7 July 2019 and 3 June 2022. The Erasmus MC Institutional Review Board approved this study (NL68955.078.19). Additionally, this study was reported in the International Clinical Trials Registry Platform (NL8695). The study procedures were performed as described in the Declaration of Helsinki. Written informed consent was obtained from every participant. Due to the explorative nature of this study, no sample size calculation was performed. The results of this study are intended as the foundation for a larger follow-up study.
2.1. Patients
Potential participants were informed about the study and the theoretical chance of finding unexpected findings. Upon informed consent, blood was drawn just before the colonoscopy was carried out via peripheral venous catheter, which is required for colonoscopy sedation; first, blood was collected in one mock EDTA tube, and subsequently, it was collected in two Streck Cell-Free BCT tubes (La Vista, NE, USA) [30]. The tubes were kept at room temperature and transferred to the laboratory as soon as possible (after a maximum of four days) for plasma isolation, as recommended by the manufacturer’s instructions. Patients with AA (adenomas ≥ 10 mm in size with a villous component or high-grade dysplasia) or CRC (advanced neoplasia) identified in the colonoscopy were analyzed. CRCs were routinely tested for the presence of MSI-H as the standard of care. However, some adenomas were stained for the presence of DNA mismatch repair proteins (MMR IHC). For readability, only the terms MSI-H and microsatellite-stable were used. Lesions with all four DNA MMR proteins present were classified as MSI-stable, whereas the absence of at least one DNA MMR protein was classified as MSI-H.
2.2. Plasma and cfDNA Handling
Plasma isolation was performed as described previously [31]. Plasma samples were stored in up to six 2 mL vials per participant at −80 °C. Only in cases in which participants were diagnosed with an advanced adenoma or CRC, the participants were included for cfDNA analyses. The cfDNA of these patients was isolated from stored plasma, as described previously, using the QIAamp Circulating Nucleic Acid Kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany) [29]. the obtained cfDNA was analyzed for CNVs, tumor-specific SNVs, and genome-wide methylation profiles.
2.3. CNV, SNV, and MeD-Seq Analyses
2.3.1. CNVs
To assess the presence of CNVs in the cfDNA, an automated NGS workflow was performed using the VeriSeq (Illumina, Cambridge, UK) and the Microlab Star Robot (Hamilton, Gräfelfing, Germany) according to the manufacturer’s instructions, as described before [31]. In brief, 1 mL of plasma was used for analyses. A unique synthetic DNA ‘barcode’ (index) was attached to each sample, and the library product was quantified using a fluorescent dye and compared to a DNA standard curve. Lastly, shallow whole genome sequencing was performed on a NextSeq500 sequencer, yielding 2 × 36 paired-end reads in a 48-plex reaction. The SeqFF model was used as a surrogate marker to assess the percentage of short fragmented cfDNA that was likely of tumor origin (ctDNA) in the total cfDNA pool (herein called the percentage tumor cfDNA) [32]. Additionally, ctDNA in the pool of cfDNA was estimated using ichorCNA software version 0.2.0 [33]. Outcomes were analyzed with WISECONDOR statistical software version 2.0.1 [34]. Next, cfDNA fragment sizes were assessed.
2.3.2. SNVs
To assess whether SNVs were present in the cfDNA, 10 ng cfDNA was analyzed with the Next Generation Sequencing (NGS) OncomineTM Colon cfDNA Assay, as described previously, using variantCaller version v5.10.0 in cases of at least three or more unique molecules [35]. Sequencing for diagnostic purposes aimed for at least 25,000 reads. This panel covers the following genes with >240 hotspots: AKT1, APC, BRAF, CTNNB1, EGFR, ERBB2, FBXW7, GNAS, KRAS, MAP2K1, NRAS, PIK3CA, SMAD4, and TP53.
Formalin-fixed paraffin-embedded (FFPE) tissue, obtained as the standard of care, was retrieved for patients included in this study. The tissue was analyzed by means of NGS analysis to assess the presence of tumor-specific SNVs using the Ion Torrent S5 sequencing system (ThermoFisher, Waltham, MA, USA). For this, the Ion Ampliseq Library Plus Kit 2.0 was used, with a custom-made primer panel including the same 14 genes as the cfDNA Assay panel [36]. Templates were prepared using the Ion Chef with the 540 Chef kit. Data were analyzed using variantCaller v5.10.0, aiming for 1250 reads.
2.3.3. MeD-Seq
Methylation patterns in cfDNA were analyzed by MeD-seq, as described previously [28]. In short, MeD-seq uses a methylation-dependent restriction enzyme (LpnPI) to generate 32 base-pair-long (methylated) DNA fragments for sequencing. For MeD-seq analyses, up to 25 ng of input DNA was used. Input DNA was end-repaired and ligated to dual-indexed adaptors. The resulting libraries were sequenced on the Illumina HiSeq 2500 sequencer, yielding single read 50 base pair reads. All patients were analyzed with MeD-seq, except for CATCA006 and CATCA044, as there was insufficient cfDNA remaining for MeD-seq analysis. The MeD-seq results were compared to those of healthy blood donors (HBDs).
2.3.4. MeD-Seq Data Processing and Analysis
Subsequent data processing was carried out as previously described and included a filtering step based on LpnPI restriction site occurrence between 13 and 17 bp from the 5’ or 3’ end of the read, after which only reads originating from methylated DNA fragments remained [28,29]. Using all unambiguously mapped reads, count scores were assigned to each individual LpnPI site in the human genome. Subsequently, count scores for individual CpG sites were summarized into 2 kilobase (kb) regions surrounding all known transcription start sites (TSS) annotated in ENSEMBL. Only regions containing data in at least 50% of all samples were included in subsequent analyses, resulting in a total of 41,570 regions on chromosomes 1–22. Data were normalized to the total number of reads passing the LpnPI filter per sample, after which square root transformation was applied to reduce skewness in the data distribution.
Differentially methylated regions (DMRs) between patients and HBDs were identified using LIMMA, resulting in 2798 regions [37].
Principal component analysis (PCA) was performed using the singular value decomposition function (svd) in the R base package (the R project for statistical computing) on mean-centered MeD-seq values from either the most variable regions (for example, regions showing a standard deviation over all samples that is larger than the median standard deviation observed for all regions) or the DMRs. The resulting principal components were uncorrelated summary statistics, each reflecting the methylation data for a mixture of correlated genomic regions. To generate an overall score for aberrant cfDNA methylation per patient, Z-scores were calculated per region for every patient relative to our normal control panel of 12 HBDs. These Z-scores per region were squared and summed into a methylation score, as described before [28,29].
3. Results
3.1. Patients
Of the 211 patients willing to participate, ten patients were diagnosed with AA or CRC and were thus included in this study (Table 1 and Table S1). Of these patients, eight participated in the Dutch national CRC screening program and were referred to the Erasmus MC for a colonoscopy after a positive iFOBT. Two patients were LS carriers with a biennial surveillance colonoscopy (CATCA038 and CATCA099). Of these patients, five presented with CRC (with stages ranging from TxN0M0 to T3NxM1). One of the CRCs (CATCA008) was found to be MSI-H due to sporadic MLH1 promoter hypermethylation; the other four CRCs were found to be microsatellite-stable. The remaining five patients presented with AA; one of them turned out to be MSI-H (LS carrier).

Table 1.
Baseline characteristics of analyzed patients and overview of analyses performed.
3.2. Single Nucleotide Variants (SNVs) in Tissue and Corresponding Plasma cfDNA
Tumor tissue from all included patients harbored at least one mutation covered by the cfDNA Oncomine panel (Table 1 and Table 2). Two patients showed abnormal SNVs in the plasma cfDNA. In patient CATCA016, who presented with metastasized CRC, concordance was observed between tissue and cfDNA for two somatic variants (Table 1). Patient CATCA008 presented with an SNV in the cfDNA that was not present in the tissue, despite being covered in both sequencing panels. Since only five molecules were detected, this might have been an artifact. Alternatively, this observation could be due to tumor heterogeneity, clonal hematopoiesis, or sampling error [38,39]. In the remaining six patients, no alterations were detected in cfDNA.

Table 2.
Outcomes of Next-Generation Sequencing (NGS) panels on tissue and cfDNA isolated from blood plasma in each patient.
3.3. cfDNA CNV and Fragment Size Analysis
Unbalanced chromosome aberrations were convincingly detected in CATCA016 but not in any of the other patients (Supplementary Figure S1). For an overview of the CNVs, please see Table 3. Of note, cfDNA fragment size comparisons in patients with CRC or AA were hampered by the small sample size and low cfDNA levels, but they were not found to be smaller than in healthy controls (Figure S2). Nevertheless, the quantile 5% analyses showed a slightly elevated level of insert size < 130 bp in the AA group in comparison to controls (Figure S2B).

Table 3.
Copy number variations (CNVs) detected in cfDNA, isolated from blood.
3.4. Genome-Wide cfDNA Methylation Profiling
For eight patients, plasma was subjected to MeD-seq; for the remaining two patients, insufficient cfDNA was left to perform this analysis. Genome-wide cfDNA methylation profiles were successfully generated for these patients as well as for 12 healthy blood donors (HBDs). Exploratory unsupervised analyses showed a clear distinction between the two LS-associated AA and the sporadic MSI-H CRC on the one hand and the MSS lesions and HBDs on the other hand (Figure S3).
Using a supervised approach in which we only included differentially methylated regions between the eight patients included in our study on the one hand and HBDs on the other hand (2798 DMRs with an unadjusted p-value of <0.05 in LIMMA analysis, Table S2), we observed a clear separation between LS-associated and/or MSI-H lesions, versus MSS lesions versus HBDs (Figure 1).
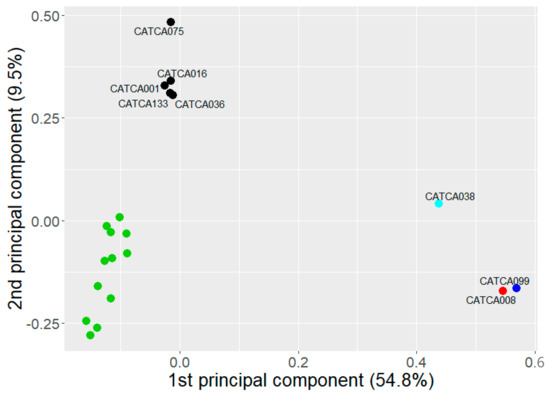
Figure 1.
Principal component analysis (PCA) for patients included in our study and healthy blood donors (HBDs, in green) based on the DMRs, with on each axis the percentage of variation that can be explained by that specific principal component. The patient with MSI-H CRC and LS carriers with AA are depicted in red and blue, respectively, while sporadic MSS lesions are shown in black.
To facilitate the use of this assay for screening or surveillance, we need to capture the methylation profile in a single score. For this purpose, we calculated a summary methylation score based on either the entire genome (genome-wide summary score) or the 2798 DMRs between patients and HBDs (DMR summary score; see the methods section for more information about this calculation). The resulting scores are shown in Figure 2 and Table S3. Genome-wide summary scores above 2.34 and DMR summary scores above 2.25 were considered elevated, which represented the upper 95% CI bound of the HBDs. As expected from the results above, the genome-wide summary score only detected the LS-associated AA and sporadic MSI-H CRC, whereas the DMR summary score was elevated in seven out of eight patients (Figure 2, Table S3).
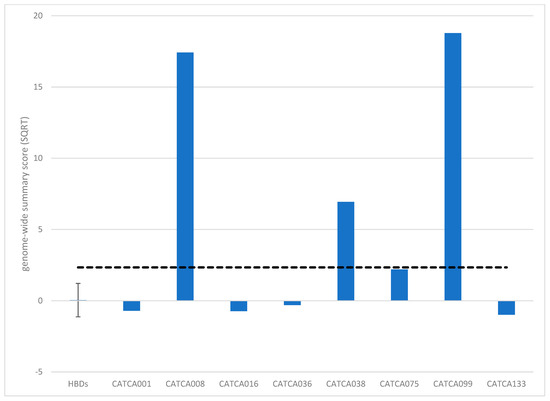
Figure 2.
Summary methylation scores upon genome-wide methylation analysis (MeD-seq) based on the entire genome (genome-wide summary score). Genome-wide summary scores above 2.34 were considered elevated (dashed line).
4. Discussion
In this proof-of-principle study, we assessed to what extent different cfDNA analysis methods could be used in screening and surveillance for AA and CRC. SNVs and CNVs were only detected in the plasma of one patient presenting with liver metastases at diagnosis. By contrast, genome-wide cfDNA methylation profiling (MeD-seq) followed by supervised data analysis could distinguish advanced neoplasia samples from HBDs. This suggests that genome-wide cfDNA methylation profiling could sensitively reflect tumor-specific anomalies, and as such, it may represent a promising and minimally invasive tool for CRC screening and surveillance. Interestingly, both LS-associated AA and the sporadic MSI-H CRC showed very distinct cfDNA methylation profiles, particularly highlighting the potential of cfDNA methylation profiling for LS surveillance.
4.1. SNVs
We did not find tumor-specific SNVs in cfDNA that were present in matched tumor tissue, except for one patient with metastasized CRC. This is in line with previous studies, which have also found that ctDNA levels increase with the CRC stage [33,40]. Even though we performed SNV analyses using a sensitive and dedicated CRC panel including unique molecular identifiers, we could not trace these SNVs in the blood, except for the patient with metastasized CRC. As SNVs were detected in the matched primary FFPE tissue of all patients included, the lack of detection in the plasma is most likely due to the extremely low levels of ctDNA present in the blood of early-stage CRCs and AA. The detection of a larger number of SNVs in a single patient is expected to increase the sensitivity, but it requires prior knowledge of the molecular characteristics of the tumor. This is followed by the design of patient-specific assays, which renders this approach infeasible for general CRC screening and high-risk population surveillance.
4.2. CNVs
Results similar to SNVs were observed for CNVs: in only one patient, an abnormal CNV pattern was identified. The literature indicates reliable CNV analysis in plasma requires at least 5% ctDNA within the total pool of cfDNA, which may explain our results [41]. According to our data, the CNV cfDNA analyses using our current approach do not seem to be a suitable option for CRC screening or surveillance. However, more sensitive methods have emerged in the field of oncology to assess cfDNA CNVs in the last few years; with so-called fragmentomics analyses, one specifically focuses on cfDNA fragments of certain sizes to more closely assess ctDNA [42,43]. Due to the limited sample size, we were unable to assess the added value of fragmentomics in the current dataset. Nevertheless, the quantile 5% analyses showed a slightly elevated level of insert size < 130 bp in the AA group in comparison with controls. Although the current results do not support CNV analysis of cfDNA as a promising approach for CRC screening and surveillance of high-risk individuals, bioinformatics analyses are continuously being improved, possibly enabling more sensitive CNV analyses in cfDNA in future research.
4.3. Genome-Wide Methylation
In contrast to the other assays performed, genome-wide methylation profiling by MeD-seq on blood plasma-derived cfDNA discriminated MSS advanced neoplasia samples from both MSI-H and LS-driven neoplasia and HBDs. This is in line with the known differences in phenotype between MSS and MSI-driven CRC lesions (chromosomal instability, CIN, versus CpG island methylator phenotype, CIMP, often accompanied by MSI-H) [44,45,46]. Larger studies should be performed to assess to what extent this holds true and to determine and validate the most informative DMRs.
4.4. Application of MeD-Seq in Lynch Syndrome Surveillance
Interestingly, we found that MSI-H - and LS-associated advanced neoplasia clustered together based on their methylation profiles. Of note, even the advanced adenoma in the LS carrier clustered more toward the sporadic and LS-associated MSI-H lesions, despite still being microsatellite-stable. Therefore, this tool might be particularly promising in LS CRC surveillance, although larger studies are needed to validate the performance of this assay in larger groups of LS patients, and sporadic microsatellite stable and MSI-H lesions. MeD-seq cfDNA profiling might contribute to the decision of who needs further colonoscopic assessment in the context of LS colorectal cancer surveillance. Additionally, we need to explore whether extra-colonic LS-associated tumors can also be detected by MeD-seq to establish whether cfDNA methylation profiling represents a promising tool to use in surveillance for all LS-associated tumors. Previous research pointed out that methylation profiles contain information about the tissue of origin, enabling further work-up and treatment [47]. In this case, all surveillance for LS carriers could be performed by biennial cfDNA methylation assessment.
4.5. Limitations
Our study has several limitations. First, due to the proof-of-principle nature of this study, we had a small number of patients with CRC or AA with a mixed etiology (LS versus sporadic). Due to the small and heterogeneous patient group, we do not know to what extent the DMRs identified here are representative of the different populations of interest.
Future studies focusing on the added value of cfDNA surveillance in LS patients should consist of a multicenter approach to increase the number of patients. Second, because our study explored multiple techniques, for two patients, we unfortunately had too little material to perform all envisioned assays. On the other hand, this underlines the need for assays requiring a limited amount of input while generating a rich data output (for example combining methylation with SNVs and/or CNVs) [48,49]. Third, CNVs and methylation were not assessed in these patients’ tissues. However, previous research showed that for a selection of genes associated with metastasized CRC, the profiles matched the tumor tissue [29]. Last, although HBDs were obtained from the healthy population, no information was available on iFOBT results and/or recent colonoscopy, meaning we cannot rule out the presence of advanced neoplasia without complaints in these individuals.
5. Conclusions
In conclusion, this proof-of-principle study highlights the promise of cfDNA methylation profiling, specifically in the context of LS surveillance, and provides a starting point for future research in this area.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15184607/s1, Figure S1: cfDNA CNV Wisecondor profile of patient CATCA016. This patient had colorectal cancer with liver metastases; Figure S2: (A) Median insert sizes and (B) quantile 5% insert sizes in cfDNA of patients with advanced adenoma (green), colorectal cancer (blue), and controls (red); Figure S3: Principal component analysis (PCA) of patient samples analyzed with genome-wide methylation analysis (MeD-seq) for patients included in our study and healthy blood donors (HBDs, in green); Table S1: Patient characteristics of patients included for cell-free DNA analyses; Table S2: List of differentially methylated regions; Table S3: Summary methylation scores upon genome-wide methylation analysis (MeD-seq).
Author Contributions
Conceptualization: E.L.E., S.M.W., M.C.W.S. and A.W.; data curation: E.L.E. and S.M.W.; formal analysis S.M.W., M.I.S. and P.A., M.V.V.-P., R.G.B. and J.B.B.; investigation: E.L.E., S.M.W., M.C.W.S. and A.W.; methodology: E.L.E., S.M.W., T.D., M.I.S., M.V.V.-P., J.H.G., P.A., H.J.D., J.W.M.M. and A.W.; project administration: E.L.E. and A.W.; resources: S.M.W., T.D., M.I.S., M.V.V.-P., R.G.B., J.B.B., W.F.J.v.I., J.H.G., P.A., H.J.D. and J.W.M.M.; supervision: S.M.W., M.C.W.S. and A.W.; writing—original draft E.L.E., S.M.W. and A.W.; writing—review and editing E.L.E., S.M.W., T.D., M.I.S., M.V.V.-P., R.G.B., J.B.B., W.F.J.v.I., J.H.G., P.A., H.J.D., J.W.M.M., M.C.W.S. and A.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding; internal funding was obtained (MRACE: the Eramus MC MRACE internal Grant for pilot studies).
Institutional Review Board Statement
The Erasmus MC Institutional Review Board approved this study (NL68955.078.19). Additionally, this study was reported in the International Clinical Trials Registry Platform (NL8695).
Informed Consent Statement
Study procedures were performed as described in the Declaration of Helsinki. Written informed consent was obtained from every participant.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Acknowledgments
We would like to thank Agnes Reijm, Sophia van Baalen-van Daalen, and Judith E. Prins-Cornelisse for their cooperation.
Conflicts of Interest
John W.M. Martens reports a fee from Novartis and grant or co-funding in the last two decades from Johnson & Johnson, GlaxoSmithKline, Pfizer, Philips, Menarini, Cergentis, and Therawis. Ruben G. Boers, Joachim B. Boers, Wilfred F.J Van IJcken, and Joost H. Gribnau are shareholders in Methylomics B.V., a commercial company that applies MeD-seq to develop methylation markers for cancer staging. The remaining authors have no conflict of interest to declare.
Abbreviations
| AA | advanced adenoma |
| AN | advanced neoplasia |
| CNV | copy number variation |
| CRC | colorectal cancer |
| ctDNA | circulating tumor DNA |
| DMR | differentially methylated region |
| FFPE | Formalin-fixed paraffin-embedded |
| HBD | healthy blood donor |
| HGD | high-grade dysplasia |
| IFOBT | fecal immunohistochemical test |
| LS | Lynch syndrome |
| MeD-seq | genome-wide methylation |
| MMR | mismatch repair |
| MMRp | mismatch repair proficient |
| MSI-H | microsatellite instability-high |
| MSS | microsatellite stable |
| SNV | single nucleotide variant |
| SP | national colorectal cancer screening program |
| TVA | tubulovillous adenoma |
| VAF | variant allele frequency |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Toes-Zoutendijk, E.; van Leerdam, M.E.; Dekker, E.; van Hees, F.; Penning, C.; Nagtegaal, I.; van der Meulen, M.P.; van Vuuren, A.J.; Kuipers, E.J.; Bonfrer, J.M.G.; et al. Real-Time Monitoring of Results During First Year of Dutch Colorectal Cancer Screening Program and Optimization by Altering Fecal Immunochemical Test Cut-Off Levels. Gastroenterology 2017, 152, 767–775.e2. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, Y.; Sato, H.; Yamada, T.; Nagasaki, H.; Tsuchiya, A.; Abe, R.; Yuasa, Y. Germ-line mutation of the hMSH6/GTBP gene in an atypical hereditary nonpolyposis colorectal cancer kindred. Cancer Res. 1997, 57, 3920–3923. [Google Scholar] [PubMed]
- Bronner, C.E.; Baker, S.M.; Morrison, P.T.; Warren, G.; Smith, L.G.; Lescoe, M.K.; Kane, M.; Earabino, C.; Lipford, J.; Lindblom, A.; et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature 1994, 368, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Fishel, R.; Lescoe, M.K.; Rao, M.R.; Copeland, N.G.; Jenkins, N.A.; Garber, J.; Kane, M.; Kolodner, R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 1993, 75, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Nicolaides, N.C.; Papadopoulos, N.; Liu, B.; Wei, Y.F.; Carter, K.C.; Ruben, S.M.; Rosen, C.A.; Haseltine, W.A.; Fleischmann, R.D.; Fraser, C.M.; et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 1994, 371, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Ligtenberg, M.J.; Kuiper, R.P.; Chan, T.L.; Goossens, M.; Hebeda, K.M.; Voorendt, M.; Lee, T.Y.; Bodmer, D.; Hoenselaar, E.; Hendriks-Cornelissen, S.J.; et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat. Genet. 2009, 41, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Moller, P.; Seppala, T.; Bernstein, I.; Holinski-Feder, E.; Sala, P.; Evans, D.G.; Lindblom, A.; Macrae, F.; Blanco, I.; Sijmons, R.; et al. Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: First report from the prospective Lynch syndrome database. Gut 2017, 66, 464–472. [Google Scholar] [CrossRef]
- Dominguez-Valentin, M.; Sampson, J.R.; Seppala, T.T.; Ten Broeke, S.W.; Plazzer, J.P.; Nakken, S.; Engel, C.; Aretz, S.; Jenkins, M.A.; Sunde, L.; et al. Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: Findings from the Prospective Lynch Syndrome Database. Genet. Med. 2020, 22, 15–25. [Google Scholar] [CrossRef]
- Møller, P.; Seppälä, T.T.; Bernstein, I.; Holinski-Feder, E.; Sala, P.; Gareth Evans, D.; Lindblom, A.; Macrae, F.; Blanco, I.; Sijmons, R.H.; et al. Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: A report from the Prospective Lynch Syndrome Database. Gut 2018, 67, 1306–1316. [Google Scholar] [CrossRef]
- Goel, A.; Nagasaka, T.; Arnold, C.N.; Inoue, T.; Hamilton, C.; Niedzwiecki, D.; Compton, C.; Mayer, R.J.; Goldberg, R.; Bertagnolli, M.M.; et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology 2007, 132, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Taieb, J.; Svrcek, M.; Cohen, R.; Basile, D.; Tougeron, D.; Phelip, J.M. Deficient mismatch repair/microsatellite unstable colorectal cancer: Diagnosis, prognosis and treatment. Eur. J. Cancer 2022, 175, 136–157. [Google Scholar] [CrossRef] [PubMed]
- IKNL. Guideline Hereditary Colorectal Cancer. December 2015. Available online: www.oncoline.nl (accessed on 5 May 2022).
- Jarvinen, H.J.; Renkonen-Sinisalo, L.; Aktan-Collan, K.; Peltomaki, P.; Aaltonen, L.A.; Mecklin, J.P. Ten years after mutation testing for Lynch syndrome: Cancer incidence and outcome in mutation-positive and mutation-negative family members. J. Clin. Oncol. 2009, 27, 4793–4797. [Google Scholar] [CrossRef] [PubMed]
- Malla, M.; Loree, J.M.; Kasi, P.M.; Parikh, A.R. Using Circulating Tumor DNA in Colorectal Cancer: Current and Evolving Practices. J. Clin. Oncol. 2022, 40, 2846–2857. [Google Scholar] [CrossRef] [PubMed]
- Mazouji, O.; Ouhajjou, A.; Incitti, R.; Mansour, H. Updates on Clinical Use of Liquid Biopsy in Colorectal Cancer Screening, Diagnosis, Follow-Up, and Treatment Guidance. Front. Cell Dev. Biol. 2021, 9, 660924. [Google Scholar] [CrossRef] [PubMed]
- Dasari, A.; Morris, V.K.; Allegra, C.J.; Atreya, C.; Benson, A.B., 3rd; Boland, P.; Chung, K.; Copur, M.S.; Corcoran, R.B.; Deming, D.A.; et al. ctDNA applications and integration in colorectal cancer: An NCI Colon and Rectal-Anal Task Forces whitepaper. Nat. Rev. Clin. Oncol. 2020, 17, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Schraa, S.J.; van Rooijen, K.L.; Koopman, M.; Vink, G.R.; Fijneman, R.J.A. Cell-Free Circulating (Tumor) DNA before Surgery as a Prognostic Factor in Non-Metastatic Colorectal Cancer: A Systematic Review. Cancers 2022, 14, 2218. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Zhao, Q.; Wei, W.; Zheng, L.; Yi, S.; Li, G.; Wang, W.; Sheng, H.; Pu, H.; Mo, H.; et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci. Transl. Med. 2020, 12, eaax7533. [Google Scholar] [CrossRef]
- Bent, A.; Raghavan, S.; Dasari, A.; Kopetz, S. The Future of ctDNA-Defined Minimal Residual Disease: Personalizing Adjuvant Therapy in Colorectal Cancer. Clin. Color. Cancer 2022, 21, 89–95. [Google Scholar] [CrossRef]
- Zviran, A.; Schulman, R.C.; Shah, M.; Hill, S.T.K.; Deochand, S.; Khamnei, C.C.; Maloney, D.; Patel, K.; Liao, W.; Widman, A.J.; et al. Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat. Med. 2020, 26, 1114–1124. [Google Scholar] [CrossRef]
- Barault, L.; Amatu, A.; Siravegna, G.; Ponzetti, A.; Moran, S.; Cassingena, A.; Mussolin, B.; Falcomatà, C.; Binder, A.M.; Cristiano, C.; et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut 2018, 67, 1995–2005. [Google Scholar] [CrossRef] [PubMed]
- Oellerich, M.; Schütz, E.; Beck, J.; Kanzow, P.; Plowman, P.N.; Weiss, G.J.; Walson, P.D. Using circulating cell-free DNA to monitor personalized cancer therapy. Crit. Rev. Clin. Lab. Sci. 2017, 54, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Mathios, D.; Johansen, J.S.; Cristiano, S.; Medina, J.E.; Phallen, J.; Larsen, K.R.; Bruhm, D.C.; Niknafs, N.; Ferreira, L.; Adleff, V.; et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat. Commun. 2021, 12, 5060. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Toung, J.M.; Jassowicz, A.F.; Vijayaraghavan, R.; Kang, H.; Zhang, R.; Kruglyak, K.M.; Huang, H.J.; Hinoue, T.; Shen, H.; et al. Targeted methylation sequencing of plasma cell-free DNA for cancer detection and classification. Ann. Oncol. 2018, 29, 1445–1453. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra224. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhang, Y.; Hu, T.; He, X.; Zou, Y.; Deng, Q.; Ke, J.; Lian, L.; He, X.; Zhao, D.; et al. A novel cell-free DNA methylation-based model improves the early detection of colorectal cancer. Mol. Oncol. 2021, 15, 2702–2714. [Google Scholar] [CrossRef] [PubMed]
- Boers, R.; Boers, J.; de Hoon, B.; Kockx, C.; Ozgur, Z.; Molijn, A.; van IJckenvan, W.; Laven, J.; Gribnau, J. Genome-wide DNA methylation profiling using the methylation-dependent restriction enzyme LpnPI. Genome Res. 2018, 28, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Deger, T.; Boers, R.G.; de Weerd, V.; Angus, L.; van der Put, M.M.J.; Boers, J.B.; Azmani, Z.; van IJcken, W.F.J.; Grünhagen, D.J.; van Dessel, L.F.; et al. High-throughput and affordable genome-wide methylation profiling of circulating cell-free DNA by methylated DNA sequencing (MeD-seq) of LpnPI digested fragments. Clin. Epigenetics 2021, 13, 196. [Google Scholar] [CrossRef]
- Medina Diaz, I.; Nocon, A.; Mehnert, D.H.; Fredebohm, J.; Diehl, F.; Holtrup, F. Performance of Streck cfDNA Blood Collection Tubes for Liquid Biopsy Testing. PLoS ONE 2016, 11, e0166354. [Google Scholar] [CrossRef]
- van der Meij, K.R.M.; Sistermans, E.A.; Macville, M.V.E.; Stevens, S.J.C.; Bax, C.J.; Bekker, M.N.; Bilardo, C.M.; Boon, E.M.J.; Boter, M.; Diderich, K.E.M.; et al. TRIDENT-2: National Implementation of Genome-wide Non-invasive Prenatal Testing as a First-Tier Screening Test in the Netherlands. Am. J. Hum. Genet. 2019, 105, 1091–1101. [Google Scholar] [CrossRef]
- Kim, S.K.; Hannum, G.; Geis, J.; Tynan, J.; Hogg, G.; Zhao, C.; Jensen, T.J.; Mazloom, A.R.; Oeth, P.; Ehrich, M.; et al. Determination of fetal DNA fraction from the plasma of pregnant women using sequence read counts. Prenat. Diagn. 2015, 35, 810–815. [Google Scholar] [CrossRef] [PubMed]
- Adalsteinsson, V.A.; Ha, G.; Freeman, S.S.; Choudhury, A.D.; Stover, D.G.; Parsons, H.A.; Gydush, G.; Reed, S.C.; Rotem, D.; Rhoades, J.; et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 2017, 8, 1324. [Google Scholar] [CrossRef] [PubMed]
- Straver, R.; Sistermans, E.A.; Holstege, H.; Visser, A.; Oudejans, C.B.; Reinders, M.J. WISECONDOR: Detection of fetal aberrations from shallow sequencing maternal plasma based on a within-sample comparison scheme. Nucleic Acids Res. 2014, 42, e31. [Google Scholar] [CrossRef] [PubMed]
- Steendam, C.M.J.; Atmodimedjo, P.; de Jonge, E.; Paats, M.S.; van der Leest, C.; Oomen-de Hoop, E.; Jansen, M.; Del Re, M.; von der Thüsen, J.H.; Dinjens, W.N.M.; et al. Plasma Cell-Free DNA Testing of Patients with EGFR Mutant Non-Small-Cell Lung Cancer: Droplet Digital PCR Versus Next-Generation Sequencing Compared with Tissue-Based Results. JCO Precis. Oncol. 2019, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hermans, B.C.M.; Derks, J.L.; Hillen, L.M.; van der Baan, I.; van den Broek, E.C.; von der Thüsen, J.H.; van Suylen, R.J.; Atmodimedjo, P.N.; den Toom, T.D.; Coumans-Stallinga, C.; et al. In-depth molecular analysis of combined and co-primary pulmonary large cell neuroendocrine carcinoma and adenocarcinoma. Int. J. Cancer 2022, 150, 802–815. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Reinert, T.; Henriksen, T.V.; Christensen, E.; Sharma, S.; Salari, R.; Sethi, H.; Knudsen, M.; Nordentoft, I.; Wu, H.T.; Tin, A.S.; et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients with Stages I to III Colorectal Cancer. JAMA Oncol. 2019, 5, 1124–1131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yao, Y.; Xu, Y.; Li, L.; Gong, Y.; Zhang, K.; Zhang, M.; Guan, Y.; Chang, L.; Xia, X.; et al. Pan-cancer circulating tumor DNA detection in over 10,000 Chinese patients. Nat. Commun. 2021, 12, 11. [Google Scholar] [CrossRef]
- Janku, F.; Zhang, S.; Waters, J.; Liu, L.; Huang, H.J.; Subbiah, V.; Hong, D.S.; Karp, D.D.; Fu, S.; Cai, X.; et al. Development and Validation of an Ultradeep Next-Generation Sequencing Assay for Testing of Plasma Cell-Free DNA from Patients with Advanced Cancer. Clin. Cancer Res. 2017, 23, 5648–5656. [Google Scholar] [CrossRef]
- Belic, J.; Koch, M.; Ulz, P.; Auer, M.; Gerhalter, T.; Mohan, S.; Fischereder, K.; Petru, E.; Bauernhofer, T.; Geigl, J.B.; et al. Rapid Identification of Plasma DNA Samples with Increased ctDNA Levels by a Modified FAST-SeqS Approach. Clin. Chem. 2015, 61, 838–849. [Google Scholar] [CrossRef]
- Ding, S.C.; Lo, Y.M.D. Cell-Free DNA Fragmentomics in Liquid Biopsy. Diagnostics 2022, 12, 978. [Google Scholar] [CrossRef] [PubMed]
- Mouliere, F.; Chandrananda, D.; Piskorz, A.M.; Moore, E.K.; Morris, J.; Ahlborn, L.B.; Mair, R.; Goranova, T.; Marass, F.; Heider, K.; et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 2018, 10, eaat4921. [Google Scholar] [CrossRef] [PubMed]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087.e3. [Google Scholar] [CrossRef] [PubMed]
- Camps, J.; Armengol, G.; del Rey, J.; Lozano, J.J.; Vauhkonen, H.; Prat, E.; Egozcue, J.; Sumoy, L.; Knuutila, S.; Miro, R. Genome-wide differences between microsatellite stable and unstable colorectal tumors. Carcinogenesis 2006, 27, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Grady, W.M.; Carethers, J.M. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 2008, 135, 1079–1099. [Google Scholar] [CrossRef] [PubMed]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019, 570, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Katsman, E.; Orlanski, S.; Martignano, F.; Fox-Fisher, I.; Shemer, R.; Dor, Y.; Zick, A.; Eden, A.; Petrini, I.; Conticello, S.G.; et al. Detecting cell-of-origin and cancer-specific methylation features of cell-free DNA from Nanopore sequencing. Genome Biol. 2022, 23, 158. [Google Scholar] [CrossRef]
- Wei, Q.; Jin, C.; Wang, Y.; Guo, S.; Guo, X.; Liu, X.; An, J.; Xing, J.; Li, B. A computational framework to unify orthogonal information in DNA methylation and copy number aberrations in cell-free DNA for early cancer detection. Brief. Bioinform. 2022, 23, bbac200. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).