Concordance of Targeted Sequencing from Circulating Tumor DNA and Paired Tumor Tissue for Early Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Targeted Sequencing Panel
2.3. Nucleic Acid Extraction
2.4. Library Preparation and Variant Calling
3. Results
3.1. Detection Rate of Liquid Biopsy (Full Cohort)
3.2. Early-Stage Breast Cancer Cohort: Concordance between Tissue and Liquid Biopsy
3.3. Early-Stage Breast Cancer Cohort: Actionable Mutations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Gouda, M.A.; Huang, H.J.; Piha-Paul, S.A.; Call, S.G.; Karp, D.D.; Fu, S.; Naing, A.; Subbiah, V.; Pant, S.; Dustin, D.J.; et al. Longitudinal Monitoring of Circulating Tumor DNA to Predict Treatment Outcomes in Advanced Cancers. JCO Precis. Oncol. 2022, 6, e2100512. [Google Scholar] [CrossRef] [PubMed]
- Gerratana, L.; Movarek, M.; Wehbe, F.; Katam, N.; Mahalingam, D.; Donahue, J.; Shah, A.; Chae, Y.K.; Mulcahy, M.; Tsarwhas, D.; et al. Genomic Landscape of Advanced Solid Tumors in Circulating Tumor DNA and Correlation With Tissue Sequencing: A Single Institution’s Experience. JCO Precis. Oncol. 2022, 6, e2100289. [Google Scholar] [CrossRef] [PubMed]
- Page, K.; Martinson, L.J.; Fernandez-Garcia, D.; Hills, A.; Gleason, K.L.T.; Gray, M.C.; Rushton, A.J.; Nteliopoulos, G.; Hastings, R.K.; Goddard, K.; et al. Circulating Tumor DNA Profiling From Breast Cancer Screening Through to Metastatic Disease. JCO Precis. Oncol. 2021, 5, 1768–1776. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.M.; Tsui, D.W.Y. Circulating cell-free DNA for non-invasive cancer management. Cancer Genet. 2018, 228–229, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Attard, G.; Bidard, F.C.; Curigliano, G.; De Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.; Montagut, C.; et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022, 33, 750–768. [Google Scholar] [CrossRef]
- Liu, C.Y.; Huang, C.C.; Tsai, Y.F.; Chao, T.C.; Lien, P.-J.; Lin, Y.-S.; Feng, C.-J.; Chen, J.-L.; Chen, Y.-J.; Chiu, J.-H.; et al. VGH-TAYLOR: Comprehensive precision medicine study protocol on the heterogeneity of Taiwanese breast cancer patients. Futur. Oncol. 2021, 17, 4057–4069. [Google Scholar] [CrossRef]
- Huang, C.C.; Tsai, Y.F.; Liu, C.Y.; Chao, T.C.; Lien, P.-J.; Lin, Y.-S.; Feng, C.-J.; Chiu, J.-H.; Hsu, C.-Y.; Tseng, L.-M. Comprehensive molecular profiling of Taiwanese breast cancers revealed potential therapeutic targets: Prevalence of actionable mutations among 380 targeted sequencing analyses. BMC Cancer 2021, 21, 199. [Google Scholar] [CrossRef]
- Huang, C.C.; Tsai, Y.F.; Liu, C.Y.; Lien, P.J.; Lin, Y.-S.; Chao, T.-C.; Feng, C.-J.; Chen, Y.-J.; Lai, J.-I.; Phan, N.N.; et al. Prevalence of Tumor Genomic Alterations in Homologous Recombination Repair Genes Among Taiwanese Breast Cancers. Ann. Surg. Oncol. 2022, 29, 3578–3590. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.E.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.; Chakravarty, D.; Dienstmann, R.; Jezdic, S.; Gonzalez-Perez, A.; Lopez-Bigas, N.; Ng, C.; Bedard, P.; Tortora, G.; Douillard, J.-Y.; et al. A framework to rank genomic alterations as targets for cancer precision medicine: The ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann. Oncol. 2018, 29, 1895–1902. [Google Scholar] [CrossRef] [PubMed]
- Ballard, D.; Winkler-Galicki, J.; Wesoły, J. Massive parallel sequencing in forensics: Advantages, issues, technicalities, and prospects. Int. J. Leg. Med. 2020, 134, 1291–1303. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.W.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Lovejoy, A.F.; Klass, D.M.; Kurtz, D.M.; Chabon, J.J.; Scherer, F.; Stehr, H.; Liu, C.L.; Bratman, S.V.; Say, C.; et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016, 34, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; Denis, M.G.; Thress, K.S.; Ratcliffe, M.; Reck, M. Guide to detecting epidermal growth factor receptor (EGFR) mutations in ctDNA of patients with advanced non-small-cell lung cancer. Oncotarget 2017, 8, 12501–12516. [Google Scholar] [CrossRef]
- Cao, R.; Lea, K.; Jasti, M.; Schageman, J.; Hanif, K.; Li, Y.; Gu, J.; Bagai, V.; Kshatriya, P.; Wikman, H.; et al. Abstract 5123: Characterization of genetic mutation spectra and identification of gene amplification and fusion variants in cell-free nucleic acid from cultured cancer cell media and liquid biopsy specimens using Oncomine™ Pan-Cancer Cell-Free Assay. Cancer Res. 2019, 79, 5123. [Google Scholar] [CrossRef]
- Sokolova, A.O.; Shirts, B.H.; Konnick, E.Q.; Tsai, G.J.; Goulart, B.H.; Montgomery, B.; Pritchard, C.C.; Yu, E.Y.; Cheng, H.H. Complexities of Next-Generation Sequencing in Solid Tumors: Case Studies. J. Natl. Compr. Cancer Netw. 2020, 18, 1150–1155. [Google Scholar] [CrossRef]
- Zhu, C.; Guan, X.; Zhang, X.; Luan, X.; Song, Z.; Cheng, X.; Zhang, W.; Qin, J.-J. Targeting KRAS mutant cancers: From druggable therapy to drug resistance. Mol. Cancer 2022, 21, 159. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.C.; Liu, C.Y.; Huang, C.J.; Hsu, Y.C.; Lien, H.H.; Wong, J.U.; Tai, F.-C.; Ku, W.-H.; Hung, C.-F.; Lin, J.-T.; et al. Deciphering Genetic Alterations of Taiwanese Patients with Pancreatic Adenocarcinoma through Targeted Sequencing. Int. J. Mol. Sci. 2022, 23, 1579. [Google Scholar] [CrossRef] [PubMed]
- André, F.; Ciruelos, E.M.; Juric, D.; Loibl, S.; Campone, M.; Mayer, I.A.; Rubovszky, G.; Yamashita, T.; Kaufman, B.; Lu, Y.-S.; et al. Alpelisib plus fulvestrant for PIK3CA-mutated, hormone receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: Final overall survival results from SOLAR-1. Ann. Oncol. 2021, 32, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Andrikopoulou, A.; Chatzinikolaou, S.; Panourgias, E.; Kaparelou, M.; Liontos, M.; Dimopoulos, M.A.; Zagouri, F. The emerging role of capivasertib in breast cancer. Breast 2022, 63, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.H.; Casbard, A.; Carucci, M.; Cox, C.; Butler, R.; Alchami, F.; Madden, T.-A.; Bale, C.; Bezecny, P.; Joffe, J.; et al. Fulvestrant plus capivasertib versus placebo after relapse or progression on an aromatase inhibitor in metastatic, oestrogen receptor-positive, HER2-negative breast cancer (FAKTION): Overall survival, updated progression-free survival, and expanded biomarker analysis from a randomised, phase 2 trial. Lancet Oncol. 2022, 23, 851–864. [Google Scholar]
- Turner, N.; Dent, R.A.; O’Shaughnessy, J.; Kim, S.B.; Isakoff, S.J.; Barrios, C.; Saji, S.; Bondarenko, I.; Nowecki, Z.; Lian, Q.; et al. Ipatasertib plus paclitaxel for PIK3CA/AKT1/PTEN-altered hormone receptor-positive HER2-negative advanced breast cancer: Primary results from cohort B of the IPATunity130 randomized phase 3 trial. Breast Cancer Res. Treat. 2022, 191, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.X.; Luo, J.; Freedman, R.A.; Pluard, T.J.; Nangia, J.R.; Lu, J.; Valdez-Albini, F.; Cobleigh, M.; Jones, J.M.; Lin, N.U.; et al. The Phase II MutHER Study of Neratinib Alone and in Combination with Fulvestrant in HER2-Mutated, Non-amplified Metastatic Breast Cancer. Clin. Cancer Res. 2022, 28, 1258–1267. [Google Scholar] [CrossRef]
- Henry, N.L.; Somerfield, M.R.; Dayao, Z.; Elias, A.; Kalinsky, K.; McShane, L.M.; Moy, B.; Park, B.H.; Shanahan, K.M.; Sharma, P.; et al. Biomarkers for Systemic Therapy in Metastatic Breast Cancer: ASCO Guideline Update. J. Clin. Oncol. 2022, 40, 3205–3221. [Google Scholar] [CrossRef]
- Narayan, P.; Prowell, T.M.; Gao, J.J.; Fernandes, L.L.; Li, E.; Jiang, X.; Qiu, J.; Fan, J.; Song, P.; Yu, J.; et al. FDA Approval Summary: Alpelisib Plus Fulvestrant for Patients with HR-positive, HER2-negative, PIK3CA-mutated, Advanced or Metastatic Breast Cancer. Clin. Cancer Res. 2021, 27, 1842–1849. [Google Scholar] [CrossRef]
- Ciruelos, E.M.; Loibl, S.; Mayer, I.A.; Campone, M.; Rugo, H.S.; Arnedos, M.; Iwata, H.; Conte, P.F.; André, F.; Reising, A.; et al. Abstract PD2-06: Clinical outcomes of alpelisib plus fulvestrant in hormone receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer with PIK3CA alterations detected in plasma ctDNA by next-generation sequencing: Biomarker analysis from the SOLAR-1 study. Cancer Res. 2021, 81, PD2-06. [Google Scholar]

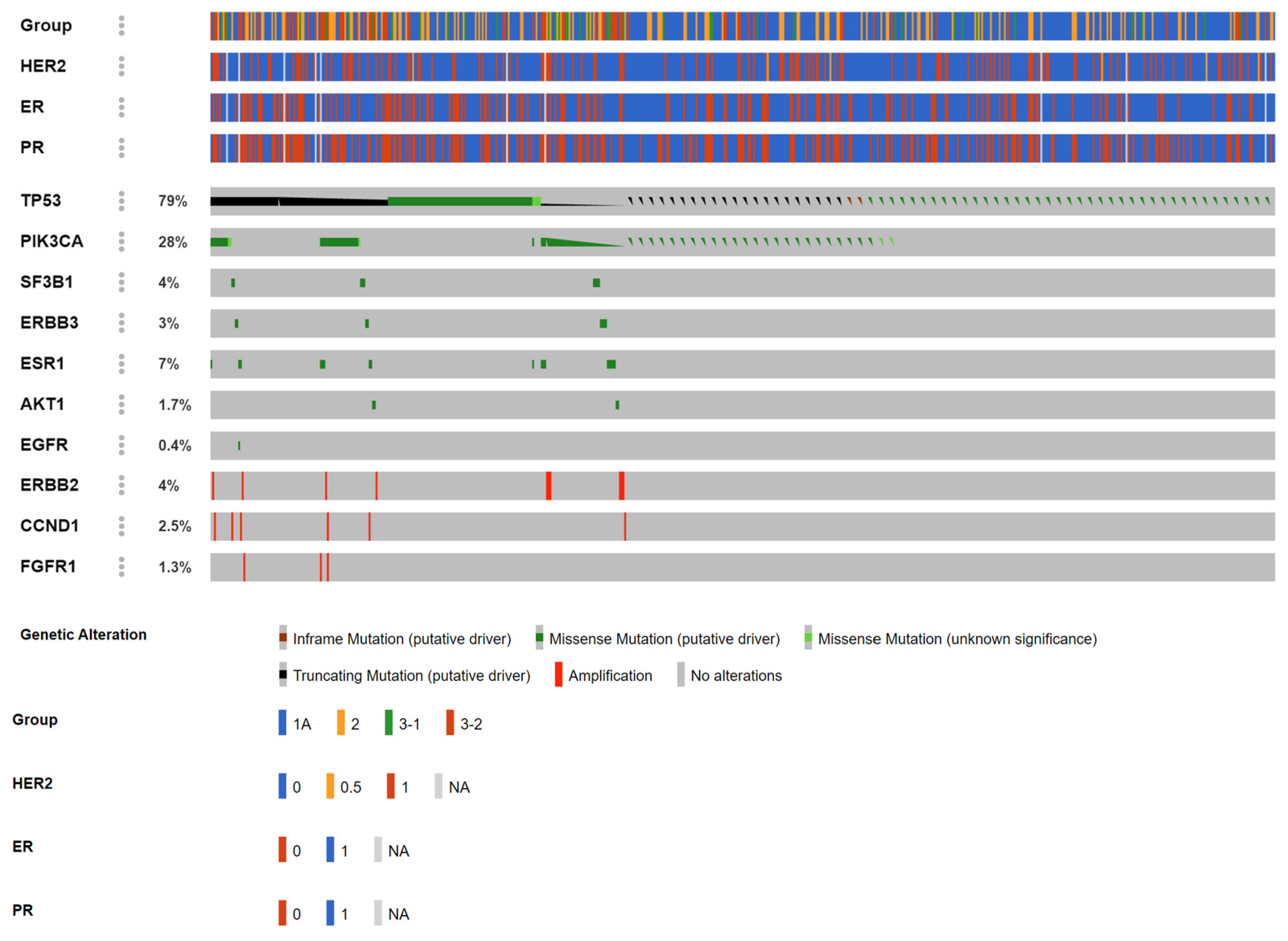
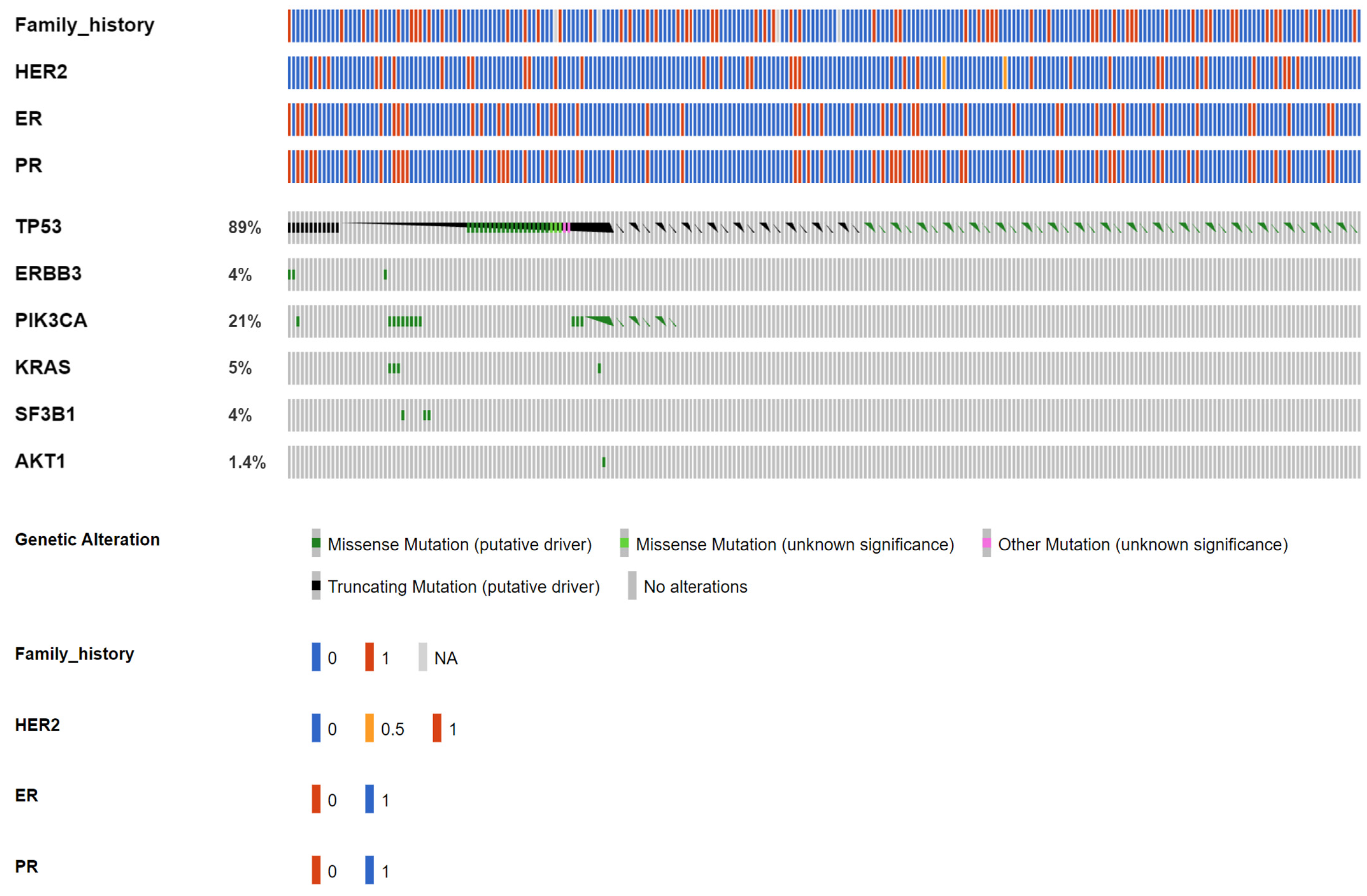
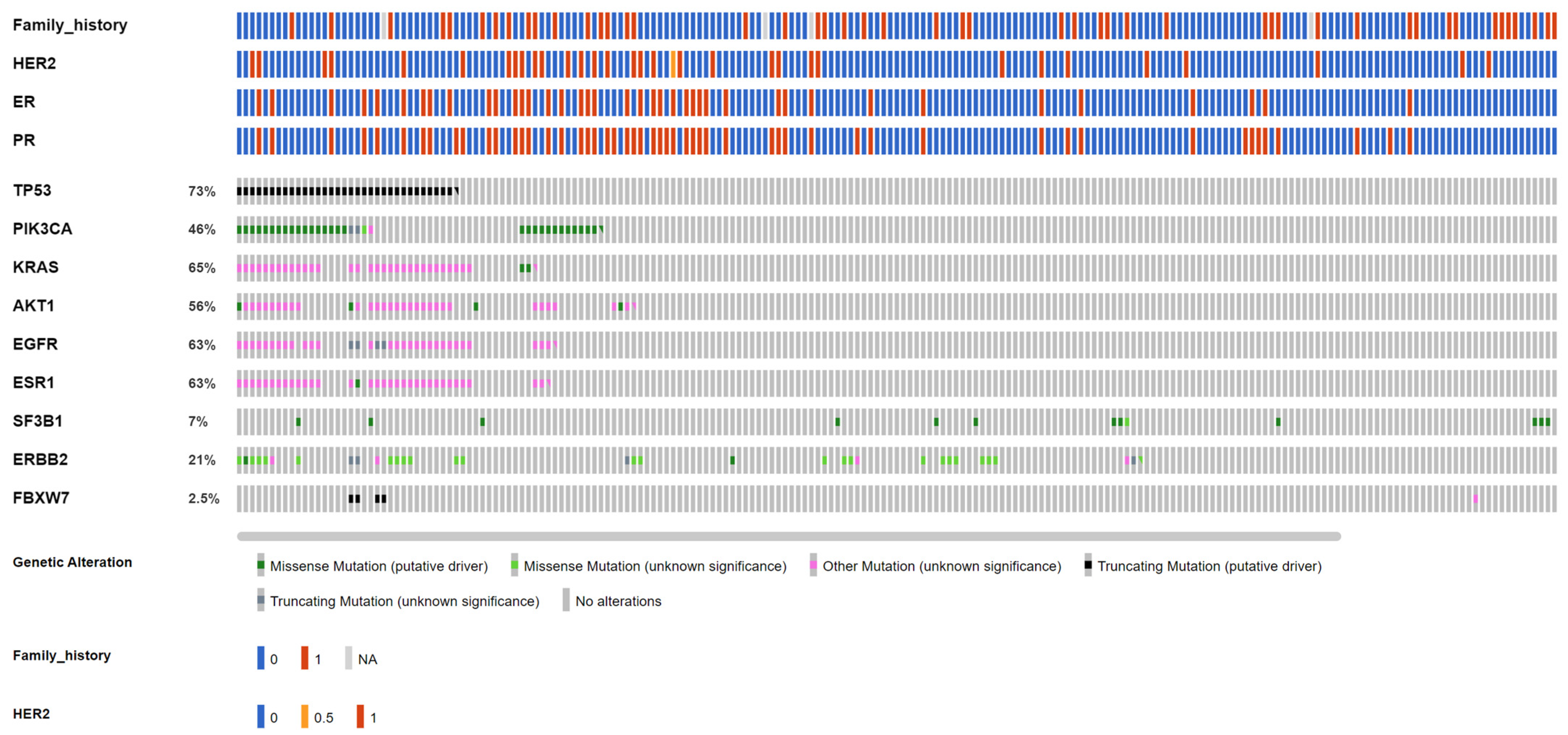
| Gene | AKT1 | CCND1 | EGFR | ERBB2 | ERBB3 | ESR1 | FGFR1 | PIK3CA | SF3B1 | TP53 |
|---|---|---|---|---|---|---|---|---|---|---|
| Altered variants | 8 | 11 | 7 | 16 | 11 | 38 | 7 | 107 | 11 | 445 |
| Altered variants collapsed to subjects | 8 | 11 | 7 | 15 | 10 | 36 | 7 | 102 | 11 | 416 |
| Amino acid change (case number) | E17K (8) | CNV(11) | L861Q(7) | CNV(16) | R103C(4), T335I(2), V104M(5) | D538G(12), E380Q(4), Y537C(1), Y537N(9), Y537S(12) | CNV(7) | C420R(3), E453K(2), E542K(11), E545G(1), E545K(28), E726K(1), G12C(4), G12D(4), G12V(1), G13D(1), H1047L(12), H1047R(28), H1047Y(1), M1043I(2), Q546R(8) | K700E(11) | A138D(1), A138V(3), A189D(1), C124*(1), C141Y(2), C176S(1), C238Y(2), C242S(1), C242Y(1), C275F(1), C275Y(1), E180*(3), E258K(1), E258Q(7), E286G(1), E286K(1), E339*(1), E339K(1), F54L(1), G154S(1), G244D(2), G244S(3), G245D(8), G245S(6), G245V(1), G266E(2), G266R(4), H178L(1), H178fs(1), H179L(4), H179R(2), H193R(2), H193fs(2), H214R(1), H365fs(23), I195T(7), I195fs(6), K132*(1), K132N(5), K132R(4), K381T(2), L137M(1), L145R(1), L14P(1), L194P(1), L323V(1), L32M(1), L35fs(1), M237I(20), M246V(2), N131K(1), P151A(5), P151S(1), P151T(2), P152L(4), P177R(1), P190L(1), P222T(1), P250R(4), P278R(1), P300T(1), P301fs(4), P318T(1), P82fs(2), P85fs(7), Q104*(2), Q136H(1), Q136fs(2), Q192*(2), Q317*(1), Q331H(1), Q331fs(9), Q38H(1), Q38R(1), R156C(1), R158C(1), R158H(2), R175G(2), R175H(11), R175L(6), R181H(1), R181L(1), R181S(1), R196*(2), R213*(10), R213L(1), R213Q(6), R248Q(31), R248W(10), R249K(1), R249S(10), R273C(10), R273H(12), R273L(1), R273P(1), R280T(7), R282W(9), R283H(1), R306*(7), R333H(1), R333fs(1), R335fs(1), R379C(1), S149Y(1), S215R(2), S241C(1), S241F(7), S378fs(4), S6P(1), S90fs(8), S94*(1), S9G(1), T140_C14(2), V143M(4), V147fs(1), V157F(1), V157I(1), V172F(1), V173E(5), V173L(8), V173M(6), V197G(1), V216L(1), V216M(2), V272M(10), V274L(2), V31I(1), V97G(1), V97fs(2), Y103H(1), Y220C(13), Y220N(1), Y234C(2) |
| Functionality (case number) | Missense (8) | Amp(11) | Missense(7) | Amp(16) | Missense(11) | Missense(38) | Amp(7) | Missense(107) | Missense(11) | Missense(338), truncting(105), inframe(2) |
| Clinical presentations | ||||||||||
| Group(1,2A,3-1,3-2) | 1:0:2:5 | 0:2:0:9 | 0:0:0:7 | 3:6:5:2 | 7:2:0:2 | 0:0:2:36 | 0:0:0:7 | 25:26:16:40 | 6:3:0:2 | 196:100:17:132 |
| ER(+/−) | 8:0 | 11:0 | NA | 2:14 | 9:2 | 24:14 | NA | 71:27 * | 9:2 | 237:157 ** |
| PR(+/−) | 8:0 | 3:8 | NA | 2:14 | 6:5 | 17:7 | NA | 49:49 * | 9:2 | 182:212 ** |
| HER2(+/−) | 0:8 | 6:5 | NA | 16:0 | 1:10 | 3:35 | NA | 40:58 * | 2:9 | 98:296 ** |
| Gene | Case Number | ||||
|---|---|---|---|---|---|
| cfDNA | Tumor Tissue | Both Detected | Both Undetected | Total Affected | |
| AKT1 | 1 | 113 | 1 | 133 | 113 (45.9%) |
| ERBB2 | 0 | 42 | 0 | 204 | 42 (17.1%) |
| KRAS | 4 | 120 | 3 | 115 | 131 (53.5%) |
| PIK3CA | 15 | 93 | 1 | 149 | 97 (39.4%) |
| SF3B1 | 3 | 14 | 1 | 230 | 16 (6.5%) |
| TP53 | 65 | 146 | 43 | 78 | 168 (68.3%) |
| Gene | AKT1 | CCND1 | EGFR | ERBB2 | ERBB3 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| ctDNA | Tissue | ctDNA | Tissue | ctDNA | Tissue | ctDNA | Tissue | ctDNA | Tissue | |
| Altered variants | 1 | 19 | - | - | - | 7 | - | 38 | 3 | - |
| Amino acid change (case number) | E17K(1) | E17K(16), L52R(2), Y18fs(1) | - | - | - | A755fs(3), N466fs(2), N756fs(1), V592I(1) | - | A763fs(1), D769Y(1), I655V(31), L841V(1), N758fs(3), V777L(1) | R103C(2), V104M(1) | - |
| Functionality (case number) | Missense(1) | Missense(18), truncating(1) | - | - | - | Missense(1), truncating(6) | - | Missense(34), truncating(4) | Missense(3) | - |
| Clinical presentations | ||||||||||
| ER(+/−) | 1:0 | 17:2 | - | - | - | 6:1 | - | 31:7 | 2:1 | - |
| PR(+/−) | 1:0 | 17:2 | - | - | - | 6:1 | - | 28:10 | 2:1 | - |
| HER2(+/−) | 0:1 | 0:19 | - | - | - | 0:7 | - | 8:30 | 0:3 | - |
| FH(case number) | 0 | 8 | - | - | - | 1 | - | 5 | 1 | - |
| Gene | ESR1 | FGFR1 | PIK3CA | SF3B1 | TP53 | |||||
| ctDNA | Tissue | ctDNA | Tissue | ctDNA | Tissue | ctDNA | Tissue | ctDNA | Tissue | |
| Altered variants | - | 4 | - | - | 16 | 103 | 3 | 14 | 94 | 207 |
| Amino acid change (case number) | - | E380Q(1), F62L(2), M297I(1) | - | - | E545K(3), H1047R(8), M1043I(1), Q546K(1), Q546R(3) | C420R(2), D1029H(1), D350N(1), D549N(1), E542K(6), E545K(18), E726K(5), E80K(1), G1049R(2), H1047L(8), H1047R(43), H419Y(1), M1043I(2), N1044K(1), N345I(1), N345K(4), Q546K(1), Q546P(1), Q546R(1), Q721fs(1), Y1021H(1), Y432fs(1) | K700E(3) | K700E(12), R625H(2), W658C(1) | C135W(1), C275Y(1), E285K(1), G244S(1), G245D(3), G245S(2), G279E(1), H179R(1), H193R(1), H214R(1), H365fs(3), I195T(1), K372E(1), L145M(1), L194R(1), M237I(4), M246V(1), N131K(1), P151A(1), P152L(2), P152fs(1), P222L(1), P222T(1), P250L(1), P278T(1), P300T(1), P4L(1), P85fs(1), Q331fs(9), R158H(1), R175C(1), R175H(2), R181H(1), R196*(3), R213*(2), R213Q(2), R248Q(2), R248W(5), R249S(2), R273C(2), R273H(2), R282W(1), R335S(1), R379C(1), S215fs(1), S378fs(1), S94*(1), V173L(1), V216M(1), V272M(1), V97G(1), W146C(1), W91L(1), Y220C(3), Y220S(1), Y234C(1) | A276fs(1), C135W(1), C176Y(1), C238S(1), C238Y(1), C242fs(1), C275F(1), C275Y(3), C275fs(3), D148fs(1), E271*(1), E285A(1), E285K(1), E286A(1), E286G(2), E336*(1), G245S(3), G266R(1), G266T(1), H168fs(1), H179R(2), H179Y(1), H193L(1), H214fs(1), I195T(2), L114fs(1), L194R(1), L348fs(1), M237I(1), M246I(1), M246R(1), M246V(1), N200fs(1), N239*(1), N268fs(1), P128fs(1), P278S(1), P278T(1), P295fs(1), P316fs(1), P390fs(1), P72R(106), Q331*(1), Q52fs(1), R110fs(1), R174W(1), R175H(5), R196*(3), R248G(1), R248Q(3), R248W(2), R249G(1), R249W(1), R273H(1), R333fs(1), R337C(1), S241F(1), S314fs(1), S94*(1), T125R(1), V218fs(1), V274D(1), V73fs(14), W91*(2), Y220C(3), Y234C(2), Y236C(1) |
| Functionality (case number) | - | Missense(4) | - | - | Missense(16) | Missense(101), truncating(2) | Missense(3) | Missense(14) | Missense(66), truncating(22), other(6) | Missense(162), truncating(45) |
| Clinical presentations | ||||||||||
| ER(+/−) | - | 4:0 | - | - | 11:5 | 86:17 | 3:0 | 14:0 | 72:22 | 154:53 |
| PR(+/−) | - | 4:0 | - | - | 8:8 | 76:27 | 2:1 | 13:1 | 65:29 | 141:66 |
| HER2(+/−) | - | 0:4 | - | - | 3:13 | 14:89 | 0:3 | 0:14 | 19:75 | 43:164 |
| FH(case number) | - | 0 | - | - | 5 | 21 | 1 | 7 | 25 | 41 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.-C.; Tsai, Y.-F.; Liu, C.-Y.; Lien, P.-J.; Lin, Y.-S.; Chao, T.-C.; Feng, C.-J.; Chen, Y.-J.; Lai, J.-I.; Cheng, H.-F.; et al. Concordance of Targeted Sequencing from Circulating Tumor DNA and Paired Tumor Tissue for Early Breast Cancer. Cancers 2023, 15, 4475. https://doi.org/10.3390/cancers15184475
Huang C-C, Tsai Y-F, Liu C-Y, Lien P-J, Lin Y-S, Chao T-C, Feng C-J, Chen Y-J, Lai J-I, Cheng H-F, et al. Concordance of Targeted Sequencing from Circulating Tumor DNA and Paired Tumor Tissue for Early Breast Cancer. Cancers. 2023; 15(18):4475. https://doi.org/10.3390/cancers15184475
Chicago/Turabian StyleHuang, Chi-Cheng, Yi-Fang Tsai, Chun-Yu Liu, Pei-Ju Lien, Yen-Shu Lin, Ta-Chung Chao, Chin-Jung Feng, Yen-Jen Chen, Jiun-I Lai, Han-Fang Cheng, and et al. 2023. "Concordance of Targeted Sequencing from Circulating Tumor DNA and Paired Tumor Tissue for Early Breast Cancer" Cancers 15, no. 18: 4475. https://doi.org/10.3390/cancers15184475
APA StyleHuang, C.-C., Tsai, Y.-F., Liu, C.-Y., Lien, P.-J., Lin, Y.-S., Chao, T.-C., Feng, C.-J., Chen, Y.-J., Lai, J.-I., Cheng, H.-F., Chen, B.-F., Hsu, C.-Y., Chiu, J.-H., & Tseng, L.-M. (2023). Concordance of Targeted Sequencing from Circulating Tumor DNA and Paired Tumor Tissue for Early Breast Cancer. Cancers, 15(18), 4475. https://doi.org/10.3390/cancers15184475





