Differences in the Clinical and Molecular Profiles of Subungual Melanoma and Acral Melanoma in Asian Patients
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Tissue Samples and NGS
2.3. Statistical Analysis
3. Results
3.1. Characteristics of Patients with SUM
3.2. Differences in the Clinicopathologic Characteristics of Patients with SUM or AM
3.3. Differences in Overall Survival of Patients with SUM or AM
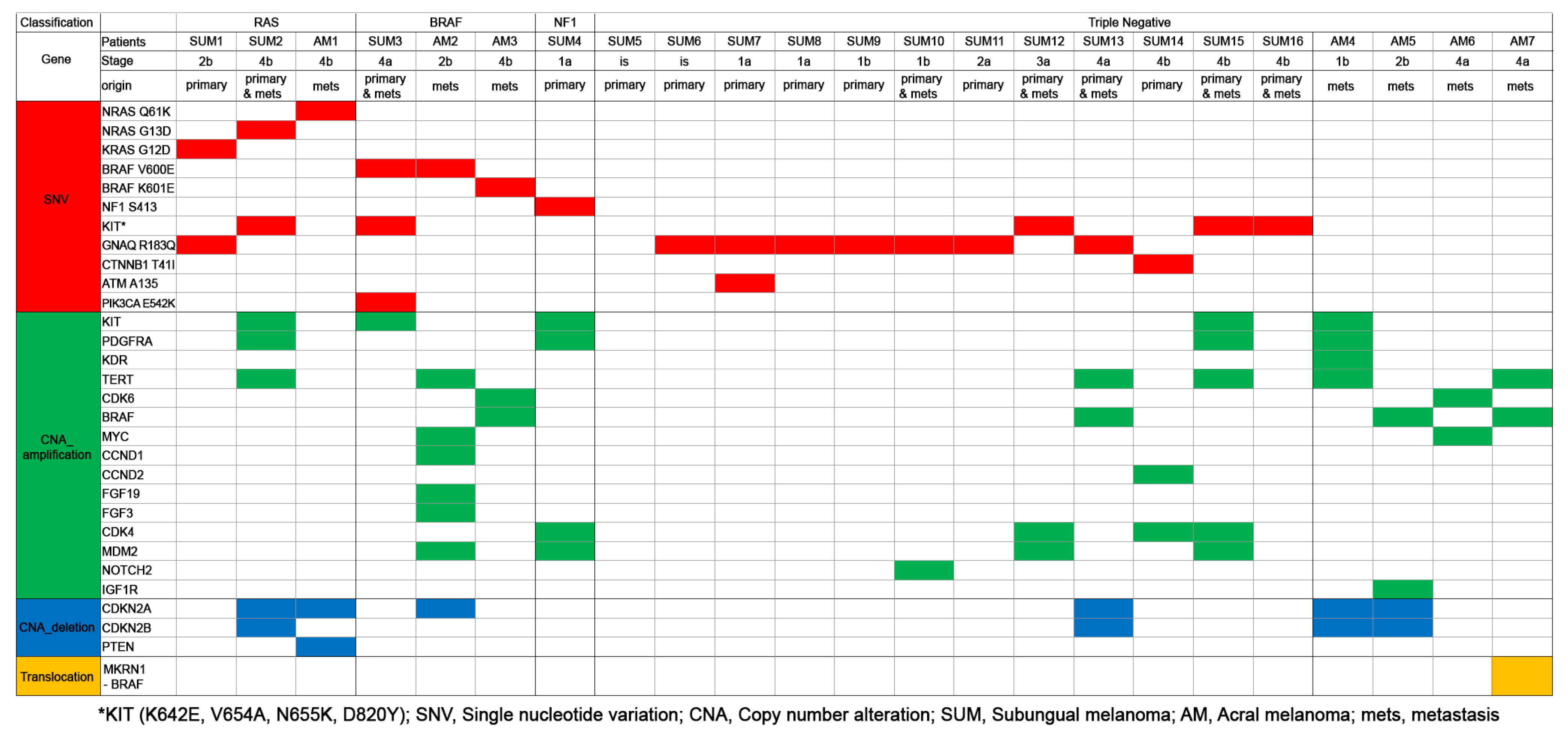
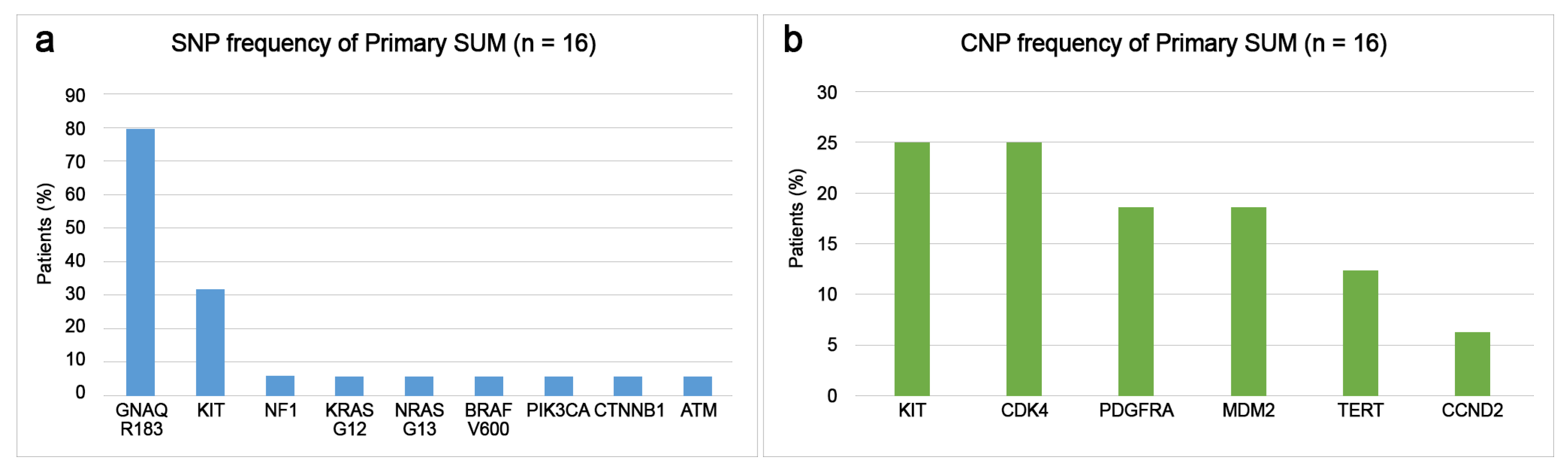
3.4. Differences of Somatic Mutational Signatures of Patients with SUM or AM
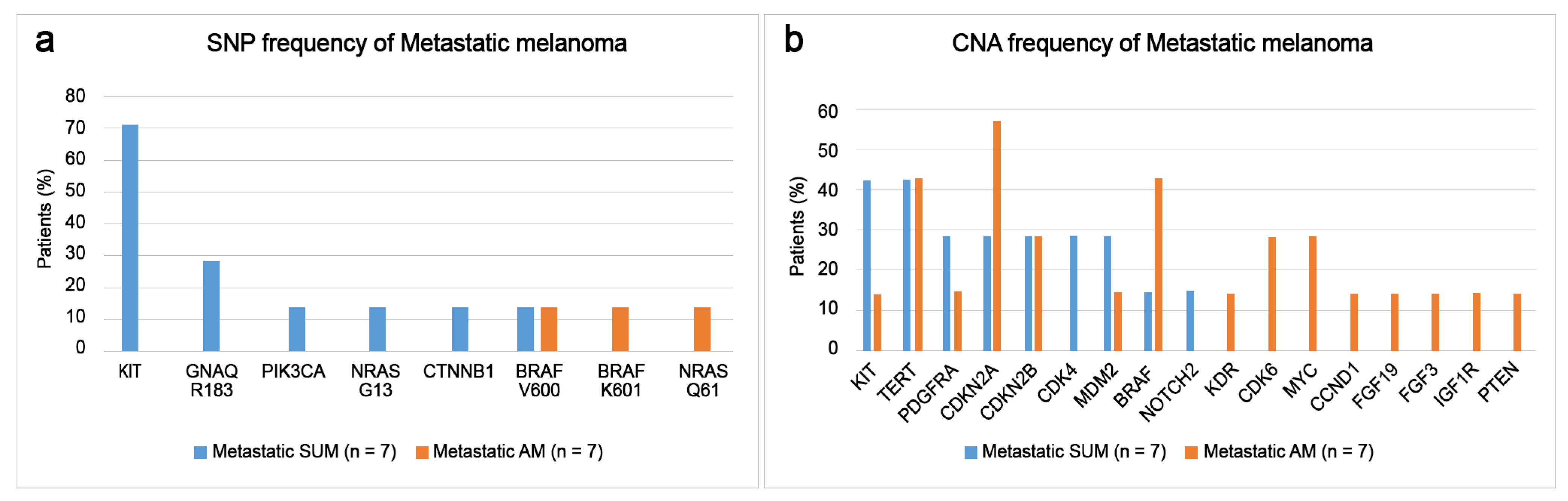
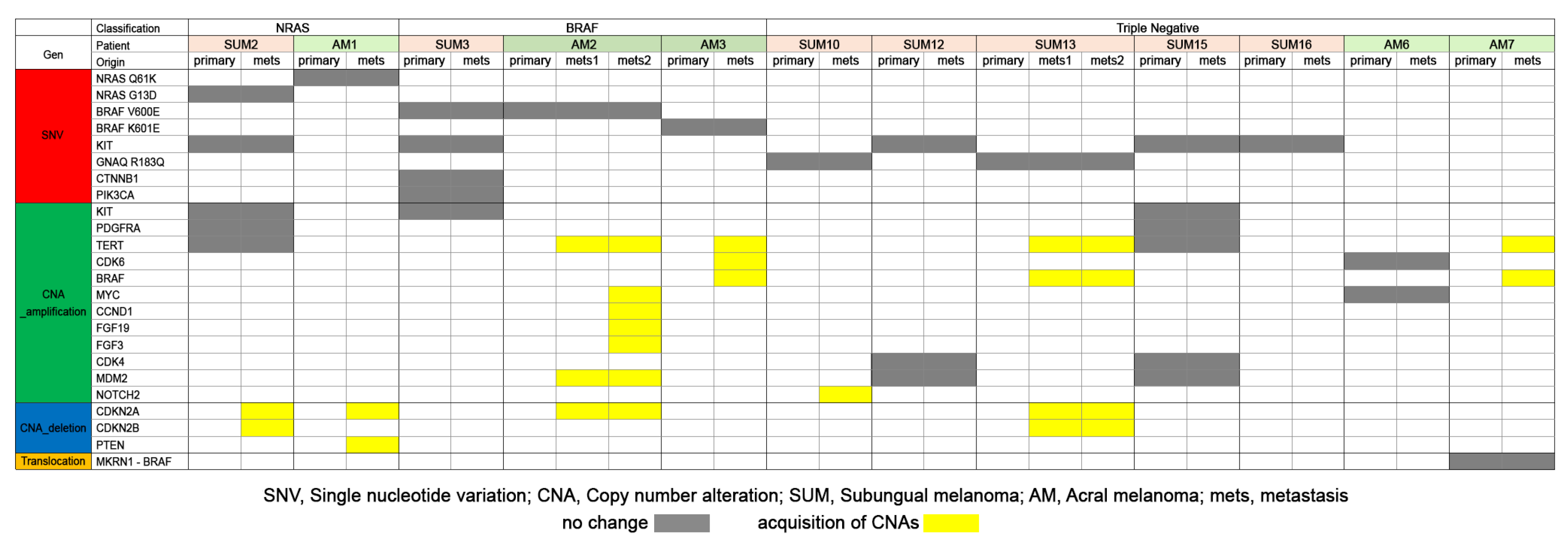
3.5. Comparison of Somatic Mutations of Primary and Metastatic Melanomas of SUM and AM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Littleton, T.W.; Murray, P.M.; Baratz, M.E. Subungual Melanoma. Orthop. Clin. N. Am. 2019, 50, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Pate, G.A.; Ragi, G.; Krysicki, J.; Schwartz, R.A.A. Subungual melanoma: A deceptive disorder. Acta Dermatovenerol. Croat. 2008, 16, 236–242. [Google Scholar]
- Kim, S.Y.; Yun, S.J. Cutaneous melanoma in Asians. Chonnam Med. J. 2016, 52, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Holman, B.N.; Van Gulick, R.J.; Amato, C.M.; MacBeth, M.L.; Davies, K.D.; Aisner, D.L.; Robinson, W.A.; Couts, K.L. Clinical and molecular features of subungual melanomas are site-specific and distinct from acral melanomas. Melanoma Res. 2020, 30, 562–573. [Google Scholar] [CrossRef]
- Newell, F.; Wilmott, J.S.; Johansson, P.A.; Nones, K.; Addala, V.; Mukhopadhyay, P.; Broit, N.; Amato, C.M.; Gulick, R.V.; Kazakoff, S.H.; et al. Whole-genome sequencing of acral melanoma reveals genomic complexity and diversity. Nat. Commun. 2020, 11, 5259. [Google Scholar] [CrossRef] [PubMed]
- Elder, D.E.; Bastian, B.C.; Cree, I.A.; Massi, D.; Scolyer, R.A. The 2018 World Health Organization classification of cutaneous, mucosal, and uveal melanoma: Detailed analysis of 9 distinct subtypes defined by their evolutionary pathway. Arch. Pathol. Lab. Med. 2020, 144, 500–522. [Google Scholar] [CrossRef] [PubMed]
- Mejbel, H.A.; Torres-Cabala, C.A.; Milton, D.R.; Ivan, D.; Nagarajan, P.; Curry, J.L.; Ciurea, A.M.; Rubin, A.I.; Hwu, W.-J.; Prieto, V.G.; et al. Prognostic significance of subungual anatomic site in acral lentiginous melanoma. Arch. Pathol. Lab. Med. 2021, 145, 943–952. [Google Scholar] [CrossRef]
- Lee, D.J.R.; Arbache, S.T.; Quaresma, M.V.; Nico, M.M.S.; Gabbi, T.V.B. Nail apparatus melanoma: Experience of 10 years in a single institution. Ski. Appendage Disord. 2019, 5, 20–26. [Google Scholar] [CrossRef]
- Cochran, A.M.; Buchanan, P.J.; Bueno, R.A., Jr.; Neumeister, M.W. Subungual melanoma: A review of current treatment. Plast. Reconst. Surg. 2014, 134, 259–273. [Google Scholar] [CrossRef]
- Gershenwald, J.E.; Scolyer, R.A.; Hess, K.R.; Thompson, J.F.; Long, G.V.; Ross, M.I.; Lazar, A.J.; Faries, M.B.; Kirkwood, J.M.; McArthur, G.A.; et al. CA Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. Cancer J. Clin. 2017, 67, 472–492. [Google Scholar] [CrossRef]
- Hovelson, D.H.; McDaniel, A.S.; Cani, A.K.; Johnson, B.; Rhodes, K.; Williams, P.D.; Bandla, S.; Bien, G.; Choppa, P.; Hyland, F.; et al. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia 2015, 17, 385–399. [Google Scholar] [CrossRef] [PubMed]
- Yeo, M.-K.; Yoon, N.; Bae, G.E. Clinicopathologic and Molecular Characteristics of Gastrointestinal MiNENs. Front. Oncol. 2021, 11, 709097. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 2015, 10, 1556–1566. [Google Scholar] [CrossRef]
- Reilly, D.; Aksakal, G.; Gilmour, R.; Gyorki, D.; Chauhan, A.; Webb, A.; Henderson, M.A. Subungual melanoma: Management in the modern era. J. Plast. Reconstr. Aesthet. Surg. 2017, 70, 1746–1752. [Google Scholar] [CrossRef] [PubMed]
- Heaton, K.M.; El-Naggar, A.; Ensign, L.G.; Ross, M.I.; Balch, C.M. Surgical management and prognostic factors in patients with subungual melanoma. Ann. Surg. 1994, 219, 197. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Lee, J.H.; Won, C.H.; Chang, S.E.; Choi, J.H.; Moon, K.C.; Lee, M.W. Nail apparatus melanoma: A comparative, clinicoprognostic study of the initial clinical and morphological characteristics of 49 patients. J. Am. Acad. Dermatol. 2015, 73, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Eberhard, P.; Kleiner, H.; Bodeker, R.H. Epidemiologie und prognose subungualer Melanome. Hautarzt 1992, 43, 286–290. [Google Scholar]
- Borkowska, A.; Szumera-Ciećkiewicz, A.; Spałek, M.; Teterycz, P.; Czarnecka, A.; Kowalik, A.; Rutkowski, P. Mutation profile of primary subungual melanomas in Caucasians. Oncotarget 2020, 11, 2404–2413. [Google Scholar] [CrossRef]
- O’Leary, J.A.; Berend, K.R.; Johnson, J.L.; Levin, L.S.; Seigler, H.F. Subungual melanoma: A review of 93 cases with identification of prognostic variables. Clin. Orthop. Relat. Res. 2000, 378, 206–212. [Google Scholar] [CrossRef]
- Nguyen, J.T.; Bakri, K.; Nguyen, E.C.; Johnson, C.H.; Moran, S.L. Surgical management of subungual melanoma: Mayo clinic experience of 124 cases. Ann. Plast. Surg. 2013, 71, 346–354. [Google Scholar] [CrossRef]
- Jung, H.J.; Kweon, S.-S.; Lee, J.-B.; Lee, S.-C.; Yun, S.J. A clinicopathologic analysis of 177 acral melanomas in Koreans: Relevance of spreading pattern and physical stress. JAMA Dermatol. 2013, 149, 1281–1288. [Google Scholar] [CrossRef] [PubMed]
- LaRocca, C.J.; Lai, L.; Nelson, R.A.; Modi, B.; Crawford, B. Subungual Melanoma: A Single Institution Experience. Med. Sci. 2021, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.-B.; Moncrieff, M.; Thompson, J.F.; McCarthy, S.W.; Shaw, H.M.; Quinn, M.J.; Li, L.-X.L.; Crotty, K.A.; Stretch, J.R.; Scolyer, R.A. Subungual Melanoma: A Study of 124 Cases Highlighting Features of Early Lesions, Potential Pitfalls in Diagnosis, and Guidelines for Histologic Reporting. Am. J. Surg. Pathol. 2007, 31, 1902–1912. [Google Scholar] [CrossRef] [PubMed]
- Haneke, E. Ungual melanoma–controversies in diagnosis and treatment. Dermatol. Ther. 2012, 25, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Dika, E.; Altimari, A.; Patrizi, A.; Gruppioni, E.; Fiorentino, M.; Piraccini, B.M.; Misciali, C.; Barisani, A.; Fanti, P.A. KIT, NRAS, and BRAF mutations in nail apparatus melanoma. Pigment. Cell Melanoma Res. 2013, 26, 758–760. [Google Scholar] [CrossRef] [PubMed]
- Saida, T.; Ohshima, Y. Clinical and histopathologic characteristics of early lesions of subungual malignant melanoma. Cancer 1989, 63, 556–560. [Google Scholar] [CrossRef] [PubMed]
- Darmawan, C.; Ohn, J.; Mun, J.H.; Kim, S.; Lim, Y.; Jo, S.; Kim, Y.-G.; Kim, B.; Seong, M.-W.; Kim, B.J.; et al. Diagnosis and treatment of nail melanoma: A review of the clinicopathologic, dermoscopic, and genetic characteristics. J. Eur. Acad. Dermatol. Venereol. 2022, 36, 651–660. [Google Scholar] [CrossRef]
- Roh, M.R.; Kim, J.; Chung, K.Y. Treatment and outcomes of melanoma in acral location in Korean patients. Yonsei Med. J. 2010, 51, 562–568. [Google Scholar] [CrossRef]
- Mole, R.J.; MacKenzie, D.N. Subungual Melanoma; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK482480/ (accessed on 2 August 2023).
- Vázquez, M.; Ramos, F.A.; Sánchez, J.L. Melanomas of volar and subungual skin in Puerto Ricans: A clinicopathologic study. J. Am. Acad. Dermatol. 1984, 10, 39–45. [Google Scholar] [CrossRef]
- Søndergaard, K. Histological type and biological behavior of primary cutaneous malignant melanoma. 2. An analysis of 86 cases located on so-called acral regions as plantar, palmar, and sub-/parungual areas. Virchows Arch. A Pathol. Anat. Histopathol. 1983, 401, 333–343. [Google Scholar] [CrossRef]
- Hayward, N.K.; Wilmott, J.S.; Waddell, N.; Johansson, P.A.; Field, M.A.; Nones, K.; Patch, A.-M.; Kakavand, H.; Alexandrov, L.B.; Burke, H.; et al. Whole-genome landscapes of major melanoma subtypes. Nature 2017, 545, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Curtin, J.A.; Fridlyand, J.; Kageshita, T.; Patel, H.N.; Busam, K.J.; Kutzner, H.; Cho, K.-H.; Aiba, S.; Brocker, E.-B.; LeBoit, P.H.; et al. Distinct sets of genetic alterations in melanoma. N. Eng. J. Med. 2005, 353, 2135–2147. [Google Scholar] [CrossRef] [PubMed]
- Haugh, A.M.; Zhang, B.; Quan, V.L.; Garfield, E.M.; Bubley, J.A.; Kudalkar, E.; Verzi, A.E.; Walton, K.; VandenBoom, T.; Merkel, E.A.; et al. Distinct Patterns of Acral Melanoma Based on Site and Relative Sun Exposure. J. Inv. Dermatol. 2018, 138, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Dai, B.; Cai, X.; Kong, Y.-Y.; Yang, F.; Shen, X.-X.; Wang, L.-W.; Kong, J.-C. Analysis of KIT expression and gene mutation in human acral melanoma: With a comparison between primary tumors and corresponding metastases/recurrences. Hum. Pathol. 2013, 44, 1472–1478. [Google Scholar] [CrossRef] [PubMed]
- Sakaizawa, K.; Ashida, A.; Uchiyama, A.; Ito, T.; Fujisawa, Y.; Ogata, D.; Matsushita, S.; Fujii, K.; Fukushima, S.; Shibayama, Y.; et al. Clinical characteristics associated with BRAF, NRAS and KIT mutations in Japanese melanoma patients. J. Dermatol. Sci. 2015, 80, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Moon, K.R.; Choi, Y.D.; Kim, J.M.; Jin, S.; Shin, M.H.; Shim, H.J.; Lee, J.-B.; Yun, S.J. Genetic Alterations in Primary Acral Melanoma and Acral Melanocytic Nevus in Korea: Common Mutated Genes Show Distinct Cytomorphological Features. J. Investig. Dermatol. 2018, 138, 933–945. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Yoon, J.; Chung, Y.J.; Lee, S.Y.; Choi, J.Y.; Shin, O.R.; Park, H.Y.; Bahk, W.-J.; Yu, D.S.; Lee, Y.B. Whole-exome sequencing reveals differences between nail apparatus melanoma and acral melanoma. J. Am. Acad. Dermatol. 2018, 79, 559–561.e1. [Google Scholar] [CrossRef]
- Sheen, Y.S.; Tan, K.T.; Tse, K.P.; Liao, Y.H.; Lin, M.H.; Chen, J.S.; Liau, J.-Y.; Tseng, Y.-J.; Lee, C.-H.; Hong, C.-H.; et al. Genetic alterations in primary melanoma in Taiwan. Br. J. Dermatol. 2020, 182, 1205–1213. [Google Scholar] [CrossRef]
- Lim, Y.; Yoon, D.; Lee, D.Y. Novel mutations identified by whole-exome sequencing in acral melanoma. J. Am. Acad. Dermatol. 2020, 83, 1792–1794. [Google Scholar] [CrossRef]
- Turajlic, S.; Furney, S.J.; Lambros, M.B.; Mitsopoulos, C.; Kozarewa, I.; Geyer, F.C.; Mackay, A.; Hakas, J.; Zvelebil, M.; Lord, C.J.; et al. Whole genome sequencing of matched primary and metastatic acral melanomas. Genome Res. 2012, 22, 196–207. [Google Scholar] [CrossRef]
- Manca, A.; Paliogiannis, P.; Colombino, M.; Casula, M.; Lissia, A.; Botti, G.; Caraco, C.; Ascierto, P.A.; Sini, M.C.; Palomba, G.; et al. Mutational concordance between primary and metastatic melanoma: A next-generation sequencing approach. J. Transl. Med. 2019, 17, 289. [Google Scholar] [CrossRef] [PubMed]
- Sanborn, J.Z.; Chung, J.; Purdom, E.; Wang, N.J.; Kakavand, H.; Wilmott, J.S.; Butler, T.; Thompson, J.F.; Mann, G.J.; Haydu, L.E.; et al. Phylogenetic analyses of melanoma reveal complex patterns of metastatic dissemination. Proc. Natl. Acad. Sci. USA 2015, 112, 10995–11000. [Google Scholar] [CrossRef] [PubMed]

| Abbreviation | Gene Name |
|---|---|
| GNAQ | G protein subunit alpha Q |
| KIT | KIT proto-oncogene receptor tyosine kinase |
| BRAF | V-raf murine sarcoma viral oncogene homolog B1 |
| RAS | Rat sarcoma virus |
| NRAS | Neuroblastoma RAS viral oncogene homolog |
| KRAS | Kirsten rat sarcoma virus |
| HRAS | Harvey rat sarcoma viral oncogene homolog |
| NF1 | Neurofibromin 1 |
| CTNNB1 | Catenin beta-1 |
| ATM | Ataxia telangiectasia mutated |
| CDK4 | Cyclin-dependent kinase inhibitor 4 |
| MDM2 | Murine double minute 2 |
| PDGFRA | Platelet derived growth factor receptor alpha |
| CCND2 | Cyclin D2 |
| PIK3CA | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| TERT | Telomerase reverse transcriptase |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A |
| CDKN2B | Cyclin-dependent kinase inhibitor 2B |
| MYC | Myelocytomatosis oncogene |
| MKRN1 | Makorin ring finger protein 1 |
| NOTCH2 | Neurogenic locus notch homolog protein 2 |
| PTEN | Phosphatase and tensin homolog |
| FGF19 | Fibroblast growth factor 19 |
| FGF3 | Fibroblast growth factor 3 |
| No. (%) | Subungual | Acral | p | |
|---|---|---|---|---|
| Sex | 0.810 | |||
| Male | 52 (49.1) | 20 (47.6) | 32 (50.0) | |
| Female | 54 (50.9) | 22 (52.4) | 32 (50.0) | |
| Age (years) | 0.009 | |||
| ≤63 | 49 (46.2) | 26 (61.9) | 23 (35.9) | |
| >63 | 57 (53.8) | 16 (38.1) | 41 (64.1) | |
| Tumor location | 0.000 | |||
| Upper | 41 (38.7) | 29 (69.0) | 12 (18.8) | |
| Lower | 65 (61.3) | 13 (31.0) | 52 (81.3) | |
| Invasion of the dermis | 0.004 | |||
| In situ melanoma | 17 (16.0) | 12 (28.6) | 5 (7.8) | |
| Invasive melanoma | 89 (84.0) | 30 (71.4) | 59 (92.2) | |
| Breslow thickness | 0.037 | |||
| In situ | 17 (16.0) | 12 (28.6) | 5 (7.8) | |
| <1 mm | 27 (25.5) | 12 (28.6) | 15 (23.4) | |
| 1.01–2.00 mm | 15 (14.2) | 4 (9.5) | 11 (17.2) | |
| 2.01–4.00 mm | 19 (17.9) | 5 (11.9) | 14 (21.9) | |
| >4.00 mm | 28 (26.4) | 9 (21.4) | 19 (29.7) | |
| Lymph node metastasis at the time of diagnosis | 0.944 | |||
| Absent | 88 (83.0) | 35 (83.3) | 53 (82.8) | |
| Present | 18 (17.0) | 7 (16.7) | 11 (17.2) | |
| Distant metastasis at the time of diagnosis | 0.821 | |||
| Absent | 103 (97.2) | 41 (97.6) | 62 (96.9) | |
| Present | 3 (2.8) | 1 (2.4) | 2 (3.1) | |
| Stage group | 0.016 | |||
| 0 | 17 (16.0) | 12 (28.6) | 5 (7.8) | |
| I–II | 70 (66.0) | 23 (54.8) | 47 (73.4) | |
| III–IV | 19 (17.9) | 7 (16.7) | 12 (18.8) | |
| Immunotherapy | 0.696 | |||
| Not carried out | 76 (71.7) | 31 (73.8) | 45 (70.3) | |
| Carried out | 30 (28.3) | 11 (26.2) | 19 (29.7) | |
| Trauma | 0.005 | |||
| Absent | 101 (95.3) | 37 (88.1) | 64 (100.0) | |
| Present | 5 (4.7) | 5 (11.9) | 0 (0.0) | |
| Ulceration | 0.226 | |||
| Absent | 71 (67.0) | 31 (73.8) | 40 (62.5) | |
| Present | 35 (33.3) | 11 (26.2) | 24 (37.5) | |
| Loco-regional recurrence | 0.056 | |||
| Absent | 93 (87.7) | 40 (95.2) | 53 (82.8) | |
| Present | 13 (12.3) | 2 (4.8) | 11 (17.2) | |
| Distant metastasis during the follow-up | 0.622 | |||
| Absent | 78 (73.6) | 32 (76.2) | 46 (71.9) | |
| Present | 28 (26.4) | 10 (23.8) | 18 (21.6) |
| No. (%) | Subungual | Acral | p | |
|---|---|---|---|---|
| Sex | 0.936 | |||
| Male | 41 (46.1) | 14 (46.7) | 27 (45.8) | |
| Female | 48 (53.9) | 16 (53.3) | 32 (54.2) | |
| Age (years) | 0.054 | |||
| ≤63 | 35 (39.3) | 16 (53.3) | 19 (32.2) | |
| >63 | 54 (30.7) | 14 (46.7) | 40 (67.8) | |
| Tumor location | 0.000 | |||
| Upper | 28 (31.5) | 18 (60.0) | 10 (16.9) | |
| Lower | 61 (68.5) | 12 (40.0) | 49 (83.1) | |
| Breslow thickness | 0.528 | |||
| <1 mm | 27 (30.3) | 12 (40.0) | 15 (25.4) | |
| 1.01–2.00 mm | 15 (16.9) | 4 (13.3) | 11 (18.6) | |
| 2.01–4.00 mm | 19 (21.3) | 5 (16.7) | 14 (23.7) | |
| >4.00 mm | 39 (31.5) | 9 (30.0) | 19 (32.2) | |
| Lymph node metastasis at the time of diagnosis | 0.603 | |||
| Absent | 71 (79.8) | 23 (76.7) | 48 (81.4) | |
| Present | 18 (20.2) | 7 (23.3) | 11 (18.6) | |
| Distant metastasis at the time of diagnosis | 0.989 | |||
| Absent | 86 (96.6) | 29 (96.7) | 57 (96.6) | |
| Present | 3 (3.4) | 1 (3.3) | 2 (3.4) | |
| Stage group | 0.745 | |||
| I–II | 70 (78.7) | 23 (76.7) | 47 (79.7) | |
| III–IV | 19 (21.3) | 7 (23.3) | 12 (20.3) | |
| Immunotherapy | 0.674 | |||
| Not carried out | 59 (66.3) | 19 (63.3) | 40 (67.8) | |
| Carried out | 30 (33.7) | 11 (36.7) | 19 (32.2) | |
| Trauma | 0.004 | |||
| Absent | 85 (95.5) | 26 (86.7) | 59 (100.0) | |
| Present | 4 (4.5) | 4 (13.3) | 0 (0.0) | |
| Ulceration | 0.714 | |||
| Absent | 54 (60.7) | 19 (63.3) | 35 (59.3) | |
| Present | 35 (39.3) | 11 (36.7) | 24 (40.7) | |
| Loco-regional recurrence | 0.179 | |||
| Absent | 77 (86.5) | 28 (93.3) | 49 (83.1) | |
| Present | 12 (13.5) | 2 (6.7) | 10 (16.9) | |
| Distant metastasis during the follow-up | 0.786 | |||
| Absent | 61 (68.5) | 20 (66.7) | 41 (69.5) | |
| Present | 28 (31.5) | 10 (33.3) | 18 (30.5) |
| Melanoma Including In Situ Lesions | p | HR | 95% CI |
|---|---|---|---|
| Subungual vs. Acral | 0.016 | 2.936 | 1.225–7.036 |
| Age (under 63 years vs. over 63 years) | 0.261 | 1.529 | 0.730–3.205 |
| Stage 0 | 0.038 | ||
| Stage I | 0.408 | 2.474 | 0.289–21.177 |
| Stage II | 0.075 | 6.592 | 0.89–52.403 |
| Stage III | 0.039 | 9.331 | 1.119–77.801 |
| Stage IV | 0.167 | 7.328 | 0.436–123.133 |
| Melanoma with Invasive Lesions | p | HR | 95% CI |
|---|---|---|---|
| Subungual vs. Acral | 0.040 | 2.462 | 1.044–5.806 |
| Age (under 63 years vs. over 63 years) | 0.279 | 1.504 | 0.718–3.151 |
| Stage I | 0.073 | ||
| Stage II | 0.030 | 2.628 | 1.099–6.282 |
| Stage III | 0.013 | 3.723 | 1.325–10.459 |
| Stage IV | 0.328 | 2.909 | 0.342–24.716 |
| Clinical and Molecular Profiles | Subungual Melanoma |
|---|---|
| Sex | Male predominance [4,7,14,15,17,21,24] Female predominance [8,14,15,16,25]. |
| Mean age (years) | 47–66 years [4,7,8,14,15,16,17,18,21]. |
| The most common tumor location | Fingernail (17–63%) [4,7,14,16,17,18,19,21,25] Toenail (26.3%, 68%, and 78%) [15,24] |
| The most common Clark level | Clark level IV (37%, 53%, and 70%) [7,15,18,21] |
| The most common Breslow thickness | ≤1 mm (63.2%) [8] 1.01–4 mm (34–50%) [7,18,21,24] >4 mm (31%, 34.9%, 50%) [14,20,25] |
| The most common stage group | 0 (in situ) (63.2%) [8]. I (83%) [17] II (32.6–53.7%) [14,15,20] III (32% and 52%) [21,24] |
| Lymph node metastasis at the time of diagnosis | 9–62.3% [15,16,17,18,21,24] |
| Distant metastasis at the time of diagnosis | 9.3% and 42.6% [18,20] |
| Trauma | 15.8% and 48% [4,8] |
| Ulceration | 47–76% [7,14,15,16,17,20,21,24] |
| 5-year disease free survival | 40% and 57% [7,18] |
| 5-year survival | 74–97% [15,17,18,26] |
| Overall mortality | 17%, 31%, and 46% [17,18,21] |
| BRAF V600E mutation | 0–40% [4,5,14,16,27] |
| NRAS mutation | 0–31% [4,14,16,27] |
| KRAS mutation | 6.5% and 11% [4,27] |
| NF1 mutation | 0–50% [4,27] |
| C-KIT mutation | 11.1%, 13%, and 16% [4,14,16,27] |
| GNAQ mutation | 0–25% [27] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahn, S.-Y.; Bae, G.-E.; Park, S.-Y.; Yeo, M.-K. Differences in the Clinical and Molecular Profiles of Subungual Melanoma and Acral Melanoma in Asian Patients. Cancers 2023, 15, 4417. https://doi.org/10.3390/cancers15174417
Ahn S-Y, Bae G-E, Park S-Y, Yeo M-K. Differences in the Clinical and Molecular Profiles of Subungual Melanoma and Acral Melanoma in Asian Patients. Cancers. 2023; 15(17):4417. https://doi.org/10.3390/cancers15174417
Chicago/Turabian StyleAhn, So-Young, Go-Eun Bae, Seung-Yeol Park, and Min-Kyung Yeo. 2023. "Differences in the Clinical and Molecular Profiles of Subungual Melanoma and Acral Melanoma in Asian Patients" Cancers 15, no. 17: 4417. https://doi.org/10.3390/cancers15174417
APA StyleAhn, S.-Y., Bae, G.-E., Park, S.-Y., & Yeo, M.-K. (2023). Differences in the Clinical and Molecular Profiles of Subungual Melanoma and Acral Melanoma in Asian Patients. Cancers, 15(17), 4417. https://doi.org/10.3390/cancers15174417







