Analyzing Flow Cytometry or Targeted Gene Expression Data Influences Clinical Discoveries—Profiling Blood Samples of Pancreatic Ductal Adenocarcinoma Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Material and Methods
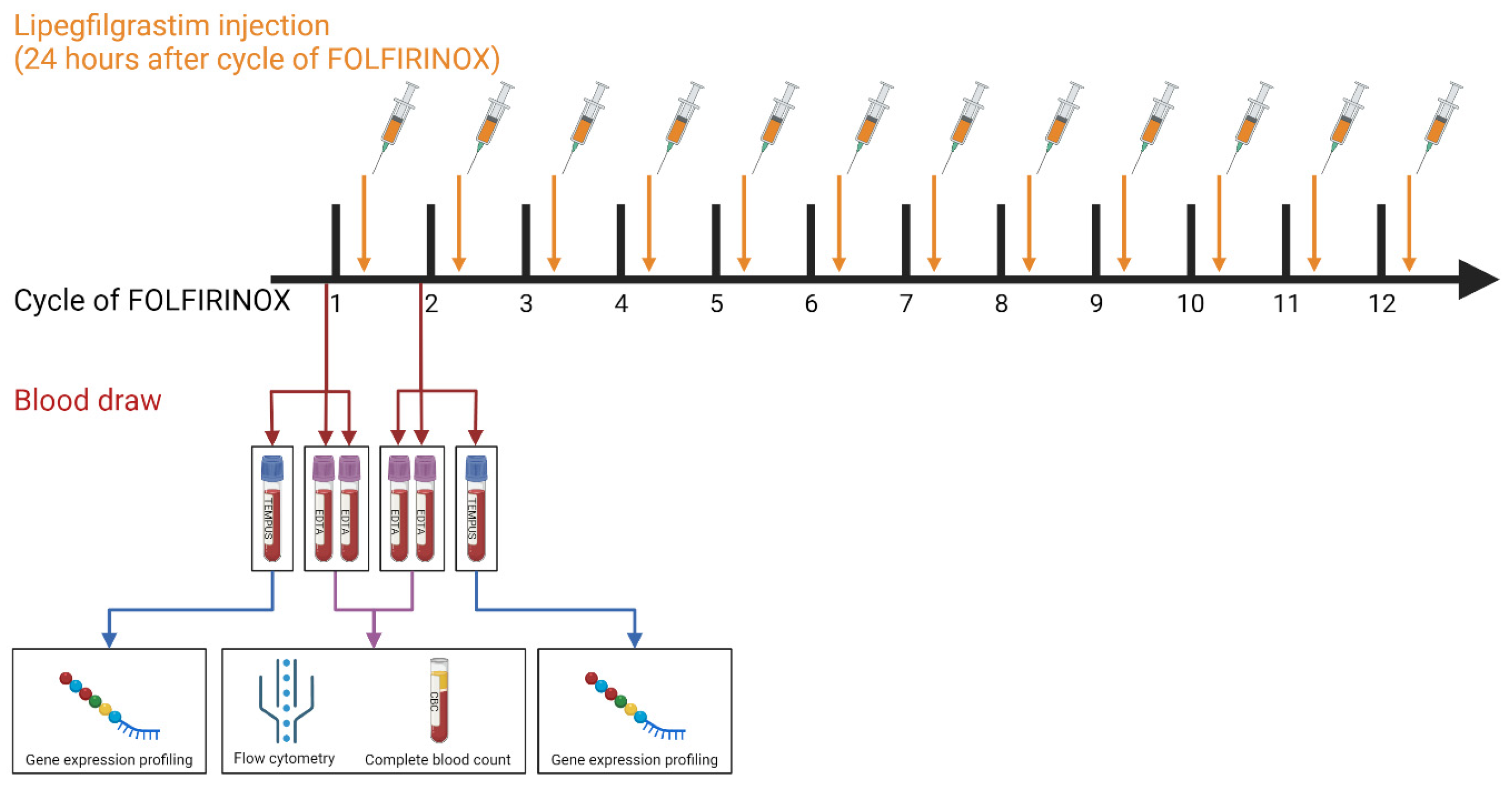
2.1. Patient Cohort and Blood Collection
2.2. Flow Cytometry and Complete Blood Count
2.3. Targeted Immune Gene Expression Profiling
2.4. Statistical Analysis
3. Results
3.1. Patients’ Characteristics
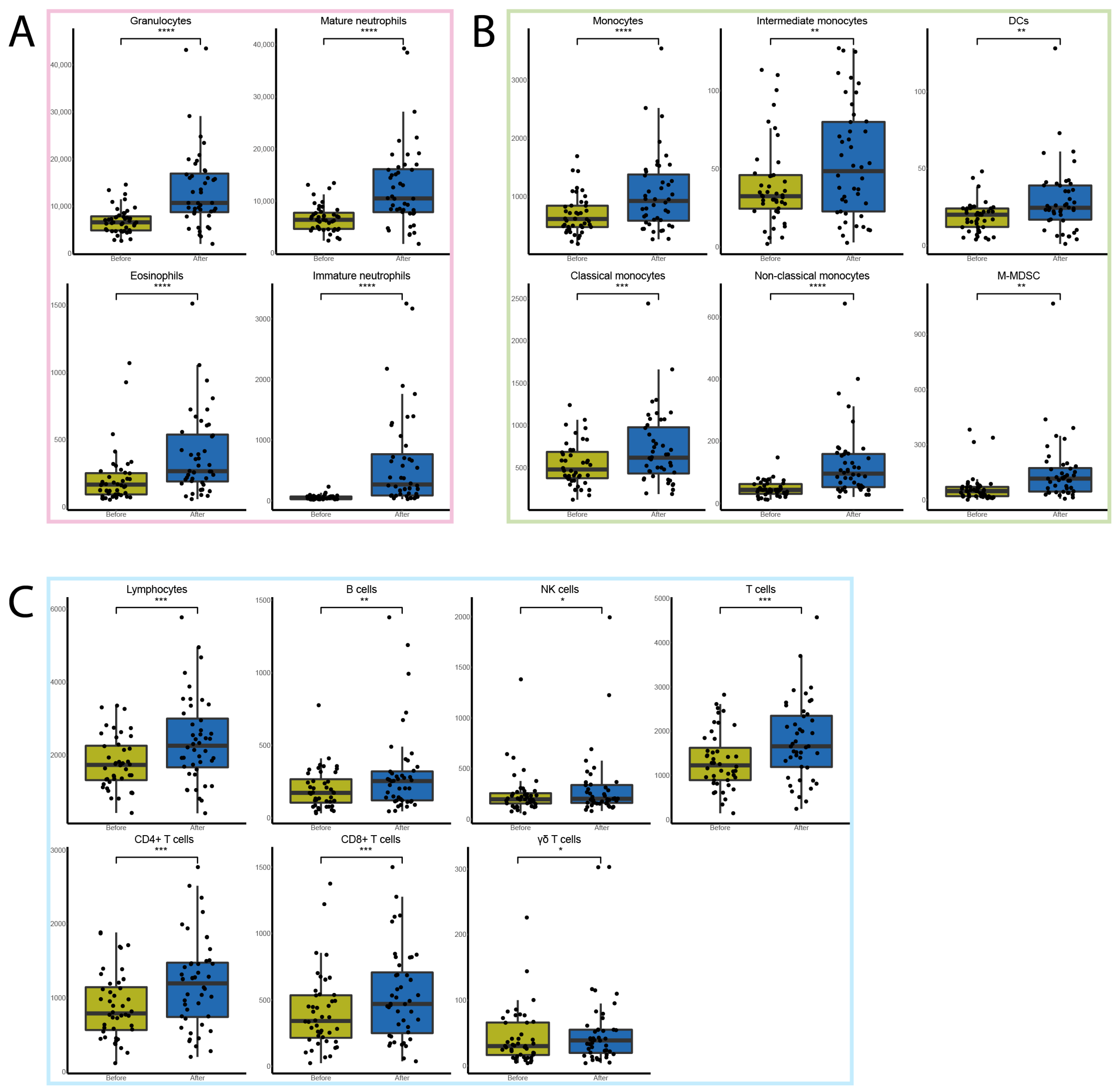
3.2. FFX-Lipeg Therapy Results in Enhanced Frequencies of Granulocytes and Monocytes in the Blood
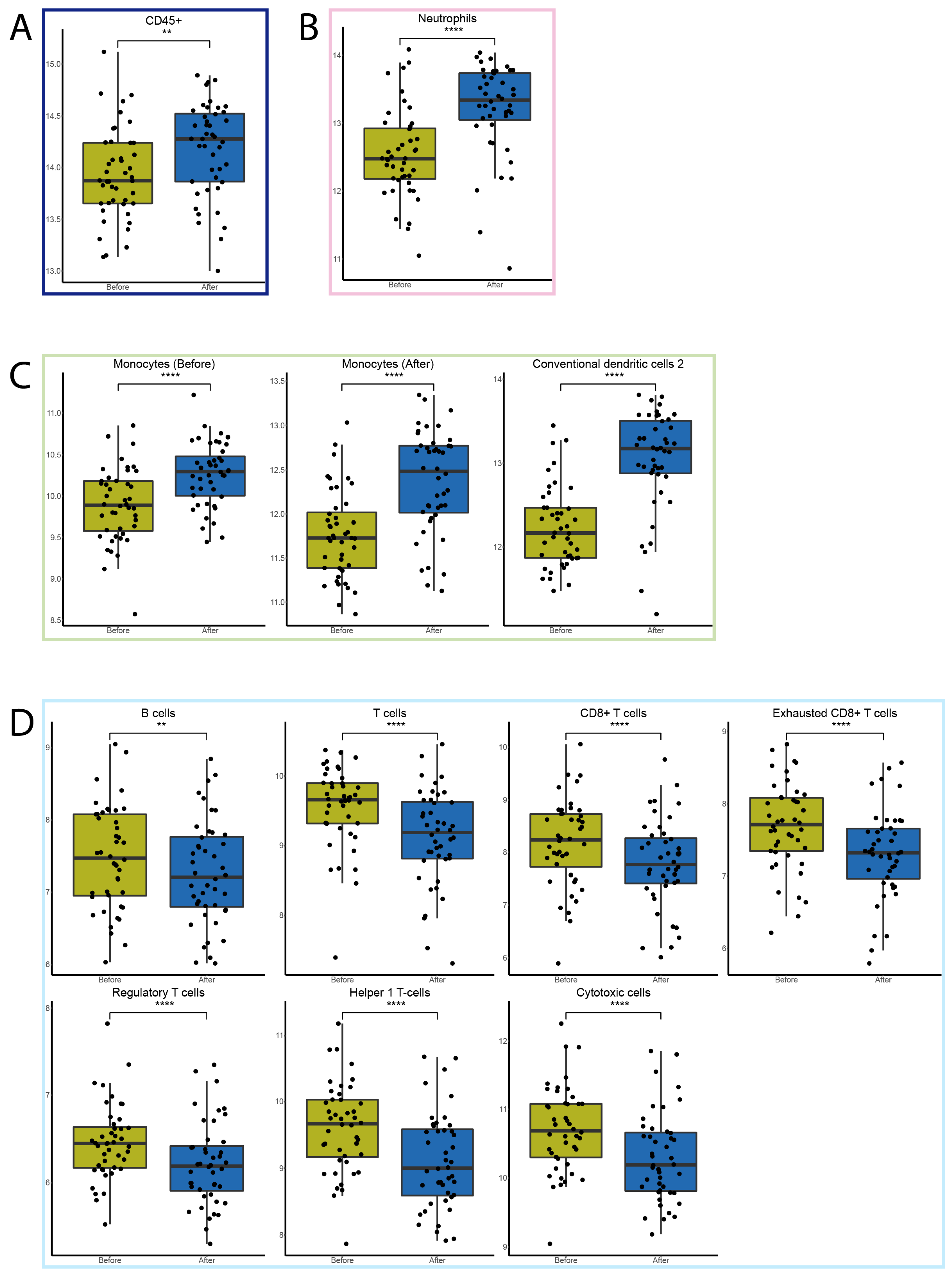
3.3. FFX-Lipeg Results in an Increased Number of Lymphocytes, Yet Pathways Associated with Lymphocyte Functions Are Downregulated
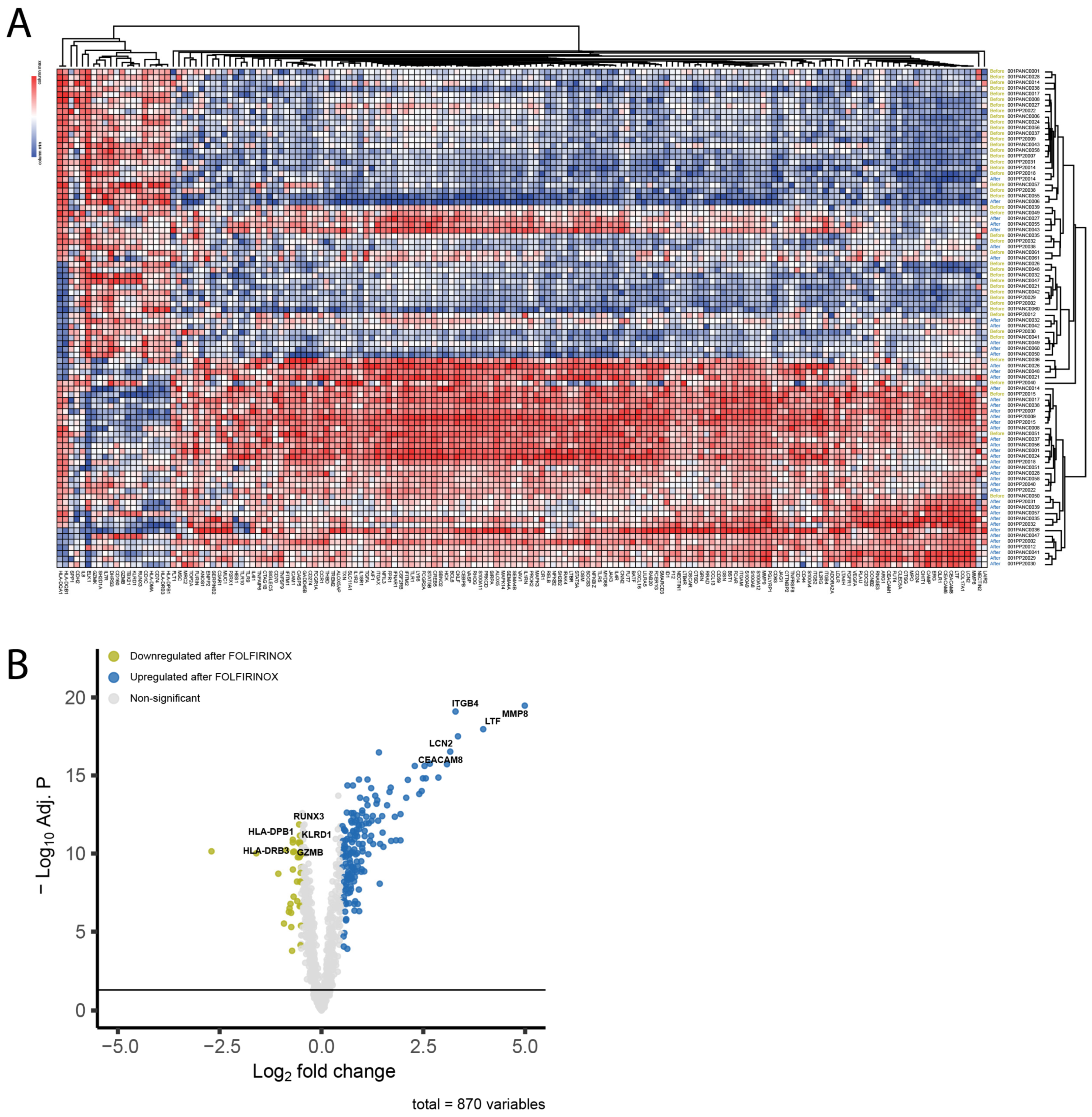
3.4. FFX-Lipeg Induces a Distinct Gene Expression Profile in the Peripheral Blood
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- van der Sijde, F.; van Dam, J.L.; Groot Koerkamp, B.; Haberkorn, B.C.M.; Homs, M.Y.V.; Mathijssen, D.; Besselink, M.G.; Wilmink, J.W.; van Eijck, C.H.J. Treatment Response and Conditional Survival in Advanced Pancreatic Cancer Patients Treated with FOLFIRINOX: A Multicenter Cohort Study. J. Oncol. 2022, 2022, 8549487. [Google Scholar] [CrossRef]
- Conroy, T.; Hammel, P.; Hebbar, M.; ben Abdelghani, M.; Wei, A.C.; Raoul, J.-L.; Choné, L.; Francois, E.; Artru, P.; Biagi, J.J.; et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2395–2406. [Google Scholar] [CrossRef]
- Thibodeau, S.; Voutsadakis, I.A. FOLFIRINOX Chemotherapy in Metastatic Pancreatic Cancer: A Systematic Review and Meta-Analysis of Retrospective and Phase II Studies. J. Clin. Med. 2018, 7, 7. [Google Scholar] [CrossRef]
- Suker, M.; Beumer, B.R.; Sadot, E.; Marthey, L.; Faris, J.E.; Mellon, E.A.; El-Rayes, B.F.; Wang-Gillam, A.; Lacy, J.; Hosein, P.J.; et al. FOLFIRINOX for Locally Advanced Pancreatic Cancer: A Systematic Review and Patient-Level Meta-Analysis. Lancet Oncol. 2016, 17, 801–810. [Google Scholar] [CrossRef]
- Chun, J.W.; Woo, S.M.; Lee, S.H.; Choi, J.H.; Park, N.; Kim, J.S.; Cho, I.R.; Paik, W.H.; Lee, W.J.; Ryu, J.K.; et al. A Real-World Analysis of Nanoliposomal-Irinotecan with 5-Fluorouracil and Folinic Acid as Third- or Later-Line Therapy in Patients with Metastatic Pancreatic Adenocarcinoma. Ther. Adv. Med. Oncol. 2022, 14, 175883592211195. [Google Scholar] [CrossRef]
- Irisawa, A.; Takeno, M.; Watanabe, K.; Takahashi, H.; Mitsunaga, S.; Ikeda, M. Incidence of and Risk Factors for Severe Neutropenia during Treatment with the Modified FOLFIRINOX Therapy in Patients with Advanced Pancreatic Cancer. Sci. Rep. 2022, 12, 15574. [Google Scholar] [CrossRef]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouché, O.; Guimbaud, R.; Bécouarn, Y.; Adenis, A.; Raoul, J.-L.; Gourgou-Bourgade, S.; de la Fouchardière, C.; et al. FOLFIRINOX versus Gemcitabine for Metastatic Pancreatic Cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef]
- Timmer-Bonte, J.N.H.; Ouwerkerk, J.; Faber, L.M.; Kerkhofs, L.G.M.; Laterveer, L.; ten Oever, D.; van Rees, B.P.; van der Linden, P.W. Lipegfilgrastim for Prophylaxis of Chemotherapy-Induced Neutropenia in Dutch Patients. Neth J. Med. 2020, 78, 270–276. [Google Scholar]
- Terazawa, T.; Goto, M.; Miyamoto, T.; Asaishi, K.; Shimamoto, F.; Kuwakado, S.; Nishitani, H.; Kii, T.; Higuchi, K. Efficacy of Prophylactic G-CSF in Patients Receiving FOLFIRINOX: A Preliminary Retrospective Study. Intern Med. 2015, 54, 2969–2973. [Google Scholar] [CrossRef][Green Version]
- Steger, G.; Pichler, P.; Airoldi, M.; Mazza, P.; Fontaine, C.; Timmer Bonte, J.; Walewski, J.A.; Katolicka, J.; Mikulova, M.; Gasparic, M. Use of Lipegfilgrastim for the Prophylaxis of Chemotherapy-Induced Neutropenia: Pan-European Non-Interventional Study. Ann. Oncol. 2018, 29, viii607–viii608. [Google Scholar] [CrossRef]
- Peng, H.; James, C.A.; Cullinan, D.R.; Hogg, G.D.; Mudd, J.L.; Zuo, C.; Takchi, R.; Caldwell, K.E.; Liu, J.; DeNardo, D.G.; et al. Neoadjuvant FOLFIRINOX Therapy Is Associated with Increased Effector T Cells and Reduced Suppressor Cells in Patients with Pancreatic Cancer. Clin. Cancer Res. 2021, 27, 6761–6771. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Chen, G.; Huang, Y.; Zhou, J.; Lin, L.Z.; Feng, J.; Wang, Z.; Shu, Y.; Shi, J.; Hu, Y.; et al. Camrelizumab plus Carboplatin and Pemetrexed versus Chemotherapy Alone in Chemotherapy-Naive Patients with Advanced Non-Squamous Non-Small-Cell Lung Cancer (CameL): A Randomised, Open-Label, Multicentre, Phase 3 Trial. Lancet Respir. Med. 2021, 9, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Principe, D.R.; Narbutis, M.; Kumar, S.; Park, A.; Viswakarma, N.; Dorman, M.J.; Kamath, S.D.; Grippo, P.J.; Fishel, M.L.; Hwang, R.F.; et al. Long-Term Gemcitabine Treatment Reshapes the Pancreatic Tumor Microenvironment and Sensitizes Murine Carcinoma to Combination Immunotherapy. Cancer Res. 2020, 80, 3101–3115. [Google Scholar] [CrossRef]
- Maecker, H.T.; McCoy, J.P.; Nussenblatt, R. Standardizing Immunophenotyping for the Human Immunology Project. Nat. Rev. Immunol. 2012, 12, 191–200. [Google Scholar] [CrossRef]
- McKinnon, K.M. Flow Cytometry: An Overview. Curr. Protoc. Immunol. 2018, 120, 5.1.1–5.1.11. [Google Scholar] [CrossRef]
- Costantini, A.; Mancini, S.; Giuliodoro, S.; Butini, L.; Regnery, C.M.; Silvestri, G.; Montroni, M. Effects of Cryopreservation on Lymphocyte Immunophenotype and Function. J. Immunol. Methods 2003, 278, 145–155. [Google Scholar] [CrossRef]
- Campbell, D.E.; Tustin, N.B.; Riedel, E.; Tustin, R.; Taylor, J.; Murray, J.; Douglas, S.D. Cryopreservation Decreases Receptor PD-1 and Ligand PD-L1 Coinhibitory Expression on Peripheral Blood Mononuclear Cell-Derived T Cells and Monocytes. Clin. Vaccine Immunol. 2009, 16, 1648–1653. [Google Scholar] [CrossRef][Green Version]
- Weinberg, A.; Song, L.Y.; Wilkening, C.; Sevin, A.; Blais, B.; Louzao, R.; Stein, D.; Defechereux, P.; Durand, D.; Riedel, E.; et al. Optimization and Limitations of Use of Cryopreserved Peripheral Blood Mononuclear Cells for Functional and Phenotypic T-Cell Characterization. Clin. Vaccine Immunol. 2009, 16, 1176–1186. [Google Scholar] [CrossRef]
- Bagaev, A.; Kotlov, N.; Nomie, K.; Svekolkin, V.; Gafurov, A.; Isaeva, O.; Osokin, N.; Kozlov, I.; Frenkel, F.; Gancharova, O.; et al. Conserved Pan-Cancer Microenvironment Subtypes Predict Response to Immunotherapy. Cancer Cell 2021, 39, 845–865.e7. [Google Scholar] [CrossRef]
- Danaher, P.; Warren, S.; Dennis, L.; D’Amico, L.; White, A.; Disis, M.L.; Geller, M.A.; Odunsi, K.; Beechem, J.; Fling, S.P. Gene Expression Markers of Tumor Infiltrating Leukocytes. J. Immunother. Cancer 2017, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- de Koning, W.; Latifi, D.; Li, Y.; van Eijck, C.H.J.; Stubbs, A.P.; Mustafa, D.A.M. Identification, Validation, and Utilization of Immune Cells in Pancreatic Ductal Adenocarcinoma Based on Marker Genes. Front. Immunol. 2021, 12, 1449. [Google Scholar] [CrossRef]
- van Eijck, C.W.F.; de Koning, W.; van der Sijde, F.; Moskie, M.; Groot Koerkamp, B.; Homs, M.Y.V.; van der Burg, S.H.; van Eijck, C.H.J.; Mustafa, D.A.M. A Multigene Circulating Biomarker to Predict the Lack of FOLFIRINOX Response after a Single Cycle in Patients with Pancreatic Ductal Adenocarcinoma. Eur. J. Cancer 2023, 181, 119–134. [Google Scholar] [CrossRef] [PubMed]
- Lyman, G.H.; Yau, L.; Nakov, R.; Krendyukov, A. Overall Survival and Risk of Second Malignancies with Cancer Chemotherapy and G-CSF Support. Ann. Oncol. 2018, 29, 1903–1910. [Google Scholar] [CrossRef] [PubMed]
- Kunert, A.; Basak, E.A.; Hurkmans, D.P.; Balcioglu, H.E.; Klaver, Y.; van Brakel, M.; Oostvogels, A.A.M.; Lamers, C.H.J.; Bins, S.; Koolen, S.L.W.; et al. CD45RA+CCR7− CD8 T Cells Lacking Co-Stimulatory Receptors Demonstrate Enhanced Frequency in Peripheral Blood of NSCLC Patients Responding to Nivolumab. J. Immunother. Cancer 2019, 7, 149. [Google Scholar] [CrossRef]
- Rijnders, M.; Robbrecht, D.G.J.; Oostvogels, A.A.M.; van Brakel, M.; Boormans, J.L.; Aarts, M.J.B.; Balcioglu, H.E.; Hamberg, P.; Voortman, J.; Westgeest, H.M.; et al. A Blood-Based Immune Marker for Resistance to Pembrolizumab in Patients with Metastatic Urothelial Cancer. Cancer Immunol. Immunother. 2022, 72, 759–767. [Google Scholar] [CrossRef]
- van der Sijde, F.; Li, Y.; Schraauwen, R.; de Koning, W.; van Eijck, C.H.J.; Mustafa, D.A.M. RNA from Stabilized Whole Blood Enables More Comprehensive Immune Gene Expression Profiling Compared to RNA from Peripheral Blood Mononuclear Cells. PLoS ONE 2020, 15, e0235413. [Google Scholar] [CrossRef]
- Geiss, G.K.; Bumgarner, R.E.; Birditt, B.; Dahl, T.; Dowidar, N.; Dunaway, D.L.; Fell, H.P.; Ferree, S.; George, R.D.; Grogan, T.; et al. Direct Multiplexed Measurement of Gene Expression with Color-Coded Probe Pairs. Nat. Biotechnol. 2008, 26, 317–325. [Google Scholar] [CrossRef]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; van Roy, N.; de Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape Plug-in to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinformatics 2009, 25, 1091. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2022. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. 2021. Available online: https://bioconductor.org/packages/devel/bioc/vignettes/EnhancedVolcano/inst/doc/EnhancedVolcano.html (accessed on 27 June 2023).
- Tukey, J.W. Exploratory Data Analysis. Addison-Wesley, Reading, MA. J. Biopharm. Stat. 1977, 1, 133–138. [Google Scholar]
- Sirniö, P.; Tuomisto, A.; Tervahartiala, T.; Sorsa, T.; Klintrup, K.; Karhu, T.; Herzig, K.-H.; Mäkelä, J.; Karttunen, T.J.; Salo, T.; et al. High-Serum MMP-8 Levels Are Associated with Decreased Survival and Systemic Inflammation in Colorectal Cancer. Br. J. Cancer 2018, 119, 213–219. [Google Scholar] [CrossRef]
- Kruzel, M.L.; Zimecki, M.; Actor, J.K. Lactoferrin in a Context of Inflammation-Induced Pathology. Front. Immunol. 2017, 8, 1438. [Google Scholar] [CrossRef]
- Ribon, M.; Mussard, J.; Semerano, L.; Singer, B.B.; Decker, P. Extracellular Chromatin Triggers Release of Soluble CEACAM8 Upon Activation of Neutrophils. Front. Immunol. 2019, 10, 1346. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Ting, S.-M.; Sun, G.; Roy-O’Reilly, M.; Mobley, A.S.; Bautista Garrido, J.; Zheng, X.; Obertas, L.; Jung, J.E.; Kruzel, M.; et al. Beneficial Role of Neutrophils Through Function of Lactoferrin After Intracerebral Hemorrhage. Stroke 2018, 49, 1241–1247. [Google Scholar] [CrossRef]
- Yamanaka, T.; Kuroki, M.; Kinugasa, T.; Matsuo, Y.; Matsuoka, Y. Preparation and Characterization of Two Human Carcinoembryonic Antigen Family Proteins of Neutrophils, CD66b and c, in Silkworm Larvae. Protein Expr. Purif. 1996, 7, 438–446. [Google Scholar] [CrossRef]
- Meng, X.; Liu, P.; Wu, Y.; Liu, X.; Huang, Y.; Yu, B.; Han, J.; Jin, H.; Tan, X. Integrin Beta 4 (ITGB4) and Its Tyrosine-1510 Phosphorylation Promote Pancreatic Tumorigenesis and Regulate the MEK1-ERK1/2 Signaling Pathway. Bosn. J. Basic Med. Sci. 2019, 20, 106. [Google Scholar] [CrossRef]
- Yang, J.; Goetz, D.; Li, J.-Y.; Wang, W.; Mori, K.; Setlik, D.; Du, T.; Erdjument-Bromage, H.; Tempst, P.; Strong, R.; et al. An Iron Delivery Pathway Mediated by a Lipocalin. Mol. Cell 2002, 10, 1045–1056. [Google Scholar] [CrossRef]
- Yamao, K.; Takenaka, M.; Yoshikawa, T.; Ishikawa, R.; Okamoto, A.; Yamazaki, T.; Nakai, A.; Omoto, S.; Kamata, K.; Minaga, K.; et al. Clinical Safety and Efficacy of Secondary Prophylactic Pegylated G-CSF in Advanced Pancreatic Cancer Patients Treated with MFOLFIRINOX: A Single-Center Retrospective Study. Intern. Med. 2019, 58, 1993. [Google Scholar] [CrossRef]
- Carvalho de Brito, A.B.; Riechelmann, R.P.; Fonseca de Jesus, V.H. Impact of Granulocyte Colony-Stimulating Factor (G-CSF) on the Outcomes of Patients with Metastatic Pancreatic Adenocarcinoma (MPA) During First-Line Treatment with FOLFIRINOX: A Single-Center Retrospective Analysis. Cancer Control 2023, 30, 10732748221149543. [Google Scholar] [CrossRef]
- Bronte, V.; Brandau, S.; Chen, S.H.; Colombo, M.P.; Frey, A.B.; Greten, T.F.; Mandruzzato, S.; Murray, P.J.; Ochoa, A.; Ostrand-Rosenberg, S.; et al. Recommendations for Myeloid-Derived Suppressor Cell Nomenclature and Characterization Standards. Nat. Commun. 2016, 7, 12150. [Google Scholar] [CrossRef]
- Nywening, T.M.; Wang-Gillam, A.; Sanford, D.E.; Belt, B.A.; Panni, R.Z.; Cusworth, B.M.; Toriola, A.T.; Nieman, R.K.; Worley, L.A.; Yano, M.; et al. Targeting Tumour-Associated Macrophages with CCR2 Inhibition in Combination with FOLFIRINOX in Patients with Borderline Resectable and Locally Advanced Pancreatic Cancer: A Single-Centre, Open-Label, Dose-Finding, Non-Randomised, Phase 1b Trial. Lancet Oncol. 2016, 17, 651–662. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Koning, W.; van Eijck, C.W.F.; van der Sijde, F.; Strijk, G.J.; Oostvogels, A.A.M.; Debets, R.; van Eijck, C.H.J.; Mustafa, D.A.M. Analyzing Flow Cytometry or Targeted Gene Expression Data Influences Clinical Discoveries—Profiling Blood Samples of Pancreatic Ductal Adenocarcinoma Patients. Cancers 2023, 15, 4349. https://doi.org/10.3390/cancers15174349
de Koning W, van Eijck CWF, van der Sijde F, Strijk GJ, Oostvogels AAM, Debets R, van Eijck CHJ, Mustafa DAM. Analyzing Flow Cytometry or Targeted Gene Expression Data Influences Clinical Discoveries—Profiling Blood Samples of Pancreatic Ductal Adenocarcinoma Patients. Cancers. 2023; 15(17):4349. https://doi.org/10.3390/cancers15174349
Chicago/Turabian Stylede Koning, Willem, Casper W. F. van Eijck, Fleur van der Sijde, Gaby J. Strijk, Astrid A. M. Oostvogels, Reno Debets, Casper H. J. van Eijck, and Dana A. M. Mustafa. 2023. "Analyzing Flow Cytometry or Targeted Gene Expression Data Influences Clinical Discoveries—Profiling Blood Samples of Pancreatic Ductal Adenocarcinoma Patients" Cancers 15, no. 17: 4349. https://doi.org/10.3390/cancers15174349
APA Stylede Koning, W., van Eijck, C. W. F., van der Sijde, F., Strijk, G. J., Oostvogels, A. A. M., Debets, R., van Eijck, C. H. J., & Mustafa, D. A. M. (2023). Analyzing Flow Cytometry or Targeted Gene Expression Data Influences Clinical Discoveries—Profiling Blood Samples of Pancreatic Ductal Adenocarcinoma Patients. Cancers, 15(17), 4349. https://doi.org/10.3390/cancers15174349








