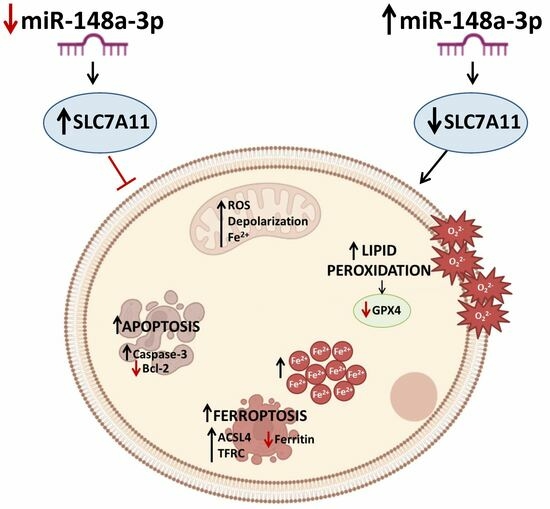
MiR-148a-3p Promotes Colorectal Cancer Cell Ferroptosis by Targeting SLC7A11
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Transfection
2.2. Quantitative Real-Time PCR (qRT-PCR)
2.3. Viability and Cytotoxicity
2.4. Apoptotic Cell Death
2.5. Mitochondrial State
2.6. Lipid Peroxidation
2.7. Ferroptotic Mechanism
2.8. Cell lysis, Immunoblotting, and Antibodies
2.9. Bioinformatics Analysis
2.10. Statistical Analysis
3. Results
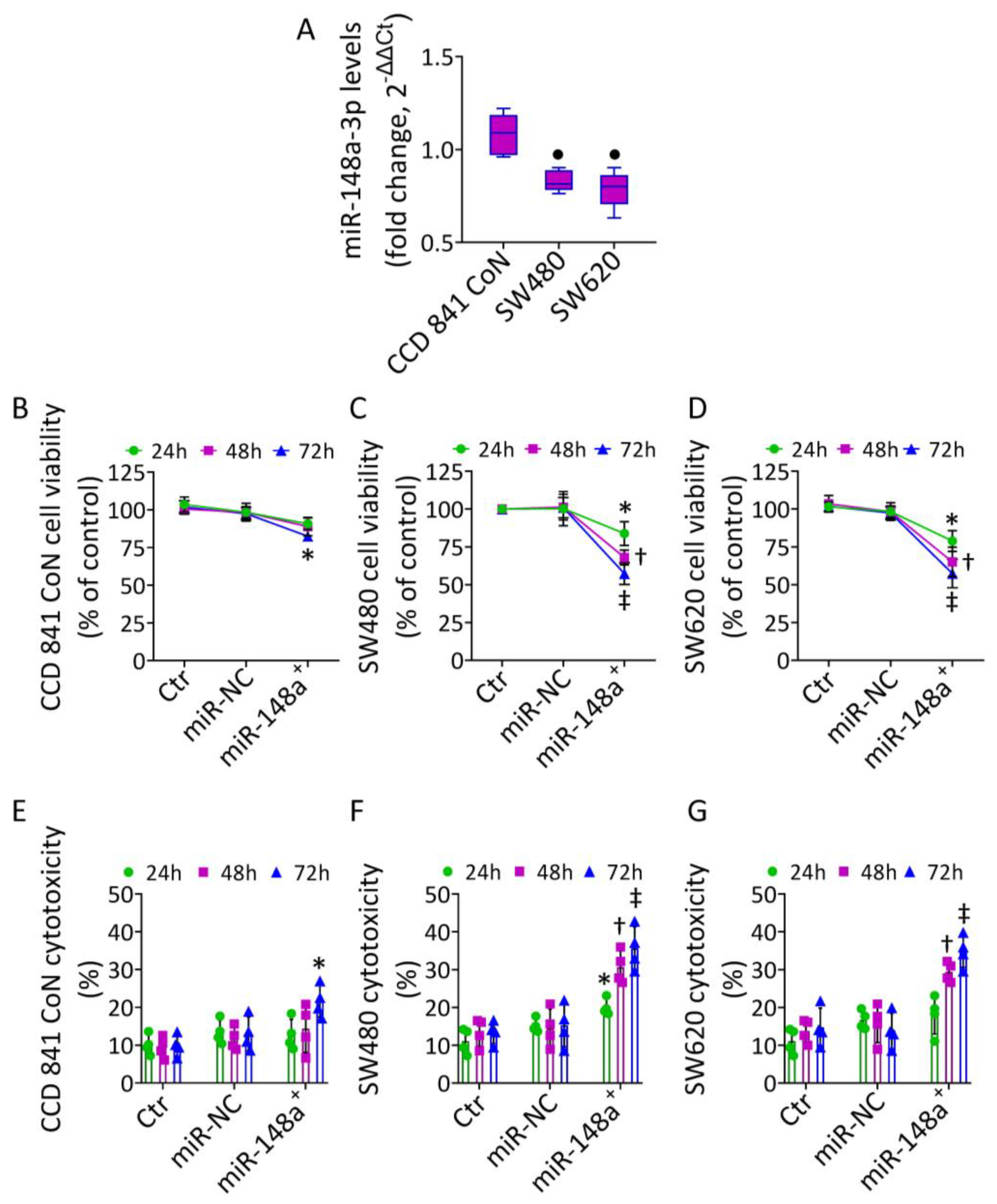
3.1. MiR-148a Levels in CRC
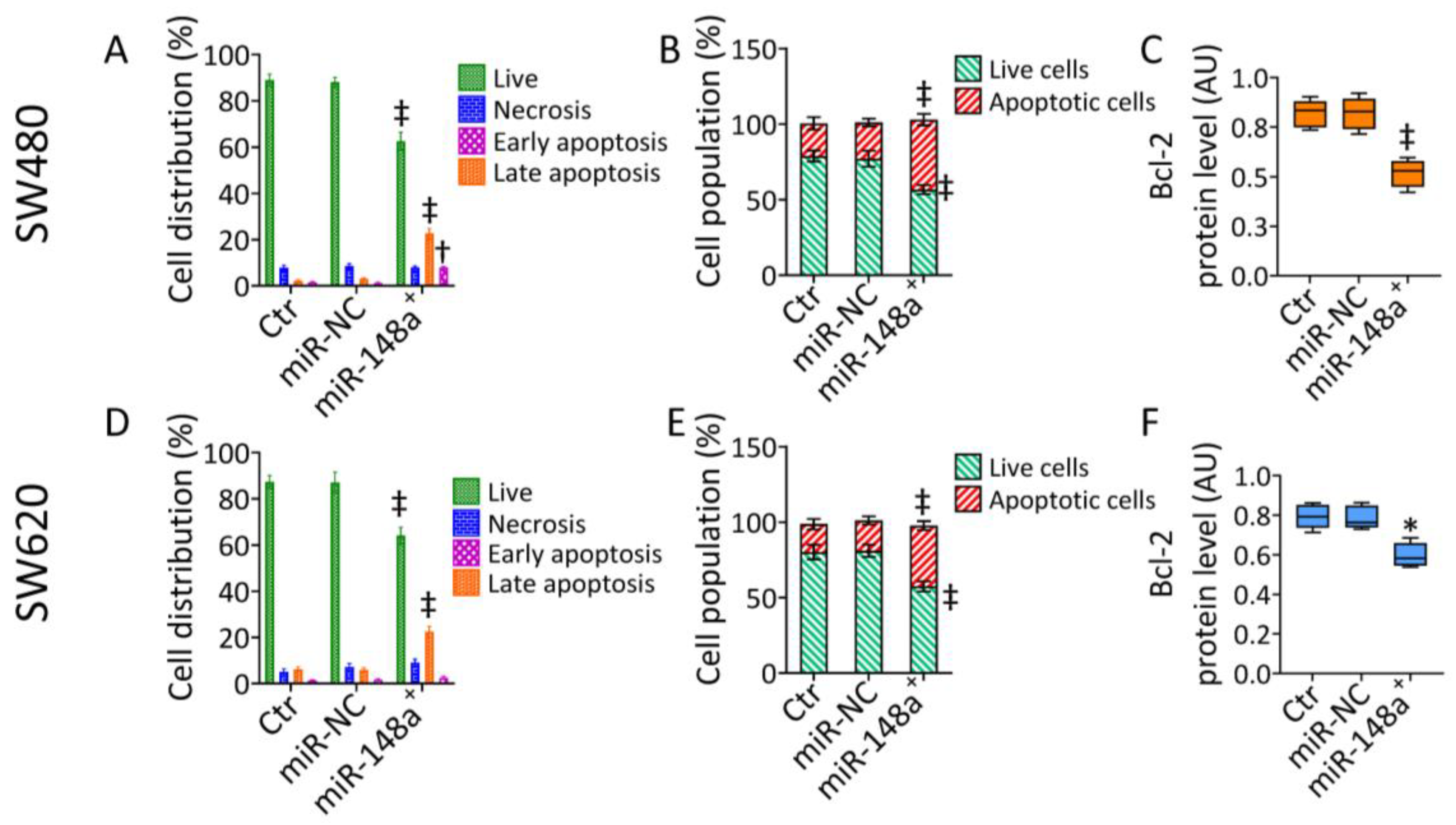
3.2. MiR-148a+ Induced Caspase-3-Dependent Apoptosis in CRC
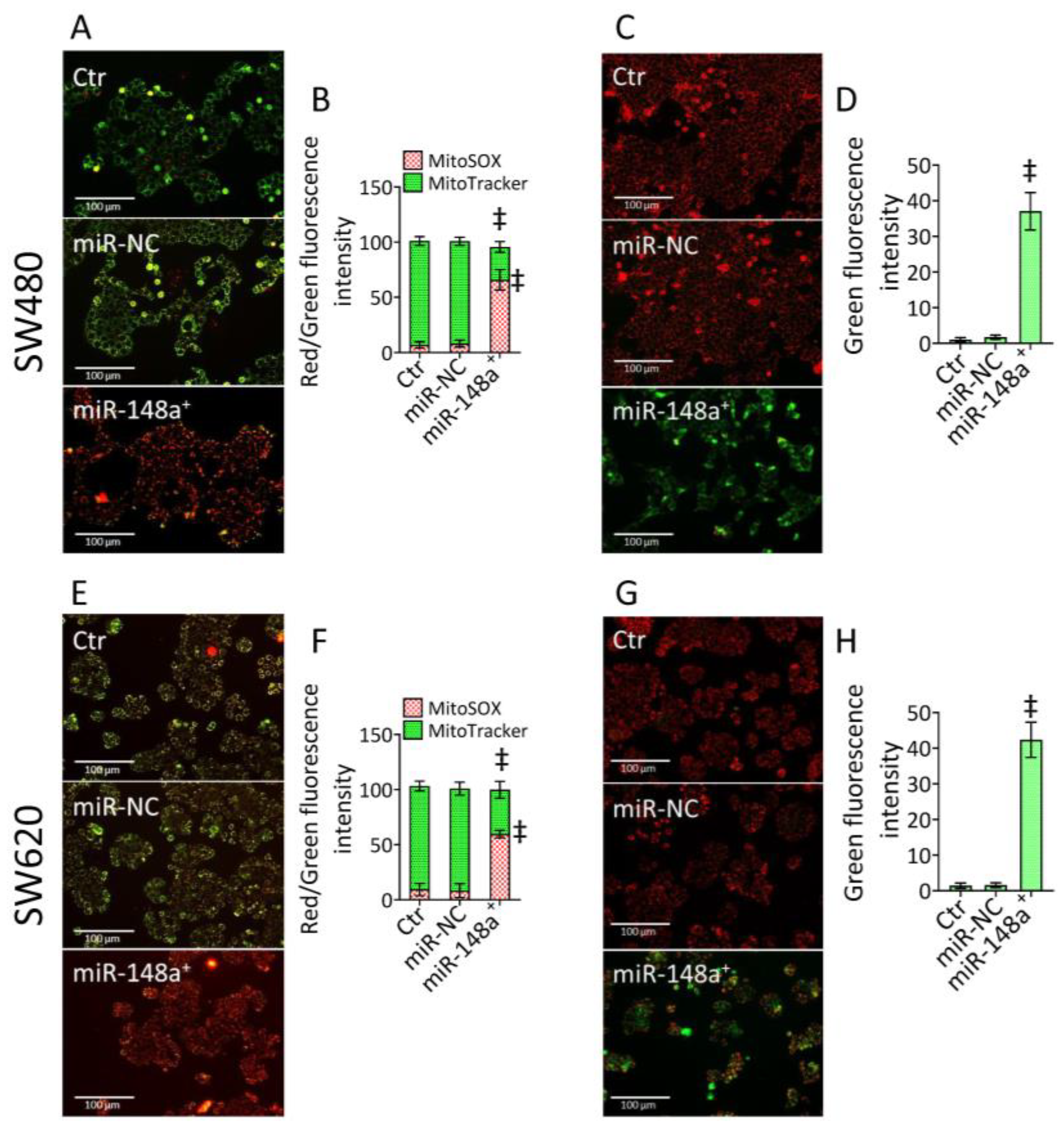
3.3. MiR-148a+ Triggered Mitochondrial Damage in CRC
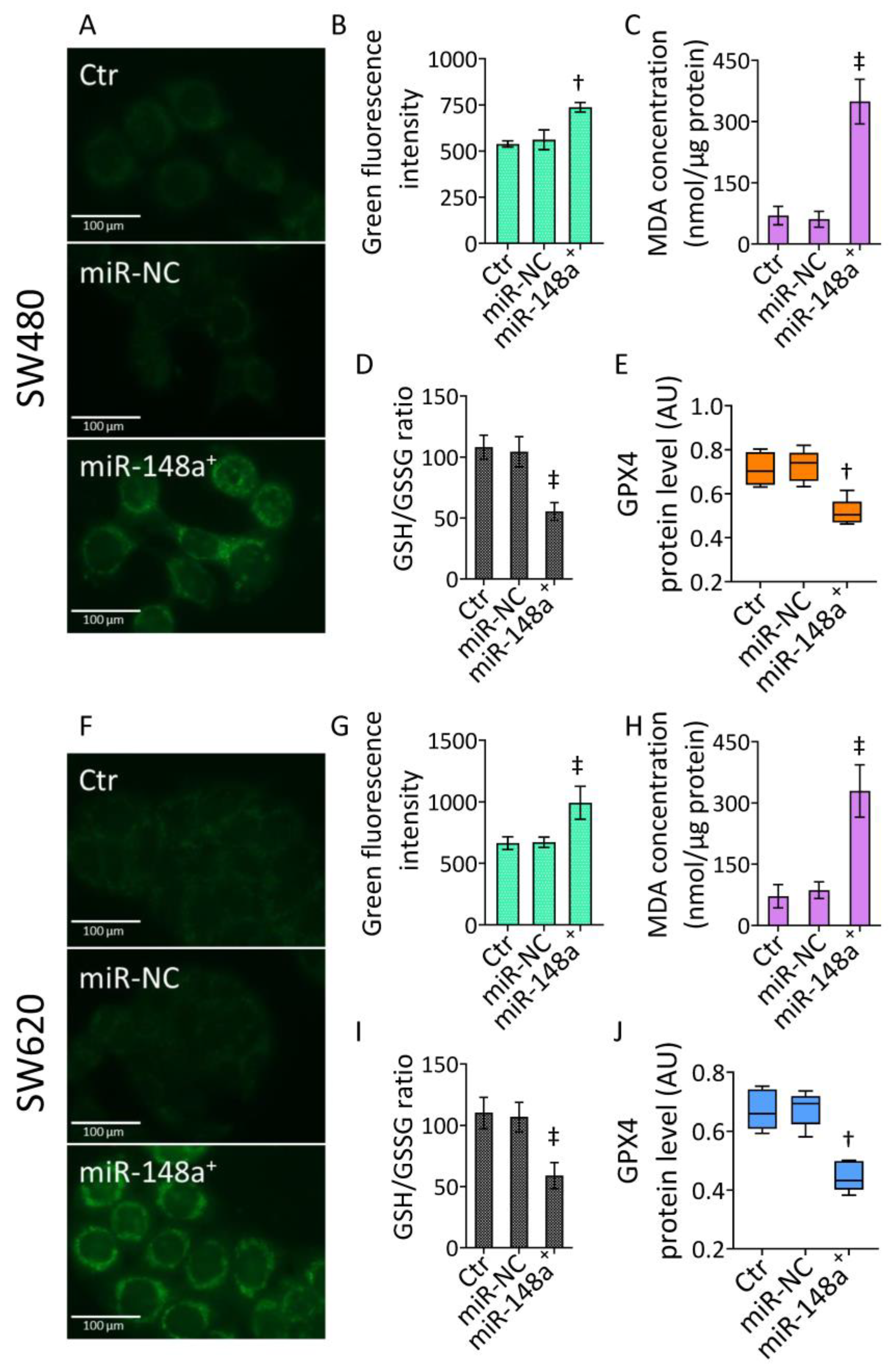
3.4. MiR-148a+ Promoted Lipid Peroxidation in CRC
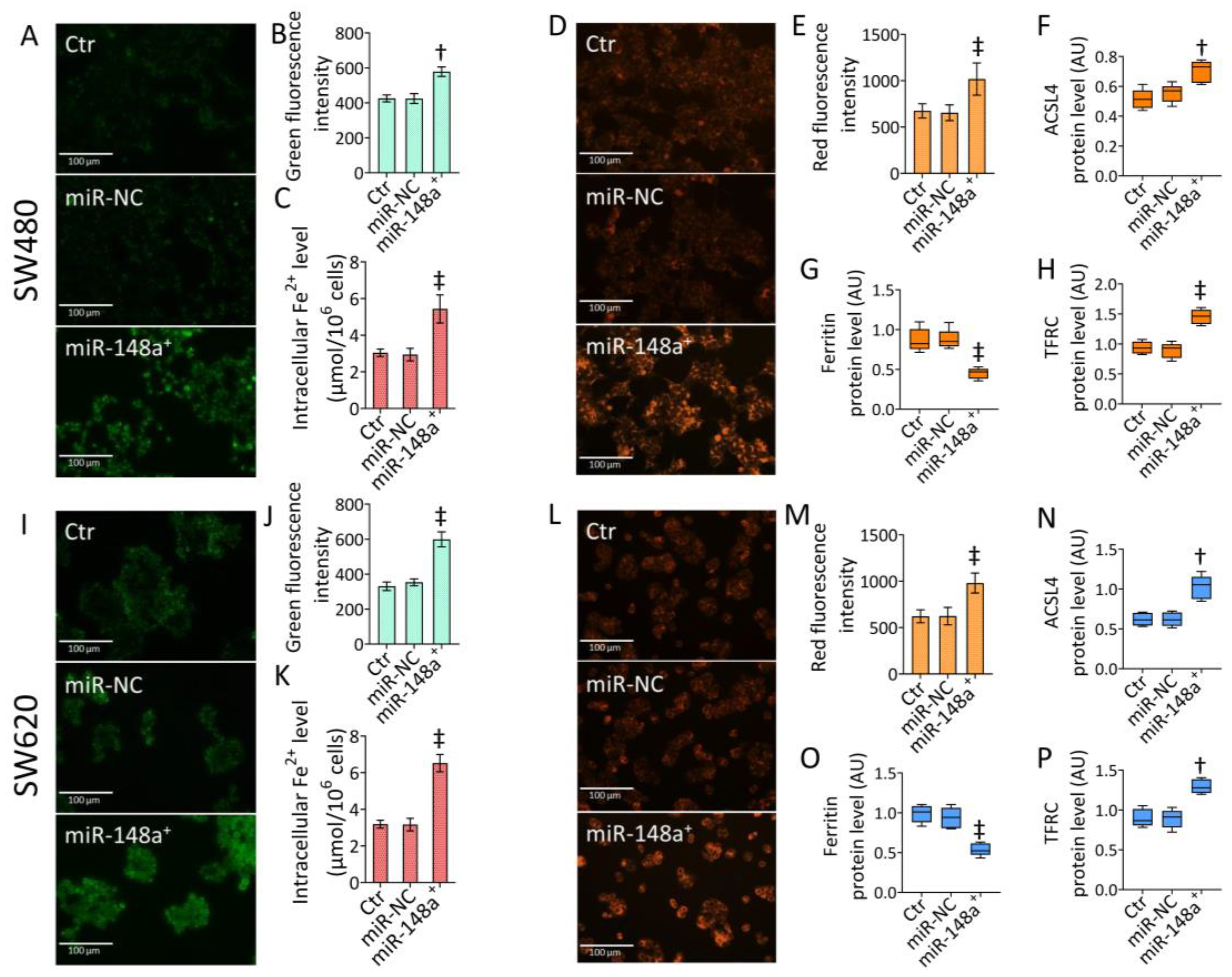
3.5. MiR-148a+ Provoked Ferroptosis in CRC
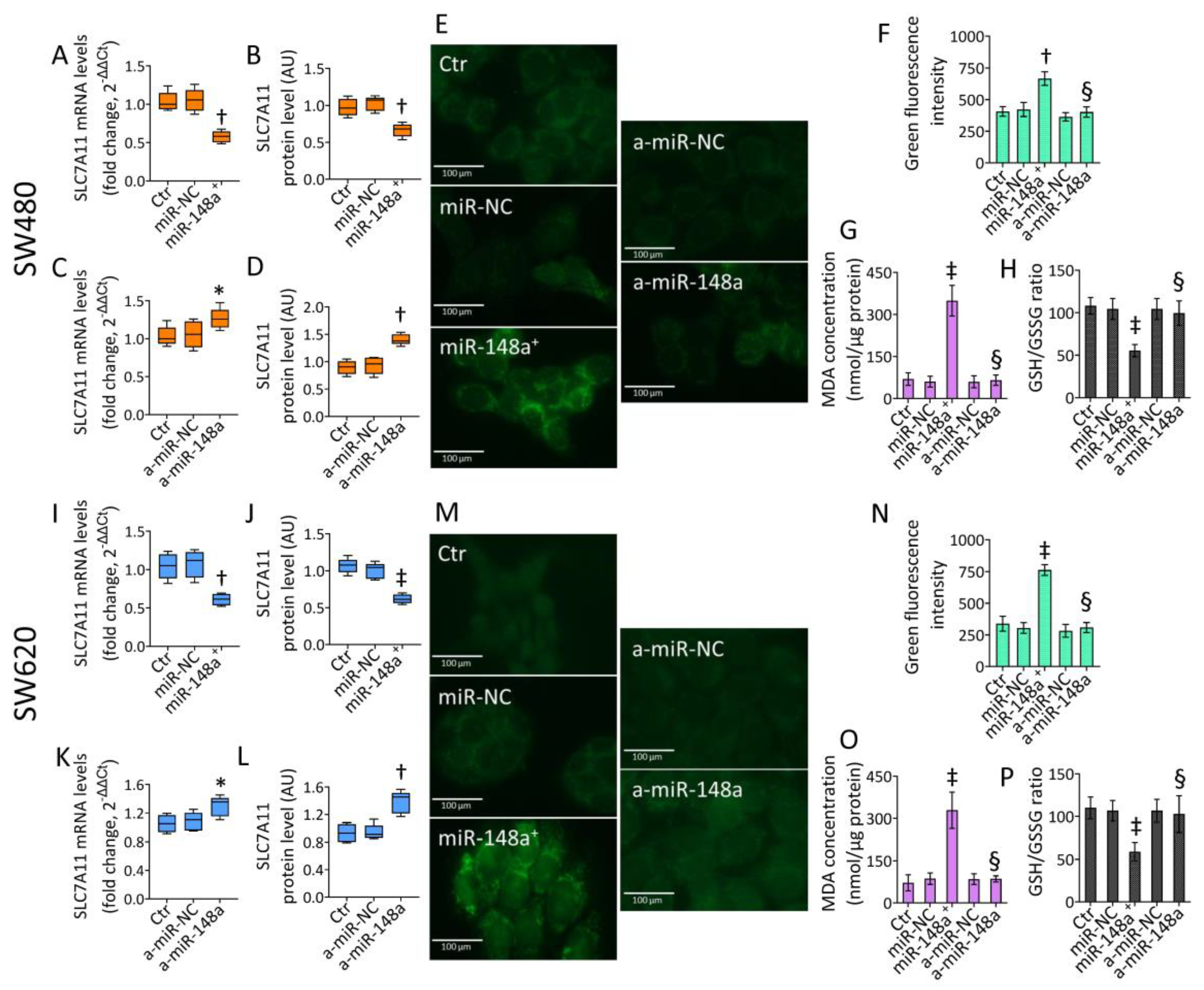
3.6. SLC7A11 as a Target of miR-148a
3.7. miR-148a Inhibition Denied Lipid Peroxidation in CRC
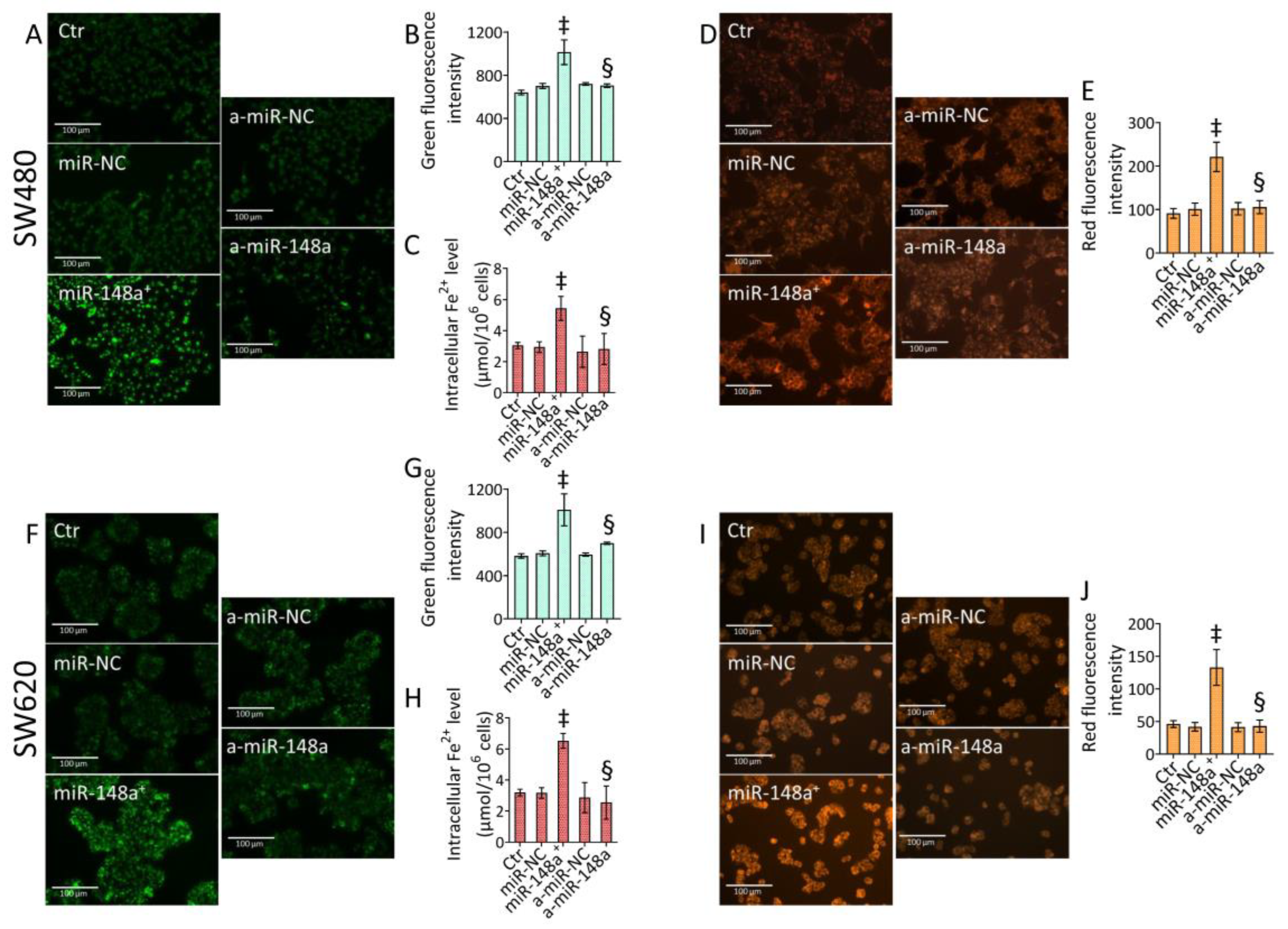
3.8. miR-148a Depletion Deleted Ferroptosis in CRC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasag, C.J.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 2023, 72, 338–344. [Google Scholar] [CrossRef]
- Chen, B.; Xia, Z.; Deng, Y.N.; Yang, Y.; Zhang, P.; Zhu, H.; Xu, N.; Liang, S. Emerging microRNA biomarkers for colorectal cancer diagnosis and prognosis. Open Biol. 2019, 9, 180212. [Google Scholar] [CrossRef]
- Cochetti, G.; Rossi de Vermandois, J.A.; Maulà, V.; Giulietti, M.; Cecati, M.; Del Zingaro, M.; Cagnani, R.; Suvieri, C.; Paladini, A.; Mearini, E. Role of miRNAs in prostate cancer: Do we really know everything? Urol. Oncol. 2020, 38, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Chakrabortty, A.; Patton, D.J.; Smith, B.F.; Agarwal, P. miRNAs: Potential as Biomarkers and Therapeutic Targets for Cancer. Genes 2023, 14, 1375. [Google Scholar] [CrossRef] [PubMed]
- Pekarek, L.; Torres-Carranza, D.; Fraile-Martinez, O.; García-Montero, C.; Pekarek, T.; Saez, M.A.; Rueda-Correa, F.; Pimentel-Martinez, C.; Guijarro, L.G.; Diaz-Pedrero, R.; et al. An Overview of the Role of MicroRNAs on Carcinogenesis: A Focus on Cell Cycle, Angiogenesis and Metastasis. Int. J. Mol. Sci. 2023, 24, 7268. [Google Scholar] [CrossRef]
- Baxter, D.E.; Allinson, L.M.; Al Amri, W.S.; Poulter, J.A.; Pramanik, A.; Thorne, J.L.; Verghese, E.T.; Hughes, T.A. MiR-195 and Its Target SEMA6D Regulate Chemoresponse in Breast Cancer. Cancers 2021, 13, 5979. [Google Scholar] [CrossRef] [PubMed]
- Martino, E.; Balestrieri, A.; Mele, L.; Sardu, C.; Marfella, R.; D’Onofrio, N.; Campanile, G.; Balestrieri, M.L. Milk Exosomal miR-27b Worsen Endoplasmic Reticulum Stress Mediated Colorectal Cancer Cell Death. Nutrients 2022, 14, 5081. [Google Scholar] [CrossRef]
- Li, J.; Chen, H.; Sun, G.; Zhang, X.; Ye, H.; Wang, P. Role of miR-21 in the diagnosis of colorectal cancer: Meta-analysis and bioinformatics. Pathol. Res. Pract. 2023, 248, 154670. [Google Scholar] [CrossRef]
- He, S.; Song, W.; Cui, S.; Li, J.; Jiang, Y.; Chen, X.; Peng, L. Modulation of miR-146b by N6-methyladenosine modification remodels tumor-associated macrophages and enhances anti-PD-1 therapy in colorectal cancer. Cell. Oncol. 2023; online ahead of print. [Google Scholar] [CrossRef]
- Fan, L.; Xu, G.; Zeng, X. M2 macrophage-derived extracellular vesicles augment immune evasion and development of colorectal cancer via a circRNA_CCDC66/microRNA-342-3p/metadherin axis. Cytotechnology 2023, 75, 293–308. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.J.; Fang, Y.X.; Tian, T.G.; Chen, W.P.; Sun, Q.; Guo, F.Q.; Gong, P.Q.; Li, C.M.; Wang, H.; Hu, Z.Q.; et al. Discovery of extracellular vesicle-delivered in the plasma of patients as an indicator for advanced adenoma and colorectal cancer. J. Transl. Med. 2023, 21, 421. [Google Scholar] [CrossRef] [PubMed]
- Christodoulou, S.; Sotiropoulou, C.D.; Vassiliu, P.; Danias, N.; Arkadopoulos, N.; Sideris, D.C. MicroRNA-675-5p Overexpression Is an Independent Prognostic Molecular Biomarker of Short-Term Relapse and Poor Overall Survival in Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 9990. [Google Scholar] [CrossRef]
- Jia, Q.; Liao, X.; Xu, B.; Li, Y.; Liang, L. MiR-128-1-5p inhibits cell proliferation and induces cell apoptosis via targeting PRKCQ in colorectal cancer. Cancer Biol. Ther. 2023, 24, 2226421. [Google Scholar] [CrossRef]
- Zor, D.S.; Hakan, M.T.; Özgür, E.; Horozoglu, C.; Yörüker, E.E.; Kulle, C.B.; Gezer, U.; Yaylim, I. Plasma Levels of Kynurenine, Soluble OX40 and Mir-138-5p Are Associated With Tumor-infiltrating Lymphocytes in Colorectal Cancer: An Exploratory Study. Anticancer Res. 2023, 43, 3281–3288. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yao, J.; Shan, Z.; Wei, Y.; You, S.; Li, D.; Zhang, Y. To Assess the Role of microRNA-451 in the Progression and Metastasis of Colorectal Cancer. Appl. Biochem. Biotechnol. 2023; online ahead of print. [Google Scholar] [CrossRef]
- Zhou, H.; He, Y.; Li, L.; Wu, C.; Hu, G. Identification novel prognostic signatures for Head and Neck Squamous Cell Carcinoma based on ceRNA network construction and immune infiltration analysis. Int. J. Med. Sci. 2021, 18, 1297–1311. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wang, X.; Qi, P.; Liu, C.; Wang, S.; Wan, Q.; Liu, Y.; Su, Y.; Jin, L.; Liu, Y.; et al. Tumor Microenvironmental Competitive Endogenous RNA Network and Immune Cells Act as Robust Prognostic Predictor of Acute Myeloid Leukemia. Front. Oncol. 2021, 11, 584884. [Google Scholar] [CrossRef]
- Chen, J.; Chen, J.G.; Sun, B.; Wu, J.H.; Du, C.Y. Integrative analysis of immune microenvironment-related CeRNA regulatory axis in gastric cancer. Math. Biosci. Eng. 2020, 17, 3953–3971. [Google Scholar] [CrossRef]
- Ardila, H.J.; Sanabria-Salas, M.C.; Meneses, X.; Rios, R.; Huertas-Salgado, A.; Serrano, M.L. Circulating miR-141-3p, miR-143-3p and miR-200c-3p are differentially expressed in colorectal cancer and advanced adenomas. Mol. Clin. Oncol. 2019, 11, 201–207. [Google Scholar] [CrossRef]
- Xu, Q.; Jiang, Y.; Yin, Y.; Li, Q.; He, J.; Jing, Y.; Qi, Y.T.; Xu, Q.; Li, W.; Lu, B.; et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J. Mol. Cell Biol. 2013, 5, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Baltruskeviciene, E.; Schveigert, D.; Stankevicius, V.; Mickys, U.; Zvirblis, T.; Bublevic, J.; Suziedelis, K.; Aleknavicius, E. Down-regulation of miRNA-148a and miRNA-625-3p in colorectal cancer is associated with tumor budding. BMC Cancer 2017, 17, 607. [Google Scholar] [CrossRef]
- Tsai, H.L.; Yang, I.P.; Huang, C.W.; Ma, C.J.; Kuo, C.H.; Lu, C.Y.; Juo, S.H.; Wang, J.Y. Clinical significance of microRNA-148a in patients with early relapse of stage II stage and III colorectal cancer after curative resection. Transl. Res. 2013, 162, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, L.; Xue, C.; Huang, R.; Hu, A.; An, X.; Shi, Y. A Novel Ferroptosis-Related Gene Signature Predicts Overall Survival of Breast Cancer Patients. Biology 2021, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Ding, W.; Ji, X.; Ao, X.; Liu, Y.; Yu, W.; Wang, J. Molecular mechanisms of ferroptosis and its role in cancer therapy. J. Cell. Mol. Med. 2019, 23, 4900–4912. [Google Scholar] [CrossRef]
- Tang, X.; Ding, H.; Liang, M.; Chen, X.; Yan, Y.; Wan, N.; Chen, Q.; Zhang, J.; Cao, J. Curcumin induces ferroptosis in non-small-cell lung cancer via activating autophagy. Thorac. Cancer 2021, 12, 1219–1230. [Google Scholar] [CrossRef]
- Bano, I.; Horky, P.; Abbas, S.Q.; Majid, M.; Bilal, A.H.M.; Ali, F.; Behl, T.; Hassan, S.S.U.; Bungau, S. Ferroptosis: A New Road towards Cancer Management. Molecules 2022, 27, 2129. [Google Scholar] [CrossRef]
- Zhao, Y.; Ma, R.; Wang, C.; Hu, R.; Wu, W.; Sun, X.; Chen, B.; Zhang, W.; Chen, Y.; Zhou, J.; et al. CAPG interference induces apoptosis and ferroptosis in colorectal cancer cells through the P53 pathway. Mol. Cell. Probes 2023, 71, 101919. [Google Scholar] [CrossRef]
- Li, H.; Feng, X.; Hu, Y.; Wang, J.; Huang, C.; Yao, X. Development of a prognostic model based on ferroptosis-related genes for colorectal cancer patients and exploration of the biological functions of NOS2 in vivo and in vitro. Front. Oncol. 2023, 13, 1133946. [Google Scholar] [CrossRef]
- Guo, C.; Liu, P.; Deng, G.; Han, Y.; Chen, Y.; Cai, C.; Shen, H.; Deng, G.; Zeng, S. Honokiol induces ferroptosis in colon cancer cells by regulating GPX4 activity. Am. J. Cancer Res. 2021, 11, 3039–3054. [Google Scholar] [PubMed]
- Wang, Y.; Zhang, Z.; Sun, W.; Zhang, J.; Xu, Q.; Zhou, X.; Mao, L. Ferroptosis in colorectal cancer: Potential mechanisms and effective therapeutic targets. Biomed. Pharmacother. 2022, 153, 113524. [Google Scholar] [CrossRef]
- Chaudhary, N.; Choudhary, B.S.; Shah, S.G.; Khapare, N.; Dwivedi, N.; Gaikwad, A.; Joshi, N.; Raichanna, J.; Basu, S.; Gurjar, M.; et al. Lipocalin 2 expression promotes tumor progression and therapy resistance by inhibiting ferroptosis in colorectal cancer. Int. J. Cancer 2021, 149, 1495–1511. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, L.; Liu, J.; Wan, Z.; Zhou, L.; Liao, H.; Wan, R. LncRNA ITGB2-AS1 promotes cisplatin resistance of non-small cell lung cancer by inhibiting ferroptosis via activating the FOSL2/NAMPT axis. Cancer Biol. Ther. 2023, 24, 2223377. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Cui, J.; Cui, M.; Jing, R. SLC7A11 promotes the progression of gastric cancer and regulates ferroptosis through PI3K/AKT pathway. Pathol. Res. Pract. 2023, 248, 154646. [Google Scholar] [CrossRef]
- Huang, F.; Huang, Z.; Wei, Q.; Liu, G.; Pu, J. E3 ubiquitin ligase HECTD3 is a tumor suppressor and mediates the polyubiquitination of SLC7A11 to promote ferroptosis in colon cancer. Exp. Cell Res. 2023, 430, 113697. [Google Scholar] [CrossRef]
- He, J.; Ding, H.; Li, H.; Pan, Z.; Chen, Q. Intra-Tumoral Expression of SLC7A11 Is Associated with Immune Microenvironment, Drug Resistance, and Prognosis in Cancers: A Pan-Cancer Analysis. Front. Genet. 2021, 12, 770857. [Google Scholar] [CrossRef]
- Bao, X.; Zhang, H.; Wu, W.; Cheng, S.; Dai, X.; Zhu, X.; Fu, Q.; Tong, Z.; Liu, L.; Zheng, Y.; et al. Analysis of the molecular nature associated with microsatellite status in colon cancer identifies clinical implications for immunotherapy. J. Immunother. Cancer 2020, 8, e001437. [Google Scholar] [CrossRef]
- D’Onofrio, N.; Martino, E.; Mele, L.; Colloca, A.; Maione, M.; Cautela, D.; Castaldo, D.; Balestrieri, M.L. Colorectal Cancer Apoptosis Induced by Dietary δ-Valerobetaine Involves PINK1/Parkin Dependent-Mitophagy and SIRT3. Int. J. Mol. Sci. 2021, 22, 8117. [Google Scholar] [CrossRef]
- D’Onofrio, N.; Prattichizzo, F.; Martino, E.; Anastasio, C.; Mele, L.; La Grotta, R.; Sardu, C.; Ceriello, A.; Marfella, R.; Paolisso, G.; et al. MiR-27b attenuates mitochondrial oxidative stress and inflammation in endothelial cells. Redox Biol. 2023, 62, 102681. [Google Scholar] [CrossRef]
- D’Onofrio, N.; Martino, E.; Balestrieri, A.; Mele, L.; Cautela, D.; Castaldo, D.; Balestrieri, M.L. Diet-derived ergothioneine induces necroptosis in colorectal cancer cells by activating the SIRT3/MLKL pathway. FEBS Lett. 2022, 596, 1313–1329. [Google Scholar] [CrossRef] [PubMed]
- Martino, E.; Balestrieri, A.; Anastasio, C.; Maione, M.; Mele, L.; Cautela, D.; Campanile, G.; Balestrieri, M.L.; D’Onofrio, N. SIRT3 Modulates Endothelial Mitochondrial Redox State during Insulin Resistance. Antioxidants 2022, 11, 1611. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Wang, H.; Chen, X.; Liang, C.; Shang, W.; Wang, L.; Li, J.; Xu, D. MiR-182-5p inhibits colon cancer tumorigenesis, angiogenesis, and lymphangiogenesis by directly downregulating VEGF-C. Cancer Lett. 2020, 488, 18–26. [Google Scholar] [CrossRef]
- Xu, S.; Lin, J.; Chen, R.; Xie, J.; Yuan, E.; Cen, F.; Kong, F. LINC00174 Promotes Colon Cancer Progression by Regulating Inflammation and Glycolysis by Targeting the MicroRNA-2467-3p/Enolase 3 Axis. Mediat. Inflamm. 2023, 2023, 8052579. [Google Scholar] [CrossRef] [PubMed]
- Gherman, A.; Balacescu, L.; Popa, C.; Cainap, C.; Vlad, C.; Cainap, S.S.; Balacescu, O. Baseline Expression of Exosomal miR-92a-3p and miR-221-3p Could Predict the Response to First-Line Chemotherapy and Survival in Metastatic Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 10622. [Google Scholar] [CrossRef] [PubMed]
- Moratalla-Navarro, F.; Díez-Villanueva, A.; Garcia-Serrano, A.; Closa, A.; Cordero, D.; Solé, X.; Guinó, E.; Sanz-Pamplona, R.; Sanjuan, X.; Santos, C.; et al. Identification of a Twelve-microRNA Signature with Prognostic Value in Stage II Microsatellite Stable Colon Cancer. Cancers 2023, 15, 3301. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Dong, J.; Wang, M.; Yao, S.; Tian, X.; Cui, X.; Fu, S.; Zhang, S. miR-148a-3p suppresses epithelial ovarian cancer progression primarily by targeting c-Met. Oncol. Lett. 2018, 15, 6131–6136. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, Y.; Guo, J.; Wang, L. miR-148a-3p Suppresses the Proliferation and Invasion of Esophageal Cancer by Targeting DNMT1. Genet. Test. Mol. Biomark. 2019, 23, 98–104. [Google Scholar] [CrossRef]
- Wang, X.; Liang, Z.; Xu, X.; Li, J.; Zhu, Y.; Meng, S.; Li, S.; Wang, S.; Xie, B.; Ji, A.; et al. miR-148a-3p represses proliferation and EMT by establishing regulatory circuits between ERBB3/AKT2/c-myc and DNMT1 in bladder cancer. Cell Death Dis. 2016, 7, e2503. [Google Scholar] [CrossRef]
- Han, L.; Yan, Y.; Zhao, L.; Liu, Y.; Lv, X.; Zhang, L.; Zhao, Y.; Zhao, H.; He, M.; Wei, M. LncRNA HOTTIP facilitates the stemness of breast cancer via regulation of miR-148a-3p/WNT1 pathway. J. Cell. Mol. Med. 2020, 24, 6242–6252. [Google Scholar] [CrossRef]
- Di Cosimo, S.; Appierto, V.; Pizzamiglio, S.; Silvestri, M.; Baselga, J.; Piccart, M.; Huober, J.; Izquierdo, M.; de la Pena, L.; Hilbers, F.S.; et al. Early Modulation of Circulating MicroRNAs Levels in HER2-Positive Breast Cancer Patients Treated with Trastuzumab-Based Neoadjuvant Therapy. Int. J. Mol. Sci. 2020, 21, 1386. [Google Scholar] [CrossRef] [PubMed]
- Hibino, Y.; Sakamoto, N.; Naito, Y.; Goto, K.; Oo, H.Z.; Sentani, K.; Hinoi, T.; Ohdan, H.; Oue, N.; Yasui, W. Significance of miR-148a in Colorectal Neoplasia: Downregulation of miR-148a Contributes to the Carcinogenesis and Cell Invasion of Colorectal Cancer. Pathobiology 2015, 82, 233–241. [Google Scholar] [CrossRef] [PubMed]
- OuYang, C.; Shu, G.; Liu, J.; Deng, S.; Lu, P.; Li, Y.; Gan, Y.; Xie, B.; Liu, J.; Yin, G. HDAC5, negatively regulated by miR-148a-3p, promotes colon cancer cell migration. Cancer Sci. 2022, 113, 2560–2574. [Google Scholar] [CrossRef] [PubMed]
- Nersisyan, S.; Galatenko, A.; Chekova, M.; Tonevitsky, A. Hypoxia-Induced miR-148a Downregulation Contributes to Poor Survival in Colorectal Cancer. Front. Genet. 2021, 12, 662468. [Google Scholar] [CrossRef]
- Hu, B.; Chen, Z.; Wang, X.; Chen, F.; Song, Z.; Cao, C. MicroRNA-148a-3p Directly Targets SERPINE1 to Suppress EMT-Mediated Colon Adenocarcinoma Progression. Cancer Manag. Res. 2021, 13, 6349–6362. [Google Scholar] [CrossRef]
- Huang, F.; Zhao, J.L.; Wang, L.; Gao, C.C.; Liang, S.Q.; An, D.J.; Bai, J.; Chen, Y.; Han, H.; Qin, H.Y. miR-148a-3p Mediates Notch Signaling to Promote the Differentiation and M1 Activation of Macrophages. Front. Immunol. 2017, 8, 1327. [Google Scholar] [CrossRef]
- Liu, J.; Sun, Q.; Sun, X.; Wang, Q.; Zou, G.; Wang, D.; Zhuang, B.; Juan, Z.; Zhang, R.; Zhang, D. Therapeutic Effects of Salvianolic Acid B on Angiotensin II-Induced Atrial Fibrosis by Regulating Atrium Metabolism via Targeting AMPK/FoxO1/miR-148a-3p Axis. J. Cardiovasc. Transl. Res. 2023, 16, 341–357. [Google Scholar] [CrossRef]
- Li, B.; Wang, W.; Li, Z.; Chen, Z.; Zhi, X.; Xu, J.; Li, Q.; Wang, L.; Huang, X.; Wang, L.; et al. MicroRNA-148a-3p enhances cisplatin cytotoxicity in gastric cancer through mitochondrial fission induction and cyto-protective autophagy suppression. Cancer Lett. 2017, 410, 212–227. [Google Scholar] [CrossRef]
- Ma, X.; Xu, M.; Zhang, X.; Wang, X.; Su, K.; Xu, Z.; Wang, X.; Yang, Y. Gambogenic acid inhibits proliferation and ferroptosis by targeting the miR-1291/FOXA2 and AMPKα/SLC7A11/GPX4 axis in colorectal cancer. Cell Biol. Int. 2023; online ahead of print. [Google Scholar] [CrossRef]
- Wang, G.; Qin, S.; Chen, L.; Geng, H.; Zheng, Y.; Xia, C.; Yao, J.; Deng, L.; Liu, L.; Yao, H.; et al. MiR-15a-3p regulates ferroptosis via targeting glutathione peroxidase GPX4 in colorectal cancer. Mol. Carcinog. 2022, 61, 301–310. [Google Scholar] [CrossRef]
- Koppula, P.; Zhuang, L.; Gan, B. Cystine transporter SLC7A11/xCT in cancer: Ferroptosis, nutrient dependency, and cancer therapy. Protein Cell 2021, 12, 599–620. [Google Scholar] [CrossRef] [PubMed]
- Shimada, K.; Skouta, R.; Kaplan, A.; Yang, W.S.; Hayano, M.; Dixon, S.J.; Brown, L.M.; Valenzuela, C.A.; Wolpaw, A.J.; Stockwell, B.R. Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat. Chem. Biol. 2016, 12, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Pi, D.; Zhou, S.; Yi, Z.; Dong, Y.; Wang, W.; Ye, H.; Chen, Y.; Zuo, Q.; Ouyang, M. Ginsenoside Rh3 induces pyroptosis and ferroptosis through the Stat3/p53/NRF2 axis in colorectal cancer cells. Acta Biochim. Biophys. Sin. 2023, 55, 587–600. [Google Scholar] [CrossRef]
- Guo, S.; Zhao, W.; Zhang, W.; Li, S.; Teng, G.; Liu, L. Vitamin D Promotes Ferroptosis in Colorectal Cancer Stem Cells via SLC7A11 Downregulation. Oxid. Med. Cell. Longev. 2023, 2023, 4772134. [Google Scholar] [CrossRef]
- Melnik, B.C.; Stadler, R.; Weiskirchen, R.; Leitzmann, C.; Schmitz, G. Potential Pathogenic Impact of Cow’s Milk Consumption and Bovine Milk-Derived Exosomal MicroRNAs in Diffuse Large B-Cell Lymphoma. Int. J. Mol. Sci. 2023, 24, 6102. [Google Scholar] [CrossRef] [PubMed]
- Ahlberg, E.; Al-Kaabawi, A.; Thune, R.; Simpson, M.R.; Pedersen, S.A.; Cione, E.; Jenmalm, M.C.; Tingö, L. Breast milk microRNAs: Potential players in oral tolerance development. Front. Immunol. 2023, 14, 1154211. [Google Scholar] [CrossRef]
- Ahlberg, E.; Martí, M.; Govindaraj, D.; Severin, E.; Duchén, K.; Jenmalm, M.C.; Tingö, L. Immune-related microRNAs in breast milk and their relation to regulatory T cells in breastfed children. Pediatr. Allergy Immunol. 2023, 34, e13952. [Google Scholar] [CrossRef]
- Simpson, M.R.; Brede, G.; Johansen, J.; Johnsen, R.; Storrø, O.; Sætrom, P.; Øien, T. Human Breast Milk miRNA, Maternal Probiotic Supplementation and Atopic Dermatitis in Offspring. PLoS ONE 2015, 10, e0143496. [Google Scholar] [CrossRef]
- Zheng, Y.; Meng, L.; Liu, H.; Sun, L.; Nie, Y.; Wu, Q.; Fan, D.; Li, M. Let food be thy medicine: The role of diet in colorectal cancer: A narrative review. J. Gastrointest. Oncol. 2022, 13, 2020–2032. [Google Scholar] [CrossRef]
- Peng, X.; Wang, J.; Zhang, C.; Liu, K.; Zhao, L.; Chen, X.; Huang, G.; Lai, Y. A three-miRNA panel in serum as a noninvasive biomarker for colorectal cancer detection. Int. J. Biol. Markers 2020, 35, 74–82. [Google Scholar] [CrossRef]
- Martino, E.; D’Onofrio, N.; Anastasio, C.; Abate, M.; Zappavigna, S.; Caraglia, M.; Balestrieri, M.L. MicroRNA-nanoparticles against cancer: Opportunities and challenges for personalized medicine. Mol. Ther. Nucleic Acids 2023, 32, 371–384. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martino, E.; Balestrieri, A.; Aragona, F.; Bifulco, G.; Mele, L.; Campanile, G.; Balestrieri, M.L.; D’Onofrio, N. MiR-148a-3p Promotes Colorectal Cancer Cell Ferroptosis by Targeting SLC7A11. Cancers 2023, 15, 4342. https://doi.org/10.3390/cancers15174342
Martino E, Balestrieri A, Aragona F, Bifulco G, Mele L, Campanile G, Balestrieri ML, D’Onofrio N. MiR-148a-3p Promotes Colorectal Cancer Cell Ferroptosis by Targeting SLC7A11. Cancers. 2023; 15(17):4342. https://doi.org/10.3390/cancers15174342
Chicago/Turabian StyleMartino, Elisa, Anna Balestrieri, Francesca Aragona, Giovanna Bifulco, Luigi Mele, Giuseppe Campanile, Maria Luisa Balestrieri, and Nunzia D’Onofrio. 2023. "MiR-148a-3p Promotes Colorectal Cancer Cell Ferroptosis by Targeting SLC7A11" Cancers 15, no. 17: 4342. https://doi.org/10.3390/cancers15174342
APA StyleMartino, E., Balestrieri, A., Aragona, F., Bifulco, G., Mele, L., Campanile, G., Balestrieri, M. L., & D’Onofrio, N. (2023). MiR-148a-3p Promotes Colorectal Cancer Cell Ferroptosis by Targeting SLC7A11. Cancers, 15(17), 4342. https://doi.org/10.3390/cancers15174342