Environmental and Lifestyle Cancer Risk Factors: Shaping Extracellular Vesicle OncomiRs and Paving the Path to Cancer Development
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Isolation and Purification of EVs and miRNA-EVs from Plasma
2.3. miRNA Analysis
2.4. Statistical Analysis
3. Results
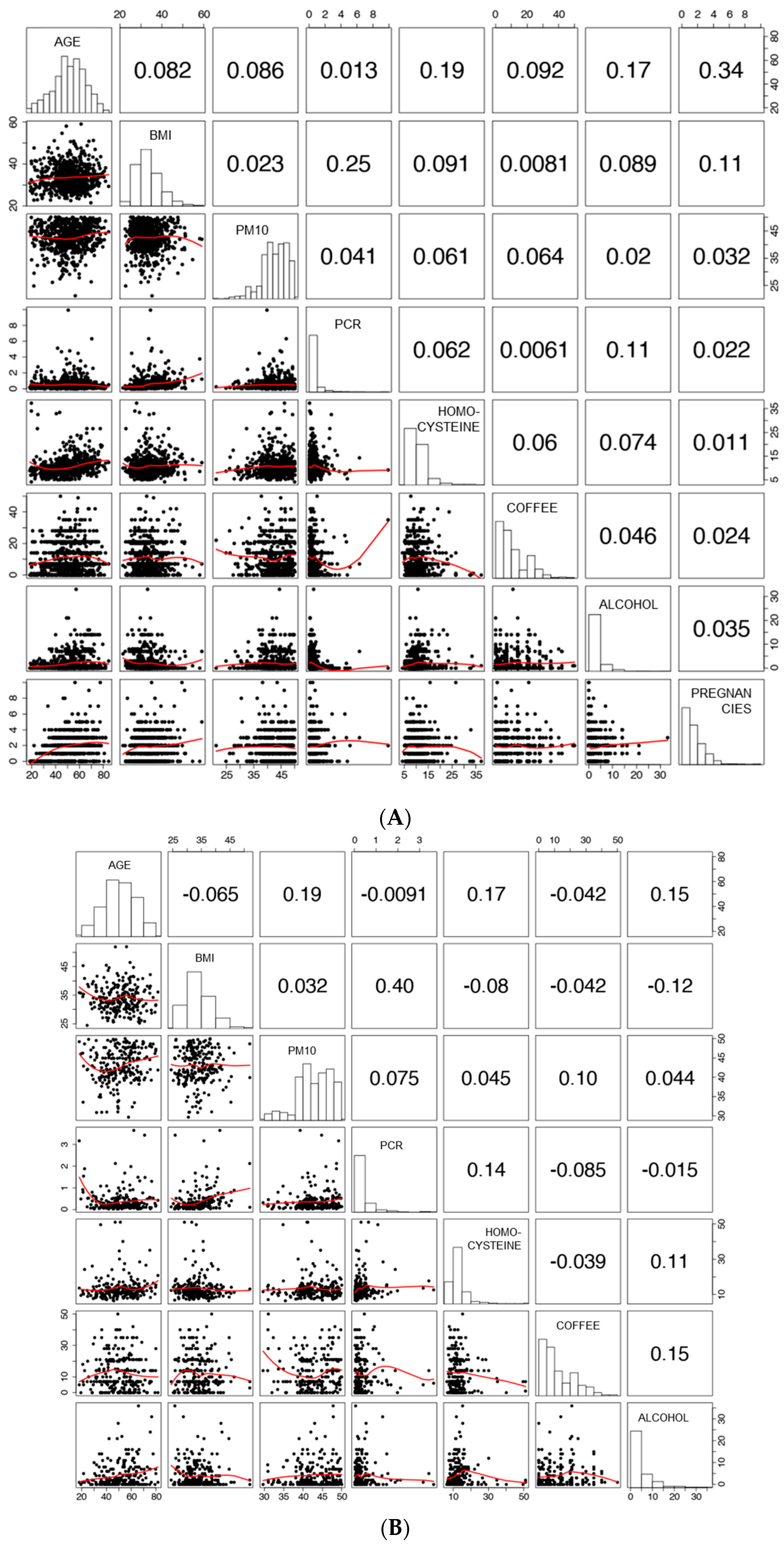
3.1. Characteristics of the Study Population
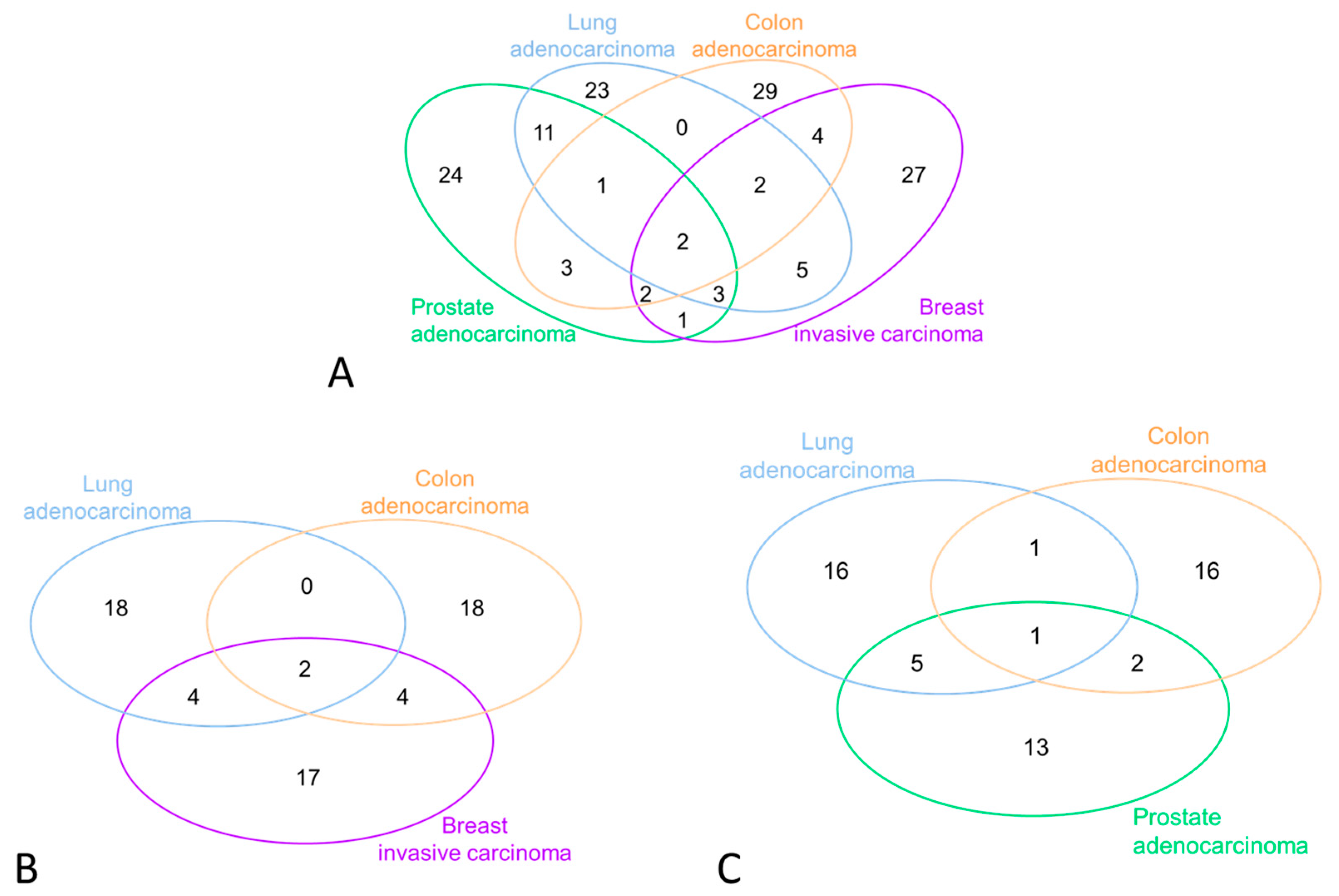
3.2. miRNA Selection and Expression Levels in the Study Subjects
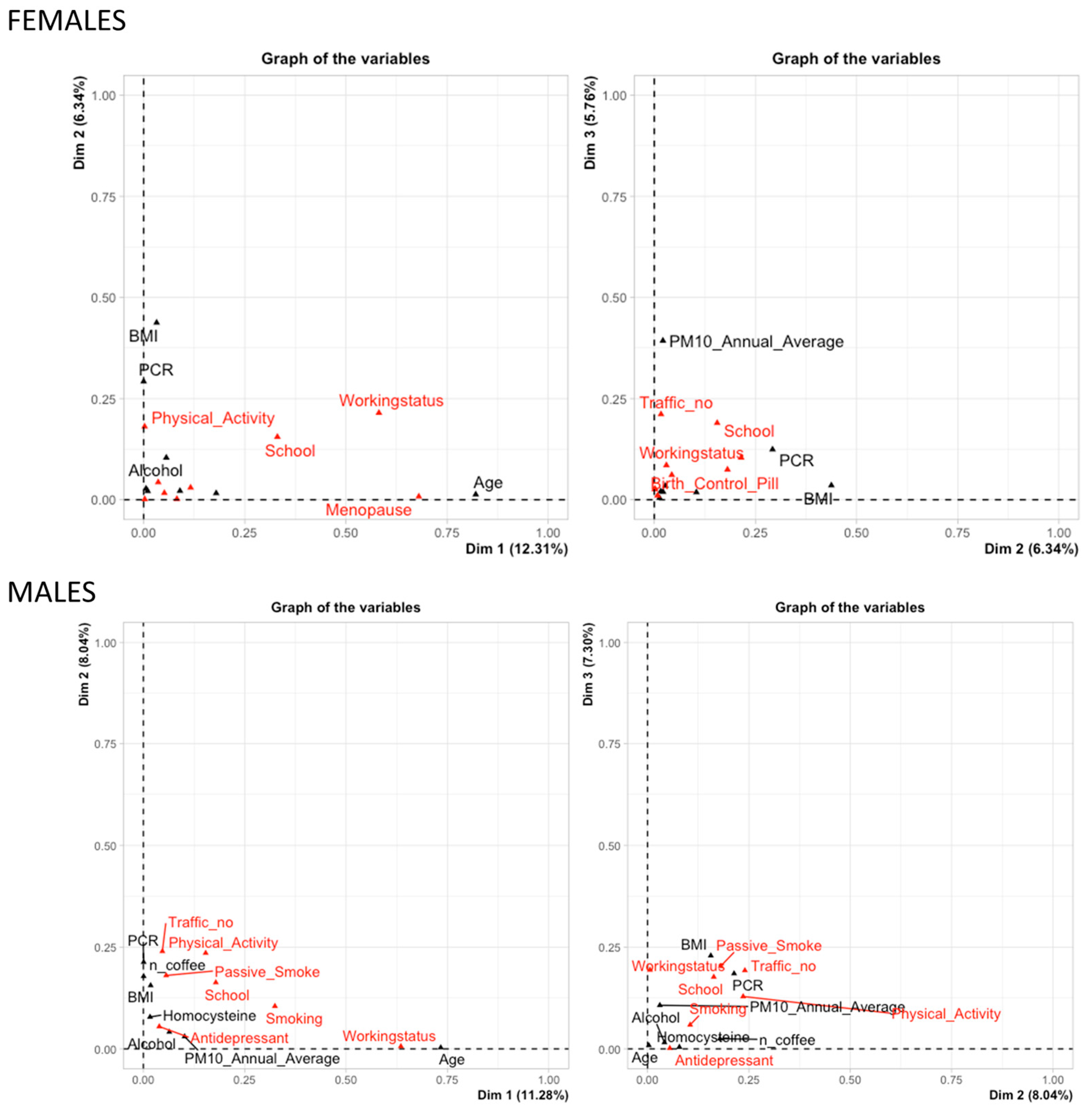
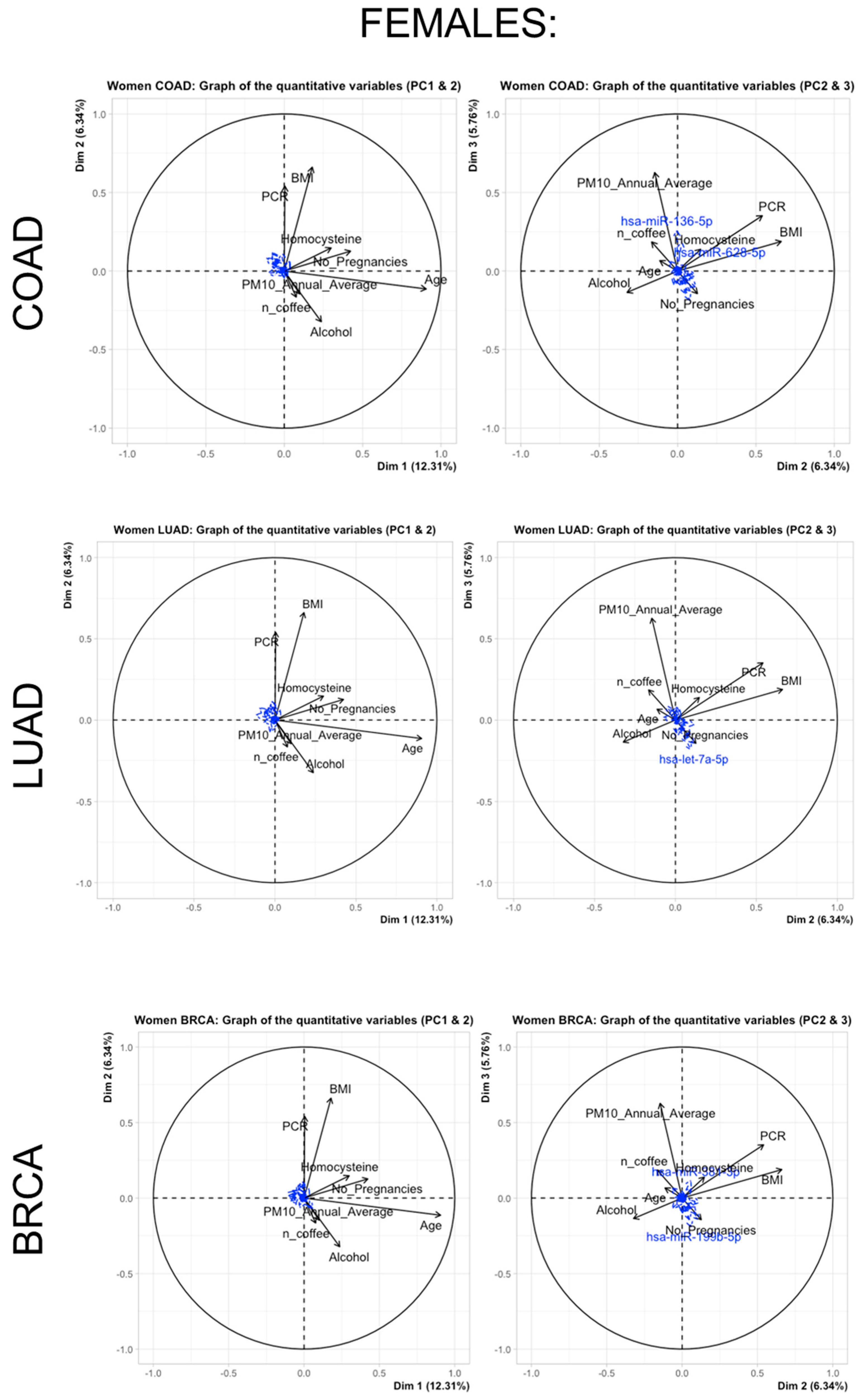
3.3. Multivariate Adaptive Regression Splines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.-B.; Pan, X.-F.; Chen, J.; Cao, A.; Zhang, Y.-G.; Xia, L.; Wang, J.; Li, H.; Liu, G.; Pan, A. Combined lifestyle factors, incident cancer, and cancer mortality: A systematic review and meta-analysis of prospective cohort studies. Br. J. Cancer 2020, 122, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Vineis, P.; Wild, C.P. Global cancer patterns: Causes and prevention. Lancet 2014, 383, 549–557. [Google Scholar] [CrossRef]
- Anand, P.; Kunnumakara, A.B.; Sundaram, C.; Harikumar, K.B.; Tharakan, S.T.; Lai, O.S.; Sung, B.; Aggarwal, B.B. Cancer is a Preventable Disease that Requires Major Lifestyle Changes. Pharm. Res. 2008, 25, 2097–2116. [Google Scholar] [CrossRef]
- Irigaray, P.; Newby, J.; Clapp, R.; Hardell, L.; Howard, V.; Montagnier, L.; Epstein, S.; Belpomme, D. Lifestyle-related factors and environmental agents causing cancer: An overview. Biomed. Pharmacother. 2007, 61, 640–658. [Google Scholar] [CrossRef] [PubMed]
- Steck, S.E.; Murphy, E.A. Dietary patterns and cancer risk. Nat. Rev. Cancer 2020, 20, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Friedenreich, C.M.; Ryder-Burbidge, C.; McNeil, J. Physical activity, obesity and sedentary behavior in cancer etiology: Epidemiologic evidence and biologic mechanisms. Mol. Oncol. 2021, 15, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Biganzoli, E.; Desmedt, C.; Fornili, M.; de Azambuja, E.; Cornez, N.; Ries, F.; Closon-Dejardin, M.-T.; Kerger, J.; Focan, C.; Di Leo, A.; et al. Recurrence dynamics of breast cancer according to baseline body mass index. Eur. J. Cancer 2017, 87, 10–20. [Google Scholar] [CrossRef] [PubMed]
- GBD 2019 Cancer Risk Factors Collaborators. The global burden of cancer attributable to risk factors, 2010–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2022, 400, 563–591. [Google Scholar] [CrossRef]
- Ward, E.; Jemal, A.; Cokkinides, V.; Singh, G.K.; Cardinez, C.; Ghafoor, A.; Thun, M. Cancer Disparities by Race/Ethnicity and Socioeconomic Status. CA A Cancer J. Clin. 2004, 54, 78–93. [Google Scholar] [CrossRef]
- Martin-Moreno, J.M.; Ruiz-Segovia, N.; Diaz-Rubio, E. Behavioural and structural interventions in cancer prevention: Towards the 2030 SDG horizon. Mol. Oncol. 2021, 15, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Catalán, V.; Avilés-Olmos, I.; Rodríguez, A.; Becerril, S.; Fernández-Formoso, J.A.; Kiortsis, D.; Portincasa, P.; Gómez-Ambrosi, J.; Frühbeck, G. Time to Consider the “Exposome Hypothesis” in the Development of the Obesity Pandemic. Nutrients 2022, 14, 1597. [Google Scholar] [CrossRef]
- Nguyen, H.-L.; Geukens, T.; Maetens, M.; Aparicio, S.; Bassez, A.; Borg, A.; Brock, J.; Broeks, A.; Caldas, C.; Cardoso, F.; et al. Obesity-associated changes in molecular biology of primary breast cancer. Nat. Commun. 2023, 14, 4418. [Google Scholar] [CrossRef]
- Deng, T.; Lyon, C.J.; Bergin, S.; Caligiuri, M.A.; Hsueh, W.A. Obesity, Inflammation, and Cancer. Annu. Rev. Pathol. Mech. Dis. 2016, 11, 421–449. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Margolis, L.; Sadovsky, Y. The biology of extracellular vesicles: The known unknowns. PLoS Biol. 2019, 17, e3000363. [Google Scholar] [CrossRef]
- van Niel, G.; Carter, D.R.F.; Clayton, A.; Lambert, D.W.; Raposo, G.; Vader, P. Challenges and directions in studying cell-cell communication by extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2022, 23, 369–382. [Google Scholar] [CrossRef]
- Yokoi, A.; Ochiya, T. Exosomes and extracellular vesicles: Rethinking the essential values in cancer biology. Semin. Cancer Biol. 2021, 74, 79–91. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [PubMed]
- Otmani, K.; Rouas, R.; Lewalle, P. OncomiRs as noncoding RNAs having functions in cancer: Their role in immune suppression and clinical implications. Front. Immunol. 2022, 13, 913951. [Google Scholar] [CrossRef]
- Bollati, V.; Iodice, S.; Favero, C.; Angelici, L.; Albetti, B.; Cacace, R.; Cantone, L.; Carugno, M.; Cavalleri, T.; De Giorgio, B.; et al. Susceptibility to particle health effects, miRNA and exosomes: Rationale and study protocol of the SPHERE study. BMC Public Health 2014, 14, 1137. [Google Scholar] [CrossRef]
- Pergoli, L.; Cantone, L.; Favero, C.; Angelici, L.; Iodice, S.; Pinatel, E.; Hoxha, M.; Dioni, L.; Letizia, M.; Albetti, B.; et al. Extracellular vesicle-packaged miRNA release after short-term exposure to particulate matter is associated with increased coagulation. Part. Fibre Toxicol. 2017, 14, 32. [Google Scholar] [CrossRef] [PubMed]
- World Cancer Research Fund International. Worldwide Cancer Data. Global Cancer Statistics for the Most Common Cancers in the World. Available online: http://wcrf.orf/cancer-trends/worldwide-cancer-data (accessed on 1 January 2023).
- Wong, N.; Chen, Y.; Chen, S.; Wang, X. OncomiR: An online resource for exploring pan-cancer microRNA dysregulation. Bio-informatics 2018, 34, 713–715. [Google Scholar] [CrossRef]
- Société Française de Statistique. Journal de la Société Française de Statistique; Société Française de Statistique: Paris, France, 1998. [Google Scholar]
- Husson, F.; Le, S.; Pagès, J. Exploratory Multivariate Analysis by Example Using R, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Kassambara, A. Ggcorrplot: Visualization of a Correlation Matrix using ‘ggplot2′_. R Package Version 0.1.4, 2022. Available online: https://CRAN.R-project.org/package=ggcorrplot (accessed on 1 June 2023).
- Hastie, T.; Tibshirani, R.; Friedman, J.H. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer: New York, NY, USA, 2009; p. xxii. 745p. [Google Scholar]
- Milborrow, S. Notes on the Earth Package. 2017. Available online: http://www.milbo.org/doc/earth-notes.pdf (accessed on 1 June 2023).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.r-project.org/ (accessed on 1 June 2023).
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Milborrow, S. Derived from mda: Mars by T. Hastie and R. Tibshirani. 2023 Earth: Multivariate Adaptive Regression Splines. R Package Version 5.3.2. Available online: https://CRAN.R-project.org/package=earth (accessed on 1 June 2023).
- Berthold, M.R.; Cebron, N.; Dill, F.; Gabriel, T.R.; Kötter, T.; Meinl, T.; Ohl, P.; Sieb, C.; Thiel, K.; Wiswedel, B. KNIME: The Konstanz Information Miner; Springer: Berlin/Heidelberg, Germany, 2008; pp. 319–326. [Google Scholar] [CrossRef]
- Hsieh, T.-H.; Hsu, C.-Y.; Tsai, C.-F.; Long, C.-Y.; Chai, C.-Y.; Hou, M.-F.; Lee, J.-N.; Wu, D.-C.; Wang, S.-C.; Tsai, E.-M. miR-125a-5p is a prognostic biomarker that targets HDAC4 to suppress breast tumorigenesis. Oncotarget 2015, 6, 494–509. [Google Scholar] [CrossRef]
- Bhattacharjya, S.; Nath, S.; Ghose, J.; Maiti, G.P.; Biswas, N.; Bandyopadhyay, S.; Panda, C.K.; Bhattacharyya, N.P.; Roychoudhury, S. miR-125b promotes cell death by targeting spindle assembly checkpoint gene MAD1 and modulating mitotic progression. Cell Death Differ. 2013, 20, 430–442. [Google Scholar] [CrossRef]
- Jiang, L.; Huang, Q.; Chang, J.; Wang, E.; Qiu, X. MicroRNA HSA-miR-125a-5p induces apoptosis by activating p53 in lung cancer cells. Exp. Lung Res. 2011, 37, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, D.; Lv, J.; Wang, S.; Zhang, Q. MiR-125a-5p suppresses bladder cancer progression through targeting FUT4. BioMedicine 2018, 108, 1039–1047. [Google Scholar] [CrossRef]
- Yang, X.; Qiu, J.; Kang, H.; Wang, Y.; Qian, J. miR-125a-5p suppresses colorectal cancer progression by targeting VEGFA. Cancer Manag. Res. 2018, 10, 5839–5853. [Google Scholar] [CrossRef]
- Makwana, K.; Patel, S.A.; Velingkaar, N.; Ebron, J.S.; Shukla, G.C.; Kondratov, R.V. Aging and calorie restriction regulate the expression of miR-125a-5p and its target genes Stat3, Casp2 and Stard13. Aging 2017, 9, 1825–1843. [Google Scholar] [CrossRef]
- Huang, N.; Wang, J.; Xie, W.; Lyu, Q.; Wu, J.; He, J.; Qiu, W.; Xu, N.; Zhang, Y. MiR-378a-3p enhances adipogenesis by targeting mitogen-activated protein kinase 1. Biochem. Biophys. Res. Commun. 2015, 457, 37–42. [Google Scholar] [CrossRef]
- Gerin, I.; Bommer, G.T.; McCoin, C.S.; Sousa, K.M.; Krishnan, V.; MacDougald, O.A.; Warren, J.S.; Oka, S.-I.; Zablocki, D.; Sadoshima, J.; et al. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am. J. Physiol. Metab. 2010, 299, E198–E206. [Google Scholar] [CrossRef]
- Eichner, L.J.; Perry, M.-C.; Dufour, C.R.; Bertos, N.; Park, M.; St-Pierre, J.; Giguère, V. miR-378* Mediates Metabolic Shift in Breast Cancer Cells via the PGC-1β/ERRγ Transcriptional Pathway. Cell Metab. 2010, 12, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Mao, C.; Quattrochi, B.; Friedline, R.H.; Zhu, L.J.; Jung, D.Y.; Kim, J.K.; Lewis, B.; Wang, Y.-X. MicroRNA-378 controls classical brown fat expansion to counteract obesity. Nat. Commun. 2014, 5, 4725. [Google Scholar] [CrossRef] [PubMed]
- Mir, F.A.; Mall, R.; Iskandarani, A.; Ullah, E.; Samra, T.A.; Cyprian, F.; Parray, A.; Alkasem, M.; Abdalhakam, I.; Farooq, F.; et al. Characteristic MicroRNAs Linked to Dysregulated Metabolic Pathways in Qatari Adult Subjects with Obesity and Metabolic Syndrome. Front. Endocrinol. 2022, 13, 937089. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shang, C.; Pan, H.; Yang, H.; Zhu, H.; Gong, F. MicroRNA Expression Profiles in the Subcutaneous Adipose Tissues of Morbidly Obese Chinese Women. Obes. Facts 2021, 14, 78–92. [Google Scholar] [CrossRef]
- Zhang, H.-G.; Wang, X.-B.; Zhao, H.; Zhou, C.-N. MicroRNA-9-5p promotes osteoporosis development through inhibiting osteogenesis and promoting adipogenesis via targeting Wnt3a. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 456–463. [Google Scholar] [CrossRef]
- Su, H.; Liang, Z.; Weng, S.; Sun, C.; Huang, J.; Zhang, T.; Wang, X.; Wu, S.; Zhang, Z.; Zhang, Y.; et al. miR-9-5p regulates immunometabolic and epigenetic pathways in β-glucan-trained immunity via IDH3α. J. Clin. Investig. 2021, 6, 144260. [Google Scholar] [CrossRef]
- Bazzoni, F.; Rossato, M.; Fabbri, M.; Gaudiosi, D.; Mirolo, M.; Mori, L.; Tamassia, N.; Mantovani, A.; Cassatella, M.A.; Locati, M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl. Acad. Sci. USA 2009, 106, 5282–5287. [Google Scholar] [CrossRef]
- Mraz, M.; Haluzik, M. The role of adipose tissue immune cells in obesity and low-grade inflammation. J. Endocrinol. 2014, 222, R113–R127. [Google Scholar] [CrossRef]
- Villard, A.; Marchand, L.; Thivolet, C.; Rome, S. Diagnostic Value of Cell-free Circulating Micrornas for Obesity and Type 2 Diabetes: A Meta-analysis. J. Mol. Biomarkers Diagn. 2015, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Assmann, T.S.; Riezu-Boj, J.I.; Milagro, F.I.; Martínez, J.A. Circulating adiposity-related microRNAs as predictors of the response to a low-fat diet in subjects with obesity. J. Cell Mol. Med. 2020, 24, 2956–2967. [Google Scholar] [CrossRef]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ye, Y.; Wang, B.; Zhao, S. miR-140-5p Aggravates Insulin Resistance via Directly Targeting GYS1 and PPP1CC in Insulin-Resistant HepG2 Cells. Diabetes Metab. Syndr. Obesity Targets Ther. 2021, 14, 2515–2524. [Google Scholar] [CrossRef] [PubMed]
- Bermudez, E.A.; Rifai, N.; Buring, J.; Manson, J.E.; Ridker, P.M. Interrelationships Among Circulating Interleukin-6, C-Reactive Protein, and Traditional Cardiovascular Risk Factors in Women. Arter. Thromb. Vasc. Biol. 2002, 22, 1668–1673. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z.; Wei, R.; Miao, X.; Sun, S.; Liang, G.; Chu, C.; Zhao, L.; Zhu, X.; Guo, Q.; et al. IL (Interleukin)-6 Contributes to Deep Vein Thrombosis and Is Negatively Regulated by miR-338-5p. Arter. Thromb. Vasc. Biol. 2020, 40, 323–334. [Google Scholar] [CrossRef]
- Barwal, T.S.; Singh, N.; Sharma, U.; Bazala, S.; Rani, M.; Behera, A.; Kumawat, R.K.; Kumar, P.; Uttam, V.; Khandelwal, A.; et al. miR-590–5p: A double-edged sword in the oncogenesis process. Cancer Treat. Res. Commun. 2022, 32, 100593. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Zhu, Y.; Wei, X.; Zhou, J.; Chang, L.; Sui, H.; Han, Y.; Piao, D.; Sha, R.; Bai, Y. MiR-590-5p inhibits colorectal cancer angiogenesis and metastasis by regulating nuclear factor 90/vascular endothelial growth factor A axis. Cell Death Dis. 2016, 7, e2413. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Luo, Y.; Cong, Z.; Mu, Y.; Qiu, Y.; Zhong, M. MicroRNA-590-5p Inhibits Intestinal Inflammation by Targeting YAP. J. Crohn’s Colitis 2018, 12, 993–1004. [Google Scholar] [CrossRef]
- Molnar, R.; Szabo, L.; Tomesz, A.; Deutsch, A.; Darago, R.; Raposa, B.L.; Ghodratollah, N.; Varjas, T.; Nemeth, B.; Orsos, Z.; et al. The Chemopreventive Effects of Polyphenols and Coffee, Based upon a DMBA Mouse Model with microRNA and mTOR Gene Expression Biomarkers. Cells 2022, 11, 1300. [Google Scholar] [CrossRef]
- Wang, A.; Wang, S.; Zhu, C.; Huang, H.; Wu, L.; Wan, X.; Yang, X.; Zhang, H.; Miao, R.; He, L.; et al. Coffee and cancer risk: A meta-analysis of prospective observational studies. Sci. Rep. 2016, 6, srep33711. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Seibold, P.; Chang-Claude, J.; Flesch-Janys, D.; Liu, J.; Czene, K.; Humphreys, K.; Hall, P. Coffee consumption modifies risk of estrogen-receptor negative breast cancer. Breast Cancer Res. BCR 2011, 13, R49. [Google Scholar] [CrossRef] [PubMed]

| Males | Females | ||
|---|---|---|---|
| (N = 238) | (N = 673) | ||
| Age | years, mean ± SD | 50.9 ± 13.3 | 51.6 ± 13.4 |
| BMI | kg/m2, mean ± SD | 33.9 ± 4.7 | 33.6 ± 5.7 |
| Education | Primary school or less | 21 (8.8%) | 85 (12.6%) |
| Secondary school | 61 (25.6%) | 178 (26.4%) | |
| High school | 117 (49.2%) | 315 (46.8%) | |
| University or more | 39 (16.4%) | 95 (14.1%) | |
| Occupation | Employee | 166 (69.8%) | 375 (55.7%) |
| Unemployed | 15 (6.3%) | 55 (8.2%) | |
| Retired | 57 (23.95%) | 160 (23.8%) | |
| Housewife | - | 83 (12.3%) | |
| Residence traffic exposure | Mild | 53 (22.2%) | 148 (22.0%) |
| Moderate | 117 (49.2%) | 329 (48.9%) | |
| Heavy | 68 (28.6%) | 196 (29.1%) | |
| PM10 annual average | from the ARPA monitoring station µg/m3, mean ± SD | 43.0 ± 4.4 | 42.7 ± 4.7 |
| Physical activity | Sedentary | 148 (62.2%) | 433 (64.4%) |
| Active | 66 (27.7%) | 206 (30.6%) | |
| Sportive | 24 (10.1%) | 34 (5.0%) | |
| Antidepressants | Yes | 15 (6.3%) | 100 (14.9%) |
| No | 223 (93.7%) | 573 (85.1%) | |
| Smoking | Never | 88 (37.0%) | 377 (56.0%) |
| Ex | 108 (45.4%) | 198 (29.4%) | |
| Current | 42 (17.6%) | 98 (14.5%) | |
| Passive smoke exposure | Yes | 110 (46.2%) | 293 (43.5%) |
| No | 128 (53.8%) | 380 (56.5%) | |
| Alcohol | weekly consumption, median [Q1, Q3] | 2 [0; 7] | 0 [0; 2] |
| Coffee | weekly consumption, median [Q1, Q3] | 9 [5; 20] | 7 [3; 16] |
| C-reactive protein | mg/L, median [Q1, Q3] | 0.22 [0.11; 0.45] | 0.31 [0.14; 0.6] |
| Homocysteine | µmol/L, median [Q1, Q3] | 12 [10.2; 14.2] | 9.8 [8.3; 12.1] |
| Birth control pill | Yes | - | 26 (3.9%) |
| No | 647 (96.1%) | ||
| Number of previous pregnancies | median [Q1, Q3] | - | 2 [1; 3] |
| Menopause | Yes | - | 355 (55.1%) |
| No | 287 (44.9%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bollati, V.; Monti, P.; Biganzoli, D.; Marano, G.; Favero, C.; Iodice, S.; Ferrari, L.; Dioni, L.; Bianchi, F.; Pesatori, A.C.; et al. Environmental and Lifestyle Cancer Risk Factors: Shaping Extracellular Vesicle OncomiRs and Paving the Path to Cancer Development. Cancers 2023, 15, 4317. https://doi.org/10.3390/cancers15174317
Bollati V, Monti P, Biganzoli D, Marano G, Favero C, Iodice S, Ferrari L, Dioni L, Bianchi F, Pesatori AC, et al. Environmental and Lifestyle Cancer Risk Factors: Shaping Extracellular Vesicle OncomiRs and Paving the Path to Cancer Development. Cancers. 2023; 15(17):4317. https://doi.org/10.3390/cancers15174317
Chicago/Turabian StyleBollati, Valentina, Paola Monti, Davide Biganzoli, Giuseppe Marano, Chiara Favero, Simona Iodice, Luca Ferrari, Laura Dioni, Francesca Bianchi, Angela Cecilia Pesatori, and et al. 2023. "Environmental and Lifestyle Cancer Risk Factors: Shaping Extracellular Vesicle OncomiRs and Paving the Path to Cancer Development" Cancers 15, no. 17: 4317. https://doi.org/10.3390/cancers15174317
APA StyleBollati, V., Monti, P., Biganzoli, D., Marano, G., Favero, C., Iodice, S., Ferrari, L., Dioni, L., Bianchi, F., Pesatori, A. C., & Biganzoli, E. M. (2023). Environmental and Lifestyle Cancer Risk Factors: Shaping Extracellular Vesicle OncomiRs and Paving the Path to Cancer Development. Cancers, 15(17), 4317. https://doi.org/10.3390/cancers15174317






