Epithelioid Mesothelioma Patients with Very Long Survival Display Defects in DNA Repair
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Patients Cohort
2.2. RNA Isolation from FFPE Tumor Samples and Gene Expression Analysis
2.3. Quantitative Real-Time PCR
2.4. Immunofluorescence (IF) Detection of Nuclear Foci on FFPE OC-PDX Samples
3. Results
3.1. Patients Cohort
3.2. Nanostring Gene Expression
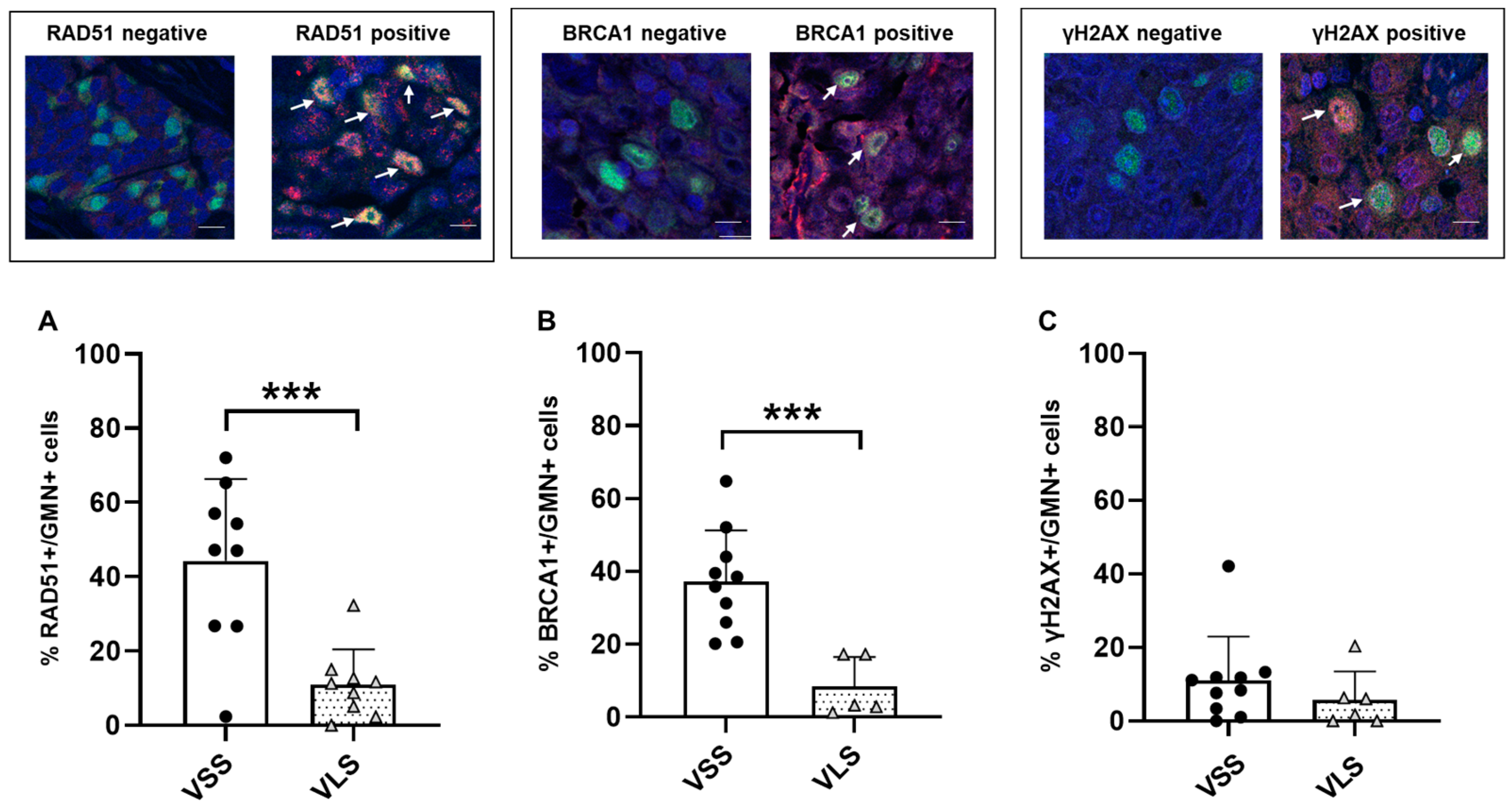
3.3. Functional Characterization of DNA Repair Status in MPM Tumor Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carbone, M.; Yang, H. Mesothelioma: Recent highlights. Ann. Transl. Med. 2017, 5, 238. [Google Scholar] [CrossRef]
- Ahmed, M.; Flannery, A.; Mujammil, I.; Breen, D. Variation in incidence trends of malignant pleural mesothelioma in Europe. Eur. Respir. J. 2018, 51, 1702384. [Google Scholar] [CrossRef]
- Davis, A.; Ke, H.; Kao, S.; Pavlakis, N. An Update on Emerging Therapeutic Options for Malignant Pleural Mesothelioma. Lung Cancer 2022, 13, 1–12. [Google Scholar] [CrossRef]
- Rondon, L.; Fu, R.; Patel, M.R. Success of Checkpoint Blockade Paves the Way for Novel Immune Therapy in Malignant Pleural Mesothelioma. Cancers 2023, 15, 2940. [Google Scholar] [CrossRef]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e6. [Google Scholar] [CrossRef] [PubMed]
- Malakoti, F.; Targhazeh, N.; Abadifard, E.; Zarezadeh, R.; Samemaleki, S.; Asemi, Z.; Younesi, S.; Mohammadnejad, R.; Hadi Hossini, S.; Karimian, A.; et al. DNA repair and damage pathways in mesothelioma development and therapy. Cancer Cell Int. 2022, 22, 176. [Google Scholar] [CrossRef]
- Panou, V.; Roe, O.D. Inherited Genetic Mutations and Polymorphisms in Malignant Mesothelioma: A Comprehensive Review. Int. J. Mol. Sci. 2020, 21, 4327. [Google Scholar] [CrossRef] [PubMed]
- Pagliuca, F.; Zito Marino, F.; Morgillo, F.; Della Corte, C.; Santini, M.; Vicidomini, G.; Guggino, G.; De Dominicis, G.; Campione, S.; Accardo, M.; et al. Inherited predisposition to malignant mesothelioma: Germline BAP1 mutations and beyond. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 4236–4246. [Google Scholar] [CrossRef]
- Hassan, R.; Morrow, B.; Thomas, A.; Walsh, T.; Lee, M.K.; Gulsuner, S.; Gadiraju, M.; Panou, V.; Gao, S.; Mian, I.; et al. Inherited predisposition to malignant mesothelioma and overall survival following platinum chemotherapy. Proc. Natl. Acad. Sci. USA 2019, 116, 9008–9013. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; DuBoff, M.; Jayakumaran, G.; Kris, M.G.; Ladanyi, M.; Robson, M.E.; Mandelker, D.; Zauderer, M.G. Novel Germline Mutations in DNA Damage Repair in Patients with Malignant Pleural Mesotheliomas. J. Thorac. Oncol. 2020, 15, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Sculco, M.; La Vecchia, M.; Aspesi, A.; Pinton, G.; Clavenna, M.G.; Casalone, E.; Allione, A.; Grosso, F.; Libener, R.; Muzio, A.; et al. Malignant pleural mesothelioma: Germline variants in DNA repair genes may steer tailored treatment. Eur. J. Cancer 2022, 163, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Konstantinopoulos, P.A.; Ceccaldi, R.; Shapiro, G.I.; D’Andrea, A.D. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov. 2015, 5, 1137–1154. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, D.M.; Krivak, T.C.; Kabil, N.; Munley, J.; Moore, K.N. PARP Inhibitors in Ovarian Cancer: A Review. Target. Oncol. 2023, 18, 471–503. [Google Scholar] [CrossRef]
- Panou, V.; Gadiraju, M.; Wolin, A.; Weipert, C.M.; Skarda, E.; Husain, A.N.; Patel, J.D.; Rose, B.; Zhang, S.R.; Weatherly, M.; et al. Frequency of Germline Mutations in Cancer Susceptibility Genes in Malignant Mesothelioma. J. Clin. Oncol. 2018, 36, 2863–2871. [Google Scholar] [CrossRef] [PubMed]
- Ghafoor, A.; Mian, I.; Wagner, C.; Mallory, Y.; Agra, M.G.; Morrow, B.; Wei, J.S.; Khan, J.; Thomas, A.; Sengupta, M.; et al. Phase 2 Study of Olaparib in Malignant Mesothelioma and Correlation of Efficacy with Germline or Somatic Mutations in BAP1 Gene. JTO Clin. Res. Rep. 2021, 2, 100231. [Google Scholar] [CrossRef] [PubMed]
- A Fennell, D.; King, S.; Mohammed, A.; Branson, C.; Brookes, L.; Darlison, A.G.; Dawson, A.; Gaba, M.; Hutka, B.; Morgan, A.; et al. Rucaparib in patients with BAP1-deficient or BRCA1-deficient mesothelioma (MiST1): An open-label, single-arm, phase 2a clinical trial. Lancet Respir. Med. 2021, 9, 593–600. [Google Scholar] [CrossRef]
- Betti, M.; Aspesi, A.; Ferrante, D.; Sculco, M.; Righi, L.; Mirabelli, D.; Napoli, F.; Rondon-Lagos, M.; Casalone, E.; Vignolo Lutati, F.; et al. Sensitivity to asbestos is increased in patients with mesothelioma and pathogenic germline variants in BAP1 or other DNA repair genes. Genes Chromosomes Cancer 2018, 57, 573–583. [Google Scholar] [CrossRef]
- Baumann, F.; Flores, E.; Napolitano, A.; Kanodia, S.; Taioli, E.; Pass, H.; Yang, H.; Carbone, M. Mesothelioma patients with germline BAP1 mutations have 7-fold improved long-term survival. Carcinogenesis 2015, 36, 76–81. [Google Scholar] [CrossRef]
- Shrestha, R.; Nabavi, N.; Lin, Y.Y.; Mo, F.; Anderson, S.; Volik, S.; Adomat, H.H.; Lin, D.; Xue, H.; Dong, X.; et al. BAP1 haploinsufficiency predicts a distinct immunogenic class of malignant peritoneal mesothelioma. Genome Med. 2019, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Dubacq, C.; Rabut, E.; Lopez, B.S.; Guirouilh-Barbat, J. Noncanonical Roles of RAD51. Cells 2023, 12, 1169. [Google Scholar] [CrossRef] [PubMed]
- Fuh, K.; Mullen, M.; Blachut, B.; Stover, E.; Konstantinopoulos, P.; Liu, J.; Matulonis, U.; Khabele, D.; Mosammaparast, N.; Vindigni, A. Homologous recombination deficiency real-time clinical assays, ready or not? Gynecol. Oncol. 2020, 159, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Guffanti, F.; Alvisi, M.F.; Anastasia, A.; Ricci, F.; Chiappa, M.; Llop-Guevara, A.; Serra, V.; Fruscio, R.; Degasperi, A.; Nik-Zainal, S.; et al. Basal expression of RAD51 foci predicts olaparib response in patient-derived ovarian cancer xenografts. Br. J. Cancer 2022, 126, 120–128. [Google Scholar] [CrossRef]
- Pellegrino, B.; Herencia-Ropero, A.; Llop-Guevara, A.; Pedretti, F.; Moles-Fernandez, A.; Viaplana, C.; Villacampa, G.; Guzman, M.; Rodriguez, O.; Grueso, J.; et al. Preclinical In Vivo Validation of the RAD51 Test for Identification of Homologous Recombination-Deficient Tumors and Patient Stratification. Cancer Res. 2022, 82, 1646–1657. [Google Scholar] [CrossRef]
- Waggott, D.; Chu, K.; Yin, S.; Wouters, B.G.; Liu, F.F.; Boutros, P.C. NanoStringNorm: An extensible R package for the pre-processing of NanoString mRNA and miRNA data. Bioinformatics 2012, 28, 1546–1548. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://media.aiom.it/userfiles/files/doc/LG/2017_LGAIOM_Mesotelioma.pdf (accessed on 20 August 2023).
- Ruff, S.E.; Logan, S.K.; Garabedian, M.J.; Huang, T.T. Roles for MDC1 in cancer development and treatment. DNA Repair 2020, 95, 102948. [Google Scholar] [CrossRef] [PubMed]
- Zimmerlin, L.; Zambidis, E.T. Pleiotropic roles of tankyrase/PARP proteins in the establishment and maintenance of human naive pluripotency. Exp. Cell Res. 2020, 390, 111935. [Google Scholar] [CrossRef]
- Janes, S.M.; Alrifai, D.; Fennell, D.A. Perspectives on the Treatment of Malignant Pleural Mesothelioma. N. Engl. J. Med. 2021, 385, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, K.; Murphy, F.A.; Duffin, R.; Poland, C.A. Asbestos, carbon nanotubes and the pleural mesothelium: A review of the hypothesis regarding the role of long fibre retention in the parietal pleura, inflammation and mesothelioma. Part. Fibre Toxicol. 2010, 7, 5. [Google Scholar] [CrossRef]
- Xu, A.; Wu, L.J.; Santella, R.M.; Hei, T.K. Role of oxyradicals in mutagenicity and DNA damage induced by crocidolite asbestos in mammalian cells. Cancer Res. 1999, 59, 5922–5926. [Google Scholar]
- Hiltbrunner, S.; Mannarino, L.; Kirschner, M.B.; Opitz, I.; Rigutto, A.; Laure, A.; Lia, M.; Nozza, P.; Maconi, A.; Marchini, S.; et al. Tumor Immune Microenvironment and Genetic Alterations in Mesothelioma. Front. Oncol. 2021, 11, 660039. [Google Scholar] [CrossRef] [PubMed]
- Betti, M.; Casalone, E.; Ferrante, D.; Aspesi, A.; Morleo, G.; Biasi, A.; Sculco, M.; Mancuso, G.; Guarrera, S.; Righi, L.; et al. Germline mutations in DNA repair genes predispose asbestos-exposed patients to malignant pleural mesothelioma. Cancer Lett. 2017, 405, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Chiappa, M.; Guffanti, F.; Bertoni, F.; Colombo, I.; Damia, G. Overcoming PARPi resistance: Preclinical and clinical evidence in ovarian cancer. Drug Resist. Updat. 2021, 55, 100744. [Google Scholar] [CrossRef]
- Borchert, S.; Wessolly, M.; Schmeller, J.; Mairinger, E.; Kollmeier, J.; Hager, T.; Mairinger, T.; Herold, T.; Christoph, D.C.; Walter, R.F.H.; et al. Gene expression profiling of homologous recombination repair pathway indicates susceptibility for olaparib treatment in malignant pleural mesothelioma in vitro. BMC Cancer 2019, 19, 108. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ma, K.; Chen, Z.; Yang, X.; Sun, F.; Jin, Y.; Shi, Y.; Jiang, W.; Wang, Q.; Zhan, C. A Nomogram to Predict Prognosis in Malignant Pleural Mesothelioma. World J. Surg. 2018, 42, 2134–2142. [Google Scholar] [CrossRef]
- Mannarino, L.; Paracchini, L.; Pezzuto, F.; Olteanu, G.E.; Moracci, L.; Vedovelli, L.; De Simone, I.; Bosetti, C.; Lupi, M.; Amodeo, R.; et al. Epithelioid Pleural Mesothelioma Is Characterized by Tertiary Lymphoid Structures in Long Survivors: Results from the MATCH Study. Int. J. Mol. Sci. 2022, 23, 5786. [Google Scholar] [CrossRef]
- Ohara, Y.; Enomoto, A.; Tsuyuki, Y.; Sato, K.; Iida, T.; Kobayashi, H.; Mizutani, Y.; Miyai, Y.; Hara, A.; Mii, S.; et al. Connective tissue growth factor produced by cancer-associated fibroblasts correlates with poor prognosis in epithelioid malignant pleural mesothelioma. Oncol. Rep. 2020, 44, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Alcala, N.; Mangiante, L.; Le-Stang, N.; Gustafson, C.E.; Boyault, S.; Damiola, F.; Alcala, K.; Brevet, M.; Thivolet-Bejui, F.; Blanc-Fournier, C.; et al. Redefining malignant pleural mesothelioma types as a continuum uncovers immune-vascular interactions. eBioMedicine 2019, 48, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Fuso Nerini, I.; Roca, E.; Mannarino, L.; Grosso, F.; Frapolli, R.; D’Incalci, M. Is DNA repair a potential target for effective therapies against malignant mesothelioma? Cancer Treat. Rev. 2020, 90, 102101. [Google Scholar] [CrossRef] [PubMed]

| n | % | |
|---|---|---|
| Age (years) | ||
| Median | 73 | |
| Range | 56–86 | |
| Gender | ||
| Female | 9 | 16.7 |
| Male | 45 | 83.3 |
| Histotype | ||
| Epitheliod | 54 | 100 |
| Chemotherapy | ||
| Yes | 23 | 42.6 |
| No | 4 | 7.4 |
| Unknown | 27 | 50 |
| Radiotherapy | ||
| Yes | 21 | 38.9 |
| No | 2 | 3.7 |
| Unknown | 31 | 57.4 |
| Survival (months) | ||
| Median (range) | 16.9 (1.3–107.7) | |
| % 1-year survival (95%CI) | 68.5% | |
| % 2-year survival (95%CI) | 37.1% | |
| Survivors subtype | ||
| Short | 26 | 48% |
| Very short | 12 | 22% |
| Long | 28 | 52% |
| Very long | 12 | 22% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganzinelli, M.; Guffanti, F.; Ianza, A.; Sobhani, N.; Crovella, S.; Zanconati, F.; Bottin, C.; Confalonieri, M.; Fumagalli, S.; Guglielmi, A.; et al. Epithelioid Mesothelioma Patients with Very Long Survival Display Defects in DNA Repair. Cancers 2023, 15, 4309. https://doi.org/10.3390/cancers15174309
Ganzinelli M, Guffanti F, Ianza A, Sobhani N, Crovella S, Zanconati F, Bottin C, Confalonieri M, Fumagalli S, Guglielmi A, et al. Epithelioid Mesothelioma Patients with Very Long Survival Display Defects in DNA Repair. Cancers. 2023; 15(17):4309. https://doi.org/10.3390/cancers15174309
Chicago/Turabian StyleGanzinelli, Monica, Federica Guffanti, Anna Ianza, Navid Sobhani, Sergio Crovella, Fabrizio Zanconati, Cristina Bottin, Marco Confalonieri, Stefano Fumagalli, Alessandra Guglielmi, and et al. 2023. "Epithelioid Mesothelioma Patients with Very Long Survival Display Defects in DNA Repair" Cancers 15, no. 17: 4309. https://doi.org/10.3390/cancers15174309
APA StyleGanzinelli, M., Guffanti, F., Ianza, A., Sobhani, N., Crovella, S., Zanconati, F., Bottin, C., Confalonieri, M., Fumagalli, S., Guglielmi, A., Generali, D., & Damia, G. (2023). Epithelioid Mesothelioma Patients with Very Long Survival Display Defects in DNA Repair. Cancers, 15(17), 4309. https://doi.org/10.3390/cancers15174309










