Minimal Infiltrative Disease Identification in Cryopreserved Ovarian Tissue of Girls with Cancer for Future Use: A Systematic Review
Abstract
Simple Summary
Abstract
1. Introduction
- Which targets are currently being used to screen for MID in cryopreserved OTs of pediatric patients?
- Which techniques were used for MID detection in OT in the relevant subgroups and may be used for assessment of the graft before autotransplantation?
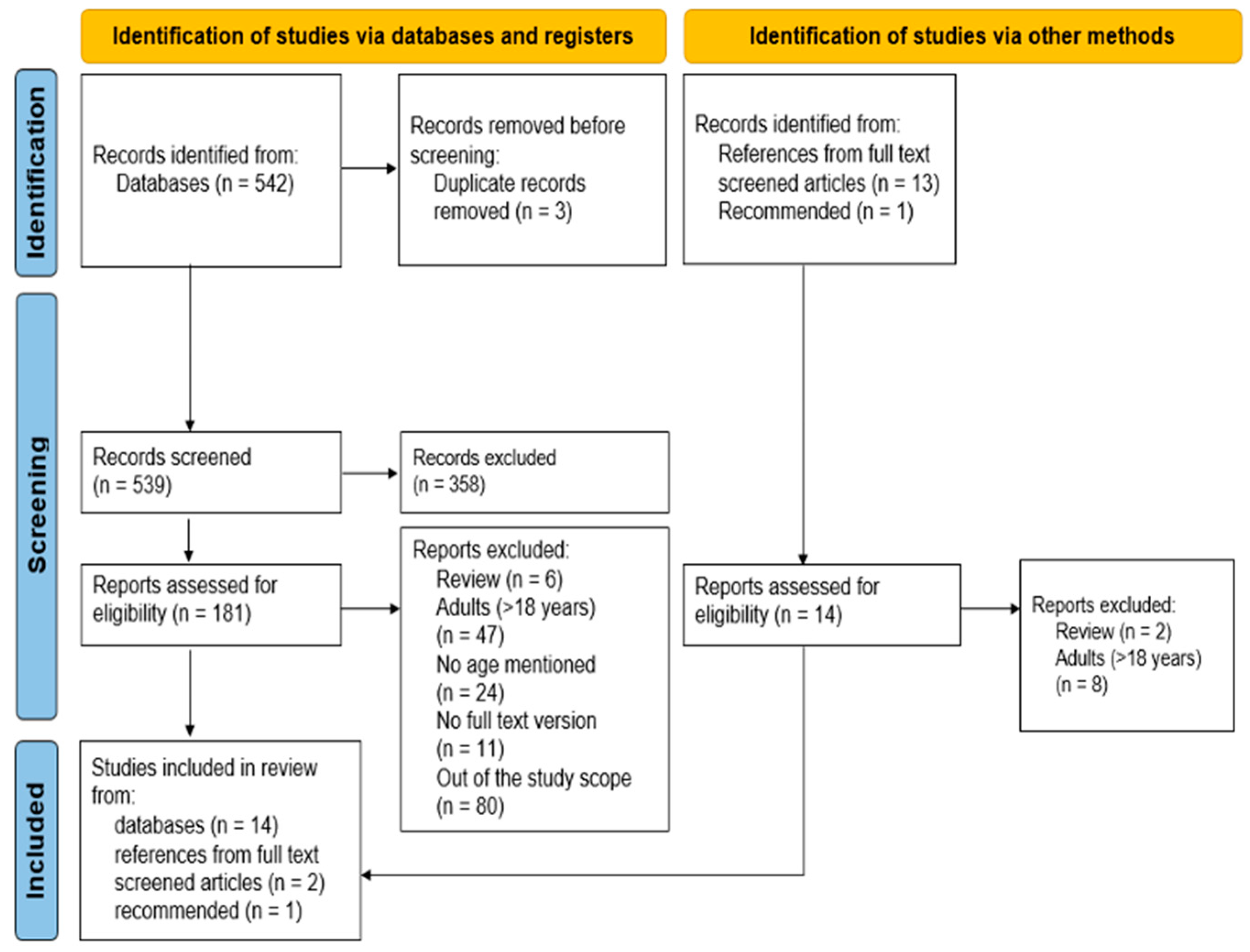
2. Materials and Methods
2.1. Search Strategy
2.2. Selection Criteria
2.3. Data Extraction
2.4. Quality of Evidence Assessment
3. Results
| Ref. | Study Type | Pt (n) | Type of Cancer | Markers and Methods (1–5) | Detected by Methods (1–5) | Positive Patients, n (%) | Potential Bias | Overall Bias | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Histology (1) | IHC (2) | FISH (3) | PCR $ (4) | Xeno (5) | SB | AB | MB | DB | SC | SA* | |||||||
| [30] | Retrospective study | 7 | Ewing sarcoma | n = 6 | CD99 n = 6 | - | EWS-FLI 1 FT n = 5 | - | 4 | method4: 1/5 (20%) |  |  |  |  |  |  |  |
| [31] | Retrospective study | 5 | Ewing sarcoma | n = 5 | - | EWSR1 n = 5 | EWS-FLI 1 FT n = 5 | - | ND | - |  |  |  |  |  |  |  |
| [32] | Case report ‡ | 1 | Ewing sarcoma | - | - | - | EWS-FLI 1 FT n = 1 | - | ND | - |  |  |  |  |  |  |  |
| [15] | Case reports | 2 | Ewing sarcoma | - | CD99 n = 2 | EWSR1 n = 2 | - | - | 2, 3 | method2: 2/2 (100%) method3: 2/2 (100%) |  |  |  |  |  |  |  |
| [24] | Retrospective study | 9 | Ewing sarcoma, synovial sarcoma, osteosarcoma | n = 9 | - | - | EWS-FLI 1 FT n = 6 | n = 9 | ND | - |  |  |  |  |  |  |  |
| [33] | Retrospective study | 2–9 † | Ewing sarcoma, osteosarcoma | n = 5 | CD99 n = 2 | - | EWS-ERG FT n = 2 | - | ND | - |  |  |  |  |  |  |  |
| [34] | Retrospective study | 16 | Ewing sarcoma, PNET, clear cell sarcoma, synovial sarcoma, rhabdomyosarcoma | n = 16 | CD99, MDM2, myogenin, S100, MyoD 1, melanoma cocktail n = 16 | - | EWS-FLI 1, EWS-ERG, PAX3-FOXO1, PAX7-FOXO1, SYT-SSX, EWS-ATF1 FTs, MyoD1 n = 12 | - | ND | - |  |  |  |  |  |  |  |
| Grade assessment | |||||||||||||||||
| Study design | Low | Retrospective cohort studies in 5/7; case reports in 2/7 | |||||||||||||||
| Study limitations | −2 | Important limitations: SB high in 7/7; AB low in 5/7, moderate in 1/7, high in 1/7; MB is low in 3/7, moderate in 3/7, high in 1/7; DB is low in 3/7, moderate in 3/7, high in 1/7; SC is low in 5/7, moderate in 2/7; SA is low in 4/7, moderate in 2/7, high in 1/7 | |||||||||||||||
| Consistency | 0 | Little inconsistency: 6/7 (1/6 studies detected MID) studies used PCR and 4/7 IHC (1/4 studies detected MID) to detect MID; FISH detected MID in 1/2, and markers used were mostly the same across all the studies for a specific method) | |||||||||||||||
| Directness | −1 | Though results are direct, population and outcomes cannot be generalized | |||||||||||||||
| Precision | −1 | Low number of patients | |||||||||||||||
| Publication bias | 0 | Unlikely | |||||||||||||||
| Effect size | 0 | NA | |||||||||||||||
| Dose–response | 0 | NA | |||||||||||||||
| Plausible confounding | +1 | Treatment as plausible confounder may seriously influence the effect | |||||||||||||||
| Quality of evidence |  | Very low | |||||||||||||||
| Conclusion | Detection of MID in ovarian tissues from solid tumor patients is unlikely despite various attempts to identify it but the results cannot be generalized (7 studies, 42–49 † participants) | ||||||||||||||||
 = low risk of bias;
= low risk of bias;  = moderate risk of bias;
= moderate risk of bias;  = high risk of bias;
= high risk of bias;  = very low quality of evidence. AB = attrition bias (the study data available (i.e., participants not lost to follow up) adequately represent the study sample), DB = detection bias (the outcome of interest is measured in a similar way for all participants), FISH = fluorescence in situ hybridization, FT = fusion transcript, IHC = immunohistochemistry, L = low risk, M = moderate risk, MB = measurement bias (the prognostic factor is measured in a similar way for all participants), ND = not detected, OT = ovarian tissue, OTC = ovarian tissue cryopreservation, PCR = polymerase chain reaction, PNET = primitive neuroectodermal tumor, Pt = patients, SA* = no statistical analysis performed, all primary outcomes are reported, SB = selection bias (the study sample adequately represents the population of interest), SC = study confounding (important potential confounding factors are appropriately accounted for), Xeno = xenotransplantation (and evaluation after). ‘$’ includes all types of PCR: PCR, RT-PCR, quantitative RT-PCR, digital droplet RT-PCR, nested PCR; ‘-’ means the method was not used in the specific study; † OS: n = 7 (mean age 15, range 13–18 y) EW: n = 4 (mean 19, range 17–21 y), it is unclear how many patients were <18 years; ‡ letter to the editor about a case report.
= very low quality of evidence. AB = attrition bias (the study data available (i.e., participants not lost to follow up) adequately represent the study sample), DB = detection bias (the outcome of interest is measured in a similar way for all participants), FISH = fluorescence in situ hybridization, FT = fusion transcript, IHC = immunohistochemistry, L = low risk, M = moderate risk, MB = measurement bias (the prognostic factor is measured in a similar way for all participants), ND = not detected, OT = ovarian tissue, OTC = ovarian tissue cryopreservation, PCR = polymerase chain reaction, PNET = primitive neuroectodermal tumor, Pt = patients, SA* = no statistical analysis performed, all primary outcomes are reported, SB = selection bias (the study sample adequately represents the population of interest), SC = study confounding (important potential confounding factors are appropriately accounted for), Xeno = xenotransplantation (and evaluation after). ‘$’ includes all types of PCR: PCR, RT-PCR, quantitative RT-PCR, digital droplet RT-PCR, nested PCR; ‘-’ means the method was not used in the specific study; † OS: n = 7 (mean age 15, range 13–18 y) EW: n = 4 (mean 19, range 17–21 y), it is unclear how many patients were <18 years; ‡ letter to the editor about a case report.3.1. Solid Tumors
3.2. Hematological Malignancies
| Ref. | Study Type | Pt (n) | Type of Cancer | Markers and Methods (1–5) | Detected by Methods (1–5) | Positive Patients, n (%) | Potential Bias | Overall Bias | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Histology (1) | IHC (2) | PCR $ (3) | Xeno (4) | MFC (5) | SB | AB | MB | DB | SC | SA* | |||||||
| [35] | Case report | 1 | CML | n = 1 | Glycophorin A, MPO, CD34, CD68, LCA/DC45, Factor VIII n = 1 | BCR-ABL FT n = 1 | - | - | 3 | 1/1 (100%) |  |  |  |  |  |  |  |
| [25] | Retrospective study | 8 | ALL, AML | n = 8 | - | ETV6-RUNX1, BCR-ABL1 FTs, IGH, TCR, FLT3 rearrangements n = 6 | n = 8 | - | 1, 3, 4 | method1: 1/8 (12.5%) method3: 5/6 (83%) method4: 2/8 (25%) |  |  |  |  |  |  |  |
| [36] | Case report | 1 | ALL | - | - | BCR-ABL FT n = 1 | - | - | ND | - |  |  |  |  |  |  |  |
| [37] | Retrospective study | 14 | ALL, AML, CML, JMML | n = 14 | CD34, CD10, CD20, CD79a, CD3, TdT, CD117, MPO, CD68 n = 14 | TEL-AML1, BCR-ABL b2a2, BCR-ABL b2a2, CBFB-MYH11 type A FTs n = 5 | - | - | 3 | 3/5 (60%) |  |  |  |  |  |  |  |
| [26] | Retrospective study | 8 | B-ALL, T-ALL, AML | - | - | - | n = 6 | CD19, CD34, CD10-, negative for myeloid markers/CD45, HLA-DR2, CD10, CD19, CD22, CD33/CD19, CD10, CD22, CD38/CD45, CD10, CD19, CD22, CD34, HLA-DR2/CD2, cyCD3, CD5, CD7, CD10, CD33, CD34, CD45RA, CD123 n = 8 | 4, 5 | method4: 1/6 (12.5%) method5: 2/8 (25%) |  |  |  |  |  |  |  |
| [38] | Retrospective study | 9 | ALL, AML, Burkitt’s lymphoma | n = 9 | - | E2A-PBX1, TEL-AML1, MLL-AF4, AML1-ETO FTs, IgK Kde, IgH, TCRD, TCRB n = 9 | - | - | 3 | 2/9 (22%) |  |  |  |  |  |  |  |
| [27] | Retrospective study | 10 | ALL, CML | n = 10 | - | BCR-ABL1, Ig and/or TCR-gama rearrangements n = 8 | n = 10 | - | 3, 4 | method3: 5/8 (62.5%) method4: 4/10 (40%) |  |  |  |  |  |  |  |
| [16] | Retrospective study | 5 | ALL | n = 5 | CD20, CD79a, CD3, TdT n = 5 | IgH rearrangements, EA2-PBX1 FT n = 4 | n = 5 | - | 3 | 4/4 (100%) |  |  |  |  |  |  |  |
| Grade assessment | |||||||||||||||||
| Study design | Low | Retrospective cohort studies in 6/8; case reports in 2/8 | |||||||||||||||
| Study limitations | −1 | Some limitations: SB high in 8/8; AB low in 8/8; MB is low in 4/8, moderate in 3/8, high in 1/8; DB is low in 4/8, moderate in 3/8, high in 1/8; SC low in 5/8, moderate in 3/8; SA low in 8/8 | |||||||||||||||
| Consistency | −1 | Though MID was detected by PCR in 6/7 studies and by xenotransplantation in 3/4, IHC detected MID in 0/3 and histology in 1/6 studies. The markers analyzed were very different across the studies | |||||||||||||||
| Directness | −1 | Though results are direct, population and outcomes cannot be generalized | |||||||||||||||
| Precision | −1 | Low number of events | |||||||||||||||
| Publication bias | 0 | Unlikely | |||||||||||||||
| Effect size | 0 | NA | |||||||||||||||
| Dose–response | 0 | NA | |||||||||||||||
| Plausible confounding | +1 | Treatment as plausible confounder may seriously influence the effect | |||||||||||||||
| Quality of evidence |  | Very low | |||||||||||||||
| Conclusion | Detection of MID in ovarian tissues from the patients of haematological malignancies is moderately likely, despite various attempts to identify it, the results cannot be generalized (8 studies, 56 participants) | ||||||||||||||||
 = low risk of bias;
= low risk of bias;  = moderate risk of bias;
= moderate risk of bias;  = high risk of bias;
= high risk of bias;  = very low quality of evidence. AB = attrition bias (the study data available (i.e., participants not lost to follow up) adequately represent the study sample), ALL = acute lymphoblastic leukemia, AML = acute myeloid leukemia, CML = chronic myeloid leukemia, DB = detection bias (the outcome of interest is measured in a similar way for all participants), FISH = fluorescence in situ hybridization, FT = fusion transcript, IHC = immunohistochemistry, JMML = juvenile myelomonocytic leukemia, L = low risk, M = moderate risk, MB = measurement bias (the prognostic factor is measured in a similar way for all participants), MID = minimally infiltrative disease, MPO = myeloperoxidase, ND = not detected, OT = ovarian tissue, OTC = ovarian tissue cryopreservation, PCR = polymerase chain reaction, Pt = patients, SA* = no statistical analysis performed, all primary outcomes are reported, SB = selection bias (the study sample adequately represents the population of interest), SC = study confounding (important potential confounding factors are appropriately accounted for), Xeno = xenotransplantation (and evaluation after). ‘-’ means the method was not used in the specific study; ‘$’ includes all types of PCR: PCR, RT-PCR, quantitative RT-PCR, digital droplet RT-PCR, nested PCR.
= very low quality of evidence. AB = attrition bias (the study data available (i.e., participants not lost to follow up) adequately represent the study sample), ALL = acute lymphoblastic leukemia, AML = acute myeloid leukemia, CML = chronic myeloid leukemia, DB = detection bias (the outcome of interest is measured in a similar way for all participants), FISH = fluorescence in situ hybridization, FT = fusion transcript, IHC = immunohistochemistry, JMML = juvenile myelomonocytic leukemia, L = low risk, M = moderate risk, MB = measurement bias (the prognostic factor is measured in a similar way for all participants), MID = minimally infiltrative disease, MPO = myeloperoxidase, ND = not detected, OT = ovarian tissue, OTC = ovarian tissue cryopreservation, PCR = polymerase chain reaction, Pt = patients, SA* = no statistical analysis performed, all primary outcomes are reported, SB = selection bias (the study sample adequately represents the population of interest), SC = study confounding (important potential confounding factors are appropriately accounted for), Xeno = xenotransplantation (and evaluation after). ‘-’ means the method was not used in the specific study; ‘$’ includes all types of PCR: PCR, RT-PCR, quantitative RT-PCR, digital droplet RT-PCR, nested PCR.3.3. Central Nervous System (CNS) Tumors
| Ref. | Study Type | Pt (n) | Type of Cancer | Markers and Methods (1–5) | Detected by Methods (1–5) | Positive Patients, n (%) | Potential Bias | Overall Bias | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Histology (1) | IHC (2) | PCR $ (3) | Xeno (4) | NGS (5) | SB | AB | MB | DB | SC | SA* | |||||||
| [29] | Prospective study | 17 | Astrocytoma, ependymoma, germinoma, glioblastomsa, medulloblastoma, PNET | n = 17 | NSE, GFAP n = 17 | GFAP, ENO2 n = 14 | n = 17 | - | ND | - |  |  |  |  |  |  |  |
| [28] | Case reports | 3 | PNET | n = 3 | NSE, GFAP n = 3 | GFAP, ENO2 n = 3 | n = 3 | n = 1 | ND | - |  |  |  |  |  |  |  |
| Grade assessment | |||||||||||||||||
| Study design | Low | Prospective cohort study in 1/1 (case report is not incorporated in the body summary of evidence since the same patients are in the prospective study) | |||||||||||||||
| Study limitations | −1 | Some limitations: SB moderate in 1/1; AB low in 1/1; MB is moderate in 1/1; DB is moderate in 1/2; SC moderate in 1/1; SA low in 1/1 | |||||||||||||||
| Consistency | NA | Only one study | |||||||||||||||
| Directness | −1 | Though results are direct, population and outcomes cannot be generalized | |||||||||||||||
| Precision | −1 | Low number of patients; only one study | |||||||||||||||
| Publication bias | 0 | Unlikely | |||||||||||||||
| Effect size | 0 | NA | |||||||||||||||
| Dose–response | 0 | NA | |||||||||||||||
| Plausible confounding | +1 | Treatment as plausible confounder may seriously influence the effect | |||||||||||||||
| Quality of evidence |  | Very low | |||||||||||||||
| Conclusion | Detection of MID in ovarian tissues from central nervous system tumors patients is unlikely despite various attempts to identify it but the results cannot be generalized (1 study, 17 participants) | ||||||||||||||||
 = low risk of bias;
= low risk of bias;  = moderate risk of bias;
= moderate risk of bias;  = high risk of bias;
= high risk of bias;  = very low quality of evidence. AB = attrition bias (the study data available (i.e., participants not lost to follow up) adequately represent the study sample), DB = detection bias (the outcome of interest is measured in a similar way for all participants), FISH = fluorescence in situ hybridization, IHC = immunohistochemistry, L = low risk, M = moderate risk, MB = measurement bias (the prognostic factor is measured in a similar way for all participants), MID = minimally infiltrative disease, ND = not detected, OT = ovarian tissue, OTC = ovarian tissue cryopreservation, PCR = polymerase chain reaction, PNET = primitive neuroectodermal tumor, Pt = patients, SA* = no statistical analysis performed, all primary outcomes are reported, SB = selection bias (the study sample adequately represents the population of interest), SC = study confounding (important potential confounding factors are appropriately accounted for), Xeno = xenotransplantation (and evaluation after). ‘-’ means the method was not used in the specific study; ‘$’ includes all types of PCR: PCR, RT-PCR, quantitative RT-PCR, digital droplet RT-PCR, nested PCR.
= very low quality of evidence. AB = attrition bias (the study data available (i.e., participants not lost to follow up) adequately represent the study sample), DB = detection bias (the outcome of interest is measured in a similar way for all participants), FISH = fluorescence in situ hybridization, IHC = immunohistochemistry, L = low risk, M = moderate risk, MB = measurement bias (the prognostic factor is measured in a similar way for all participants), MID = minimally infiltrative disease, ND = not detected, OT = ovarian tissue, OTC = ovarian tissue cryopreservation, PCR = polymerase chain reaction, PNET = primitive neuroectodermal tumor, Pt = patients, SA* = no statistical analysis performed, all primary outcomes are reported, SB = selection bias (the study sample adequately represents the population of interest), SC = study confounding (important potential confounding factors are appropriately accounted for), Xeno = xenotransplantation (and evaluation after). ‘-’ means the method was not used in the specific study; ‘$’ includes all types of PCR: PCR, RT-PCR, quantitative RT-PCR, digital droplet RT-PCR, nested PCR.4. Discussion
4.1. Strengths and Limitations
4.2. Implications for Clinical Practice and Research
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Script |
|---|
| (((((((((((((((((((((((((((((((((((((((minimal residual disease*[Title/Abstract]) OR (MRD[Title/Abstract])) OR (measurable residual disease*[Title/Abstract])) OR (residual tumor*[Title/Abstract])) OR (residual tumour*[Title/Abstract])) OR (residual minimal disease*[Title/Abstract])) OR (residual disease*[Title/Abstract])) OR (contaminat*[Title/Abstract])) OR (infiltrat*[Title/Abstract])) OR (detect*[Title/Abstract])) OR (molecular residual disease*[Title/Abstract]))) OR (aberration*[Title/Abstract])) OR (fusion transcript*[Title/Abstract])) OR (gene fusion*[Title/Abstract])) OR (translocation*[Title/Abstract])) OR (tumor-specific transcript*[Title/Abstract])) OR (PAX3[Title/Abstract])) OR (PHOX2B[Title/Abstract])) OR (CRMP1[Title/Abstract])) OR (GAP43[Title/Abstract])) OR (ISL1[Title/Abstract])) OR (PAX3- FKHR[Title/Abstract])) OR (PAX7-FKHR[Title/Abstract])) OR (FKHR[Title/Abstract])) OR (PAX7[Title/Abstract])) OR (FOXO1[Title/Abstract])) OR (RASSF1A[Title/Abstract])) OR (CTNNB1[Title/Abstract])) OR (onfFN[Title/Abstract])) OR (EWS-FLI1[Title/Abstract])) OR (EWSR1[Title/Abstract])) OR (FLI1[Title/Abstract])) OR (ETV1[Title/Abstract])) OR (ETV6[Title/Abstract])) OR (CHOP[Title/Abstract])) OR (GADD153[Title/Abstract])) OR (ATF1[Title/Abstract])) AND ((((((((((((((((((cancer*[Title/Abstract]) OR (cancer cell*[Title/Abstract]) OR (tumor*[Title/Abstract])) OR (tumour*[Title/Abstract])) OR (malignan*[Title/Abstract])) OR (neoplasm*[Title/Abstract])) OR (carcinoma*[Title/Abstract])) OR (oncolog*[Title/Abstract])) OR (sarcoma*[Title/Abstract])) OR (blastoma*[Title/Abstract]))) OR (osteosarcoma*[Title/Abstract])) OR (neuroblastoma*[Title/Abstract])) OR (rhabdomyosarcoma*[Title/Abstract])) OR (nephroblastoma*[Title/Abstract])) OR (retinoblastoma*[Title/Abstract])) OR (hepatoblastoma*[Title/Abstract])) OR (Wilms*[Title/Abstract])) OR (ewing[Title/Abstract]))) AND (((((ovarian[Title/Abstract]) OR (ovary[Title/Abstract])) OR (ovaries[Title/Abstract])) OR (gonad*[Title/Abstract])) AND ((((((cryopresev*[Title/Abstract]) OR (frozen[Title/Abstract])) OR (freeze[Title/Abstract])) OR (harvest*[Title/Abstract])) OR (frozen-thawed[Title/Abstract])) OR (thawed[Title/Abstract]))) |
| Inclusion | Exclusion |
|---|---|
|
|
References
- Corkum, K.S.; Rhee, D.S.; Wafford, Q.E.; Demeestere, I.; Dasgupta, R.; Baertschiger, R.; Malek, M.M.; Aldrink, J.H.; Heaton, T.E.; Weil, B.R.; et al. Fertility and hormone preservation and restoration for female children and adolescents receiving gonadotoxic cancer treatments: A systematic review. J. Pediatr. Surg. 2019, 54, 2200–2209. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, F.; Frederiksen, L.E.; Bonaventure, A.; Mader, L.; Hasle, H.; Robison, L.L.; Winther, J.F. Childhood cancer: Survival, treatment modalities, late effects and improvements over time. Cancer Epidemiol. 2021, 71, 101733. [Google Scholar] [CrossRef] [PubMed]
- The ESHRE Guideline Group on Female Fertility Preservation; Anderson, R.A.; Amant, F.; Braat, D.; D’Angelo, A.; Lopes, S.M.C.d.S.; Demeestere, I.; Dwek, S.; Frith, L.; Lambertini, M.; et al. ESHRE guideline: Female fertility preservation. Hum. Reprod. Open 2020, 2020, hoaa052. [Google Scholar] [CrossRef] [PubMed]
- Marin, L.; Bedoschi, G.; Kawahara, T.; Oktay, K.H. History, Evolution and Current State of Ovarian Tissue Auto-Transplantation with Cryopreserved Tissue: A Successful Translational Research Journey from 1999 to 2020. Reprod. Sci. 2020, 27, 955–962. [Google Scholar] [CrossRef] [PubMed]
- van der Perk, M.M.; van der Kooi, A.-L.L.; Bos, A.M.; Broer, S.L.; Veening, M.A.; van Leeuwen, J.; van Santen, H.M.; van Dorp, W.; van den Heuvel-Eibrink, M.M. Oncofertility Perspectives for Girls with Cancer. J. Pediatr. Adolesc. Gynecol. 2022, 35, 523–526. [Google Scholar] [CrossRef]
- Gellert, S.E.; Pors, S.E.; Kristensen, S.G.; Bay-Bjørn, A.M.; Ernst, E.; Andersen, C.Y. Transplantation of frozen-thawed ovarian tissue: An update on worldwide activity published in peer-reviewed papers and on the Danish cohort. J. Assist. Reprod. Genet. 2018, 35, 561–570. [Google Scholar] [CrossRef]
- McLaughlin, M.; Albertini, D.F.; Wallace, W.H.B.; Anderson, R.A.; Telfer, E.E. Metaphase II oocytes from human unilaminar follicles grown in a multi-step culture system. Mol. Hum. Reprod. 2018, 24, 135–142. [Google Scholar] [CrossRef]
- Telfer, E.E. Fertility Preservation: Progress and prospects for developing human immature oocytes in vitro. Reproduction 2019, 158, F45–F54. [Google Scholar] [CrossRef]
- Dolmans, M.-M.; Amorim, C.A. FERTILITY PRESERVATION: Construction and use of artificial ovaries. Reproduction 2019, 158, F15–F25. [Google Scholar] [CrossRef]
- Clarkson, Y.L.; McLaughlin, M.; Waterfall, M.; Dunlop, C.E.; Skehel, P.A.; Anderson, R.A.; Telfer, E.E. Initial characterisation of adult human ovarian cell populations isolated by DDX4 expression and aldehyde dehydrogenase activity. Sci. Rep. 2018, 8, 6953. [Google Scholar] [CrossRef]
- Silvestris, E.; Cafforio, P.; D’oronzo, S.; Felici, C.; Silvestris, F.; Loverro, G. In vitro differentiation of human oocyte-like cells from oogonial stem cells: Single-cell isolation and molecular characterization. Hum. Reprod. 2018, 33, 464–473. [Google Scholar] [CrossRef]
- Dolmans, M.-M.; Demylle, D.; Martinez-Madrid, B.; Donnez, J. Efficacy of in vitro fertilization after chemotherapy. Fertil. Steril. 2005, 83, 897–901. [Google Scholar] [CrossRef] [PubMed]
- Bastings, L.; Beerendonk, C.C.M.; Westphal, J.R.; Massuger, L.F.A.G.; Kaal, S.E.J.; van Leeuwen, F.E.; Braat, D.D.M.; Peek, R. Autotransplantation of cryopreserved ovarian tissue in cancer survivors and the risk of reintroducing malignancy: A systematic review. Hum. Reprod. Updat. 2013, 19, 483–506. [Google Scholar] [CrossRef]
- Dolmans, M.-M.; Luyckx, V.; Donnez, J.; Andersen, C.Y.; Greve, T. Risk of transferring malignant cells with transplanted frozen-thawed ovarian tissue. Fertil. Steril. 2013, 99, 1514–1522. [Google Scholar] [CrossRef] [PubMed]
- Schifflers, S.; Delbecque, K.; Galant, C.; Francotte, N.; Philippet, P.; Chantrain, C.F. Microscopic Infiltration of Cryopreserved Ovarian Tissue in 2 Patients with Ewing Sarcoma. J. Pediatr. Hematol. Oncol. 2018, 40, e167–e170. [Google Scholar] [CrossRef]
- Díaz-García, C.; Herraiz, S.; Such, E.; Andrés, M.D.M.; Villamón, E.; Mayordomo-Aranda, E.; Cervera, J.V.; Sanz, M.A.; Pellicer, A. Dexamethasone does not prevent malignant cell reintroduction in leukemia patients undergoing ovarian transplant: Risk assessment of leukemic cell transmission by a xenograft model. Hum. Reprod. 2019, 34, 1485–1493. [Google Scholar] [CrossRef]
- Mulder, C.L.; Eijkenboom, L.L.; Beerendonk, C.C.M.; Braat, D.D.M.; Peek, R. Enhancing the safety of ovarian cortex autotransplantation: Cancer cells are purged completely from human ovarian tissue fragments by pharmacological inhibition of YAP/TAZ oncoproteins. Hum. Reprod. 2019, 34, 506–518. [Google Scholar] [CrossRef]
- Eijkenboom, L.; Mulder, C.; van der Reijden, B.; van Mello, N.; van Leersum, J.; Koorenhof-Scheele, T.; Braat, D.; Beerendonk, C.; Peek, R. Purging human ovarian cortex of contaminating leukaemic cells by targeting the mitotic catastrophe signalling pathway. J. Assist. Reprod. Genet. 2021, 38, 1571–1588. [Google Scholar] [CrossRef]
- Moghassemi, S.; Dadashzadeh, A.; Camboni, A.; Feron, O.; Azevedo, R.B.; Amorim, C.A. Photodynamic therapy using OR141-loaded nanovesicles for eradication of leukemic cells from ovarian tissue. Photodiagnosis Photodyn. Ther. 2022, 40, 103139. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A web and mobile app for systematic reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, 71. [Google Scholar]
- Hayden, J.A.; Van Der Windt, D.A.; Cartwright, J.L.; Côté, P.; Bombardier, C. Assessing Bias in Studies of Prognostic Factors. Ann. Intern. Med. 2013, 158, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Schünemann, H.J.; Oxman, A.D.; Brozek, J.; Glasziou, P.; Jaeschke, R.; Vist, G.E.; Williams, J.W., Jr.; Kunz, R.; Craig, J.; Montori, V.M.; et al. Grading quality of evidence and strength of recommendations for diagnostic tests and strategies. BMJ 2008, 336, 1106–1110. [Google Scholar] [CrossRef]
- Greve, T.; Wielenga, V.T.; Grauslund, M.; Sørensen, N.; Christiansen, D.B.; Rosendahl, M.; Andersen, C.Y. Ovarian tissue cryopreserved for fertility preservation from patients with Ewing or other sarcomas appear to have no tumour cell contamination. Eur. J. Cancer 2013, 49, 1932–1938. [Google Scholar] [CrossRef] [PubMed]
- Soares, M.; Saussoy, P.; Maskens, M.; Reul, H.; Amorim, C.A.; Donnez, J.; Dolmans, M.-M. Eliminating malignant cells from cryopreserved ovarian tissue is possible in leukaemia patients. Br. J. Haematol. 2017, 178, 231–239. [Google Scholar] [CrossRef]
- Zver, T.; Frontczak, S.; Poirot, C.; Rives-Feraille, A.; Leroy-Martin, B.; Koscinski, I.; Arbez-Gindre, F.; Garnache-Ottou, F.; Roux, C.; Amiot, C. Minimal residual disease detection by multicolor flow cytometry in cryopreserved ovarian tissue from leukemia patients. J. Ovarian Res. 2022, 15, 9. [Google Scholar] [CrossRef]
- Dolmans, M.-M.; Marinescu, C.; Saussoy, P.; Van Langendonckt, A.; Amorim, C.; Donnez, J. Reimplantation of cryopreserved ovarian tissue from patients with acute lymphoblastic leukemia is potentially unsafe. Blood 2010, 116, 2908–2914. [Google Scholar] [CrossRef]
- Nguyen, T.Y.T.; Camboni, A.; Masciangelo, R.; Donnez, J.; Dolmans, M.-M. Is Ovarian Tissue Transplantation Safe in Patients with Central Nervous System Primitive Neuroectodermal Tumors? J. Clin. Med. 2020, 9, 4101. [Google Scholar] [CrossRef]
- Nguyen, T.Y.T.; Cacciottola, L.; Camboni, A.; Ravau, J.; De Vos, M.; Demeestere, I.; Donnez, J.; Dolmans, M.-M. Ovarian tissue cryopreservation and transplantation in patients with central nervous system tumours. Hum. Reprod. 2021, 36, 1296–1309. [Google Scholar] [CrossRef]
- Abir, R.; Feinmesser, M.; Yaniv, I.; Fisch, B.; Cohen, I.J.; Ben-Haroush, A.; Meirow, D.; Felz, C.; Avigad, S. Occasional involvement of the ovary in Ewing sarcoma. Hum. Reprod. 2010, 25, 1708–1712. [Google Scholar] [CrossRef][Green Version]
- Chaput, L.; Grèze, V.; Halle, P.; Radosevic-Robin, N.; Pereira, B.; Véronèse, L.; Lejeune, H.; Durand, P.; Martin, G.; Sanfilippo, S.; et al. Sensitive and Specific Detection of Ewing Sarcoma Minimal Residual Disease in Ovarian and Testicular Tissues in an In Vitro Model. Cancers 2019, 11, 1807. [Google Scholar] [CrossRef]
- Andersen, C.Y.; Ernst, E.; Bærentzen, S.; Birkebæk, N.H.; Clausen, N. No malignancy detected in surplus ovarian tissue from a former Ewing sarcoma patient who experienced relapse four years after being grafted with frozen/thawed ovarian tissue. J. Assist. Reprod. Genet. 2014, 31, 1567–1568. [Google Scholar] [CrossRef][Green Version]
- Hoekman, E.J.; Smit, V.T.; Fleming, T.P.; Louwe, L.A.; Fleuren, G.J.; Hilders, C.G. Searching for metastases in ovarian tissue before autotransplantation: A tailor-made approach. Fertil. Steril. 2015, 103, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Dolmans, M.; Iwahara, Y.; Donnez, J.; Soares, M.; Vaerman, J.; Amorim, C.; Poirel, H. Evaluation of minimal disseminated disease in cryopreserved ovarian tissue from bone and soft tissue sarcoma patients. Hum. Reprod. 2016, 31, 2292–2302. [Google Scholar] [CrossRef] [PubMed]
- Abir, R.; Aviram, A.; Feinmesser, M.; Stein, J.; Yaniv, I.; Parnes, D.; Ben-Haroush, A.; Meirow, D.; Rabizadeh, E.; Fisch, B. Ovarian minimal residual disease in chronic myeloid leukaemia. Reprod. Biomed. Online 2014, 28, 255–260. [Google Scholar] [CrossRef]
- Rodriguez-Wallberg, K.A.; Milenkovic, M.; Papaikonomou, K.; Keros, V.; Gustafsson, B.; Sergouniotis, F.; Wikander, I.; Perot, R.; Borgström, B.; Ljungman, P.; et al. Successful pregnancies after transplantation of ovarian tissue retrieved and cryopreserved at time of childhood acute lymphoblastic leukemia—A case report. Haematologica 2021, 106, 2783–2787. [Google Scholar] [CrossRef] [PubMed]
- Rosendahl, M.; Andersen, M.T.; Ralfkiær, E.; Kjeldsen, L.; Andersen, M.K.; Andersen, C.Y. Evidence of residual disease in cryopreserved ovarian cortex from female patients with leukemia. Fertil. Steril. 2010, 94, 2186–2190. [Google Scholar] [CrossRef]
- Asadi-Azarbaijani, B.; Sheikhi, M.; Nurmio, M.; Tinkanen, H.; Juvonen, V.; Dunkel, L.; Hovatta, O.; Oskam, I.C.; Jahnukainen, K. Minimal residual disease of leukemia and the quality of cryopreserved human ovarian tissue in vitro. Leuk. Lymphoma 2016, 57, 700–707. [Google Scholar] [CrossRef]
- Lam, W.A.; Rosenbluth, M.J.; Fletcher, D.A. Chemotherapy exposure increases leukemia cell stiffness. Blood 2007, 109, 3505–3508. [Google Scholar] [CrossRef]
- Paterson, E. Distant metastases from medulloblastoma of the cerebellum. Brain 1961, 84, 301–309. [Google Scholar] [CrossRef]
- Uemura, S.; Ishida, T.; Thwin, K.K.M.; Yamamoto, N.; Tamura, A.; Kishimoto, K.; Hasegawa, D.; Kosaka, Y.; Nino, N.; Lin, K.S.; et al. Dynamics of Minimal Residual Disease in Neuroblastoma Patients. Front. Oncol. 2019, 9, 455. [Google Scholar] [CrossRef] [PubMed]
- Riva, G.; Nasillo, V.; Ottomano, A.M.; Bergonzini, G.; Paolini, A.; Forghieri, F.; Lusenti, B.; Barozzi, P.; Lagreca, I.; Fiorcari, S.; et al. Multiparametric Flow Cytometry for MRD Monitoring in Hematologic Malignancies: Clinical Applications and New Challenges. Cancers 2021, 13, 4582. [Google Scholar] [CrossRef] [PubMed]
- Rosendahl, M.; Greve, T.; Andersen, C.Y. The safety of transplanting cryopreserved ovarian tissue in cancer patients: A review of the literature. J. Assist. Reprod. Genet. 2013, 30, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.; Andersson, M.; Eksborg, S.; Soder, O.; Jahnukainen, K. Xenotransplantation of testicular tissue into nude mice can be used for detecting leukemic cell contamination. Hum. Reprod. 2007, 22, 1899–1906. [Google Scholar] [CrossRef]
- Greve, T.; Clasen-Linde, E.; Andersen, M.T.; Andersen, M.K.; Sørensen, S.D.; Rosendahl, M.; Ralfkiær, E.; Andersen, C.Y. Cryopreserved ovarian cortex from patients with leukemia in complete remission contains no apparent viable malignant cells. Blood 2012, 120, 4311–4316. [Google Scholar] [CrossRef]
- Chevillon, F.; Clappier, E.; Arfeuille, C.; Cayuela, J.-M.; Dalle, J.H.; Kim, R.; Caye-Eude, A.; Chalas, C.; Abdo, C.; Drouineaud, V.; et al. Minimal residual disease quantification in ovarian tissue collected from patients in complete remission of acute leukemia. Blood 2021, 137, 1697–1701. [Google Scholar] [CrossRef]
- Zver, T.; Mouloungui, E.; Berdin, A.; Roux, C.; Amiot, C. Validation of an automated technique for ovarian cortex dissociation: Isolation of viable ovarian cells and their qualification by multicolor flow cytometry. J. Ovarian Res. 2017, 10, 38. [Google Scholar] [CrossRef]
- Tembhare, P.R.; Narula, G.; Khanka, T.; Ghogale, S.; Chatterjee, G.; Patkar, N.V.; Prasad, M.; Badrinath, Y.; Deshpande, N.; Gudapati, P.; et al. Post-induction Measurable Residual Disease Using Multicolor Flow Cytometry Is Strongly Predictive of Inferior Clinical Outcome in the Real-Life Management of Childhood T-Cell Acute Lymphoblastic Leukemia: A Study of 256 Patients. Front. Oncol. 2020, 10, 577. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, Y.; Huang, X.; Zhang, Y.; Qian, J.; Li, J.; Li, C.; Li, X.; Lou, Y.; Zhu, Q.; et al. Minimal residual disease level determined by flow cytometry provides reliable risk stratification in adults with T-cell acute lymphoblastic leukaemia. Br. J. Haematol. 2021, 193, 1096–1104. [Google Scholar] [CrossRef]
- Pfister, S.M.; Reyes-Múgica, M.; Chan, J.K.; Hasle, H.; Lazar, A.J.; Rossi, S.; Ferrari, A.; Jarzembowski, J.A.; Pritchard-Jones, K.; Hill, D.A.; et al. A Summary of the Inaugural WHO Classification of Pediatric Tumors: Transitioning from the Optical into the Molecular Era. Cancer Discov. 2022, 12, 331–355. [Google Scholar] [CrossRef]
- Ren, W.; Gu, G. Prognostic implications of RB1 tumour suppressor gene alterations in the clinical outcome of human osteosarcoma: A meta-analysis. Eur. J. Cancer Care 2017, 26, e12401. [Google Scholar] [CrossRef]
- Dolmans, M.-M.; Donnez, J. Fertility preservation in women for medical and social reasons: Oocytes vs ovarian tissue. Best Pract. Res. Clin. Obstet. Gynaecol. 2021, 70, 63–80. [Google Scholar] [CrossRef] [PubMed]
- Eijkenboom, L.; Saedt, E.; Zietse, C.; Braat, D.; Beerendonk, C.; Peek, R. Strategies to safely use cryopreserved ovarian tissue to restore fertility after cancer: A systematic review. Reprod. Biomed. Online 2022, 45, 763–778. [Google Scholar] [CrossRef]
- Kourta, D.; Kanbar, M.; Amorim, C.A.; Wyns, C. Cancer cell contamination and decontamination methods for ovaries and testes: Special focus on prepubertal gonads with a view to safe fertility restoration. Hum. Reprod. 2023, 38, 780–798. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, R.; Ayala, R.; Martínez-López, J. Minimal Residual Disease Monitoring with Next-Generation Sequencing Methodologies in Hematological Malignancies. Int. J. Mol. Sci. 2019, 20, 2832. [Google Scholar] [CrossRef]
- Gough, S.M.; Slape, C.I.; Aplan, P.D. NUP98 gene fusions and hematopoietic malignancies: Common themes and new biologic insights. Blood 2011, 118, 6247–6257. [Google Scholar] [CrossRef] [PubMed]
- Ruteshouser, E.C.; Robinson, S.M.; Huff, V. Wilms tumor genetics: Mutations inWT1, WTX, andCTNNB1 account for only about one-third of tumors. Genes Chromosom. Cancer 2008, 47, 461–470. [Google Scholar] [CrossRef]
- Hesson, L.B.; Cooper, W.N.; Latif, F. The Role of RASSF1A Methylation in Cancer. Dis. Markers 2007, 23, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Leroy, B.; Ballinger, M.L.; Baran-Marszak, F.; Bond, G.L.; Braithwaite, A.; Concin, N.; Donehower, L.A.; El-Deiry, W.S.; Fenaux, P.; Gaidano, G.; et al. Recommended Guidelines for Validation, Quality Control, and Reporting of TP53 Variants in Clinical Practice. Cancer Res. 2017, 77, 1250–1260. [Google Scholar] [CrossRef]
- Calandrini, C.; Schutgens, F.; Oka, R.; Margaritis, T.; Candelli, T.; Mathijsen, L.; Ammerlaan, C.; van Ineveld, R.L.; Derakhshan, S.; de Haan, S.; et al. An organoid biobank for childhood kidney cancers that captures disease and tissue heterogeneity. Nat. Commun. 2020, 11, 1310. [Google Scholar] [CrossRef]
- South, A.P.; Cho, R.J.; Aster, J.C. The double-edged sword of Notch signaling in cancer. Semin. Cell Dev. Biol. 2012, 23, 458–464. [Google Scholar] [CrossRef] [PubMed]
- Mikhailova, E.; Illarionova, O.; Komkov, A.; Zerkalenkova, E.; Mamedov, I.; Shelikhova, L.; Olshanskaya, Y.; Miakova, N.; Novichkova, G.; Karachunskiy, A.; et al. Reliable Flow-Cytometric Approach for Minimal Residual Disease Monitoring in Patients with B-Cell Precursor Acute Lymphoblastic Leukemia after CD19-Targeted Therapy. Cancers 2022, 14, 5445. [Google Scholar] [CrossRef] [PubMed]
- Dondero, A.; Morini, M.; Cangelosi, D.; Mazzocco, K.; Serra, M.; Spaggiari, G.M.; Rotta, G.; Tondo, A.; Locatelli, F.; Castellano, A.; et al. Multiparametric flow cytometry highlights B7-H3 as a novel diagnostic/therapeutic target in GD2neg/low neuroblastoma variants. J. Immunother. Cancer 2021, 9, e002293. [Google Scholar] [CrossRef] [PubMed]
- Garcia, D.R.N.; Arancibia, A.M.; Ribeiro, R.C.; Land, M.G.P.; Silva, M.L.M. Intrachromosomal amplification of chromosome 21 (iAMP21) detected by ETV6/RUNX1 FISH screening in childhood acute lymphoblastic leukemia: A case report. Rev. Bras. Hematol. Hemoter. 2013, 35, 369–371. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Meyer, C.; Burmeister, T.; Gröger, D.; Tsaur, G.; Fechina, L.; Renneville, A.; Sutton, R.; Venn, N.C.; Emerenciano, M.; Pombo-De-Oliveira, M.S.; et al. The MLL recombinome of acute leukemias in 2017. Leukemia 2018, 32, 273–284. [Google Scholar] [CrossRef]
- Roberts, K.G.; Li, Y.; Payne-Turner, D.; Harvey, R.C.; Yang, Y.-L.; Pei, D.; McCastlain, K.; Ding, L.; Lu, C.; Song, G.; et al. Targetable Kinase-Activating Lesions in Ph-like Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2014, 371, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhu, G.; Wang, N.; Zhou, X.; Yang, Y.; Zhou, S.; Xiong, J.; He, J.; Jiang, L.; Li, C.; et al. SIL-TAL1 Rearrangement is Related with Poor Outcome: A Study from a Chinese Institution. PLoS ONE 2013, 8, e73865. [Google Scholar] [CrossRef]
- Hurteau, G.J.; Broome, J.D.; Brock, G.J. Accurate detection of Asparagine Synthetase (ASNS) using quantitative real-time PCR (qRT-PCR), without requiring DNaseI treatment. Leukemia 2005, 19, 2368–2370. [Google Scholar] [CrossRef][Green Version]
- Junmei, Z.; Fengkuan, Y.; Yongping, S.; Baijun, F.; Yuzhang, L.; Lina, L.; Qinglan, Z. Coexistence of P190 and P210 BCR/ABL transcripts in chronic myeloid leukemia blast crisis resistant to imatinib. Springerplus 2015, 4, 170. [Google Scholar] [CrossRef][Green Version]
- Chatterjee, T.; Gupta, S.; Sharma, A.; Sharma, S.; Ganguli, P. Distribution of Different PML/RARα bcr Isoforms in Indian Acute Promyelocytic Leukemia (APL) Patients and Clinicohema-tological Correlation. Mediterr. J. Hematol. Infect. Dis. 2014, 6, e2014004. [Google Scholar] [CrossRef]
- Talami, A.; Bettelli, F.; Pioli, V.; Giusti, D.; Gilioli, A.; Colasante, C.; Galassi, L.; Giubbolini, R.; Catellani, H.; Donatelli, F.; et al. How to Improve Prognostication in Acute Myeloid Leukemia with CBFB-MYH11 Fusion Transcript: Focus on the Role of Molecular Measurable Residual Disease (MRD) Monitoring. Biomedicines 2021, 9, 953. [Google Scholar] [CrossRef]
- Cho, E.K.; Bang, S.M.; Ahn, J.Y.; Yoo, S.M.; Park, P.W.; Seo, Y.H.; Shin, D.B.; Lee, J.H. Prognostic Value of AML1/ETO Fusion Transcripts in Patients with Acute Myelogenous Leukemia. Korean J. Intern. Med. 2003, 18, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Wilhelmsson, M.; Glosli, H.; Ifversen, M.; Abrahamsson, J.; Winiarski, J.; Jahnukainen, K.; Hasle, H.; Nordic Society of Pediatric Hematology and Oncology (NOPHO). Long-term health outcomes in survivors of childhood AML treated with allogeneic HSCT: A NOPHO–AML Study. Bone Marrow Transplant. 2019, 54, 726–736. [Google Scholar] [CrossRef] [PubMed]
- Noort, S.; Zimmermann, M.; Reinhardt, D.; Cuccuini, W.; Pigazzi, M.; Smith, J.; Ries, R.E.; Alonzo, T.A.; Hirsch, B.; Tomizawa, D.; et al. Prognostic impact of t(16;21)(p11;q22) and t(16;21)(q24;q22) in pediatric AML: A retrospective study by the I-BFM Study Group. Blood 2018, 132, 1584–1592. [Google Scholar] [CrossRef]
- Kim, H.J.; Baek, S.K.; Maeng, C.H.; Kim, S.-Y.; Park, T.S.; Han, J.J. Acute myeloid leukemia with t(11;19)(q23;p13.1) in a patient with a gastrointestinal stromal tumor undergoing imatinib therapy: A case report. World J. Clin. Cases 2020, 8, 1251–1256. [Google Scholar] [CrossRef]
- Dastugue, N.; Lafage-Pochitaloff, M.; Pagès, M.-P.; Radford, I.; Bastard, C.; Talmant, P.; Mozziconacci, M.J.; Léonard, C.; Bilhou-Nabéra, C.; Cabrol, C.; et al. Cytogenetic profile of childhood and adult megakaryoblastic leukemia (M7): A study of the Groupe Francais de Cytogenetique Hematologique (GFCH). Blood 2002, 100, 618–626. [Google Scholar] [CrossRef]
- Smith, M.L.; Cavenagh, J.D.; Lister, T.A.; Fitzgibbon, J. Mutation of CEBPA in Familial Acute Myeloid Leukemia. N. Engl. J. Med. 2004, 351, 2403–2407. [Google Scholar] [CrossRef]
- Noort, S.; van Oosterwijk, J.; Ma, J.; Garfinkle, E.A.; Nance, S.; Walsh, M.; Song, G.; Reinhardt, D.; Pigazzi, M.; Locatelli, F.; et al. Analysis of rare driving events in pediatric acute myeloid leukemia. Haematologica 2023, 108, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Hitzler, J.K.; Cheung, J.; Li, Y.; Scherer, S.W.; Zipursky, A. GATA1 mutations in transient leukemia and acute megakaryoblastic leukemia of Down syndrome. Blood 2003, 101, 4301–4304. [Google Scholar] [CrossRef] [PubMed]
- Goemans, B.F.; Noort, S.; Blink, M.; Wang, Y.-D.; Peters, S.T.C.J.; van Wouwe, J.P.; Kaspers, G.; de Haas, V.; Kollen, W.J.; van der Velden, V.H.J.; et al. Sensitive GATA1 mutation screening reliably identifies neonates with Down syndrome at risk for myeloid leukemia. Leukemia 2021, 35, 2403–2406. [Google Scholar] [CrossRef]
- Damm-Welk, C.; Klapper, W.; Oschlies, I.; Gesk, S.; Röttgers, S.; Bradtke, J.; Siebert, R.; Reiter, A.; Woessmann, W. Distribution of NPM1-ALK and X-ALK fusion transcripts in paediatric anaplastic large cell lymphoma: A molecular-histological correlation. Br. J. Haematol. 2009, 146, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Chuang, S.-S.; Lin, C.-N.; Shen, F.-C.; Liao, P.-J.; Liao, Y.-L.; Chang, J.H.; Tsai, Y.-C.; Cho, C.-Y.; Huang, W. Detecting Clonal Rearrangement in Non-Hodgkin’s Lymphomas in Taiwan by Polymerase Chain Reaction. Leuk. Lymphoma 2003, 44, 117–121. [Google Scholar] [CrossRef]
- Green, M.R.; Monti, S.; Rodig, S.J.; Juszczynski, P.; Currie, T.; O’Donnell, E.; Chapuy, B.; Takeyama, K.; Neuberg, D.; Golub, T.R.; et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 2010, 116, 3268–3277. [Google Scholar] [CrossRef] [PubMed]
- Almire, C.; Bertrand, P.; Ruminy, P.; Maingonnat, C.; Wlodarska, I.; Martín-Subero, J.I.; Siebert, R.; Tilly, H.; Bastard, C. PVRL2 is translocated to theTRA@ locus in t(14;19)(q11;q13)-positive peripheral T-cell lymphomas. Genes Chromosom. Cancer 2007, 46, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Wegert, J.; Ishaque, N.; Vardapour, R.; Geörg, C.; Gu, Z.; Bieg, M.; Ziegler, B.; Bausenwein, S.; Nourkami, N.; Ludwig, N.; et al. Mutations in the SIX1/2 Pathway and the DROSHA/DGCR8 miRNA Microprocessor Complex Underlie High-Risk Blastemal Type Wilms Tumors. Cancer Cell 2015, 27, 298–311. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.D.; Al-Saadi, R.; Chagtai, T.; Popov, S.; Messahel, B.; Sebire, N.; Gessler, M.; Wegert, J.; Graf, N.; Leuschner, I.; et al. Subtype-Specific FBXW7 Mutation and MYCN Copy Number Gain in Wilms’ Tumor. Clin. Cancer Res. 2010, 16, 2036–2045. [Google Scholar] [CrossRef]
- Hol, J.A.; Kuiper, R.P.; van Dijk, F.; Waanders, E.; van Peer, S.E.; Koudijs, M.J.; Bladergroen, R.; van Reijmersdal, S.V.; Morgado, L.M.; Bliek, J.; et al. Prevalence of (Epi)genetic Predisposing Factors in a 5-Year Unselected National Wilms Tumor Cohort: A Comprehensive Clinical and Genomic Characterization. J. Clin. Oncol. 2022, 40, 1892–1902. [Google Scholar] [CrossRef]
- Ooms, A.H.; Vujanić, G.M.; D’hooghe, E.; Collini, P.; L’herminé-Coulomb, A.; Vokuhl, C.; Graf, N.; Heuvel-Eibrink, M.M.v.D.; de Krijger, R.R. Renal Tumors of Childhood—A Histopathologic Pattern-Based Diagnostic Approach. Cancers 2020, 12, 729. [Google Scholar] [CrossRef]
- Roy, A.; Kumar, V.; Zorman, B.; Fang, E.; Haines, K.M.; Doddapaneni, H.; Hampton, O.A.; White, S.; Bavle, A.A.; Patel, N.R.; et al. Recurrent internal tandem duplications of BCOR in clear cell sarcoma of the kidney. Nat. Commun. 2015, 6, 8891. [Google Scholar] [CrossRef]
- Gooskens, S.L.; Gadd, S.; Auvil, J.M.G.; Gerhard, D.S.; Khan, J.; Patidar, R.; Meerzaman, D.; Chen, Q.-R.; Hsu, C.H.; Yan, C.; et al. TCF21 hypermethylation in genetically quiescent clear cell sarcoma of the kidney. Oncotarget 2015, 6, 15828–15841. [Google Scholar] [CrossRef]
- Xin, J.; Xu, R.; Lin, S.; Xin, M.; Cai, W.; Zhou, J.; Fu, C.; Zhen, G.; Lai, J.; Li, Y.; et al. Clinical potential of TCF21 methylation in the diagnosis of renal cell carcinoma. Oncol. Lett. 2016, 12, 1265–1270. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, W.; Hu, H.; Zhang, Y.; Wang, Y.; Gu, H.; Huang, D. Case Analysis of 14 Children with Malignant Rhabdoid Tumor of the Kidney. Cancer Manag. Res. 2021, 13, 4865–4872. [Google Scholar] [CrossRef] [PubMed]
- Zhanghuang, C.; Chen, S.; Li, L.; Yang, Z.; Xie, Y.; Li, J.; Tang, H.; He, X.; Dong, L.; Yan, B. Clinical and Molecular Differentiation Between Malignant Rhabdoid Tumor of the Kidney and Normal Tissue: A Two-Case Report. Front. Oncol. 2021, 11, 659709. [Google Scholar] [CrossRef]
- Argani, P.; Zhong, M.; Reuter, V.E.; Fallon, J.T.; Epstein, J.I.; Netto, G.J.; Antonescu, C.R. TFE3-Fusion Variant Analysis Defines Specific Clinicopathologic Associations Among Xp11 Translocation Cancers. Am. J. Surg. Pathol. 2016, 40, 723–737. [Google Scholar] [CrossRef]
- Cajaiba, M.M.; Dyer, L.M.; Geller, J.I.; Jennings, L.J.; George, D.; Kirschmann, D.; Rohan, S.M.; Cost, N.G.; Khanna, G.; Mullen, E.A.; et al. The classification of pediatric and young adult renal cell carcinomas registered on the children’s oncology group (COG) protocol AREN03B2 after focused genetic testing. Cancer 2018, 124, 3381–3389. [Google Scholar] [CrossRef]
- Cajaiba, M.M.; Jennings, L.J.; Rohan, S.M.; Perez-Atayde, A.R.; Marino-Enriquez, A.; Fletcher, J.A.; Geller, J.I.; Leuer, K.M.C.; Bridge, J.A.; Perlman, E.J. ALK-rearranged renal cell carcinomas in children. Genes Chromosom. Cancer 2016, 55, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Somasundaram, D.B.; Aravindan, S.; Yu, Z.; Jayaraman, M.; Tran, N.T.B.; Li, S.; Herman, T.S.; Aravindan, N. Droplet digital PCR as an alternative to FISH for MYCN amplification detection in human neuroblastoma FFPE samples. BMC Cancer 2019, 19, 106. [Google Scholar] [CrossRef]
- Hung, Y.P.; Lee, J.P.; Bellizzi, A.M.; Hornick, J.L. PHOX2B reliably distinguishes neuroblastoma among small round blue cell tumours. Histopathology 2017, 71, 786–794. [Google Scholar] [CrossRef]
- Lee, N.H.; Son, M.H.; Choi, Y.B.; Yi, E.; Lee, J.W.; Yoo, K.H.; Sung, K.W.; Koo, H.H. Clinical Significance of Tyrosine Hydroxylase mRNA Transcripts in Peripheral Blood at Diagnosis in Patients with Neuroblastoma. Cancer Res. Treat. 2016, 48, 1399–1407. [Google Scholar] [CrossRef]
- van Zogchel, L.M.J.; Zappeij-Kannegieter, L.; Javadi, A.; Lugtigheid, M.; Gelineau, N.U.; Lak, N.S.M.; Zwijnenburg, D.A.; Koster, J.; Stutterheim, J.; van der Schoot, C.E.; et al. Specific and Sensitive Detection of Neuroblastoma mRNA Markers by Multiplex RT-qPCR. Cancers 2021, 13, 150. [Google Scholar] [CrossRef]
- Inomistova, M.V.; Svergun, N.M.; Khranovska, N.M.; Skachkova, O.V.; Gorbach, O.I.; Klymnyuk, G.I. Prognostic significance of MDM2 gene expression in childhood neuroblastoma. Exp. Oncol. 2015, 37, 111–115. [Google Scholar] [CrossRef]
- Cheung, N.-K.V.; Zhang, J.; Lu, C.; Parker, M.; Bahrami, A.; Tickoo, S.K.; Heguy, A.; Pappo, A.S.; Federico, S.; Dalton, J.; et al. Association of Age at Diagnosis and Genetic Mutations in Patients with Neuroblastoma. JAMA 2012, 307, 1062–1071. [Google Scholar] [CrossRef]
- Mossé, Y.P.; Laudenslager, M.; Longo, L.; Cole, K.A.; Wood, A.; Attiyeh, E.F.; Laquaglia, M.J.; Sennett, R.; Lynch, J.E.; Perri, P.; et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 2008, 455, 930–935. [Google Scholar] [CrossRef]
- Kenny, C.; Bausenwein, S.; Lazaro, A.; Furtwängler, R.; Gooskens, S.L.; van den Heuvel Eibrink, M.; Vokuhl, C.; Leuschner, I.; Graf, N.; Gessler, M.; et al. Mutually exclusive BCOR internal tandem duplications and YWHAE-NUTM2 fusions in clear cell sarcoma of kidney: Not the full story. J. Pathol. 2016, 238, 617–620. [Google Scholar] [CrossRef]
- Lak, N.S.; Voormanns, T.L.; Zappeij-Kannegieter, L.; van Zogchel, L.M.; Fiocco, M.; van Noesel, M.M.; Merks, J.H.; van der Schoot, C.E.; Tytgat, G.A.; Stutterheim, J. Improving Risk Stratification for Pediatric Patients with Rhabdomyosarcoma by Molecular Detection of Disseminated Disease. Clin. Cancer Res. 2021, 27, 5576–5585. [Google Scholar] [CrossRef]
- Butel, T.; Karanian, M.; Pierron, G.; Orbach, D.; Ranchere, D.; Cozic, N.; Galmiche, L.; Coulomb, A.; Corradini, N.; Lacour, B.; et al. Integrative clinical and biopathology analyses to understand the clinical heterogeneity of infantile rhabdomyosarcoma: A report from the French MMT committee. Cancer Med. 2020, 9, 2698–2709. [Google Scholar] [CrossRef]
- Arden, K.C.; Anderson, M.J.; Finckenstein, F.G.; Czekay, S.; Cavenee, W.K. Detection of the t(2;13) chromosomal translocation in alveolar rhabdomyosarcoma using the reverse transcriptase-polymerase chain reaction. Genes Chromosom. Cancer 1996, 16, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Van Paemel, R.; De Koker, A.; Vandeputte, C.; van Zogchel, L.; Lammens, T.; Laureys, G.; Vandesompele, J.; Schleiermacher, G.; Chicard, M.; Van Roy, N.; et al. Minimally invasive classification of paediatric solid tumours using reduced representation bisulphite sequencing of cell-free DNA: A proof-of-principle study. Epigenetics 2021, 16, 196–208. [Google Scholar] [CrossRef]
- Orr, B.A. Pathology, diagnostics, and classification of medulloblastoma. Brain Pathol. 2020, 30, 664–678. [Google Scholar] [CrossRef] [PubMed]
- Timmerman, D.M.; Eleveld, T.F.; Sriram, S.; Dorssers, L.C.; Gillis, A.J.; Schmidtova, S.; Kalavska, K.; van de Werken, H.J.; Oing, C.; Honecker, F.; et al. Chromosome 3p25.3 Gain Is Associated with Cisplatin Resistance and Is an Independent Predictor of Poor Outcome in Male Malignant Germ Cell Tumors. J. Clin. Oncol. 2022, 40, 3077–3087. [Google Scholar] [CrossRef] [PubMed]
| Incidence Rate | Types of Cancer in Adults | Types of Cancer in Children |
|---|---|---|
| High risk >11% | Leukemia, Burkitt’s lymphoma, neuroblastoma | Ewing sarcoma, acute lymphoblastic leukemia, acute myeloid leukemia, chronic myeloid leukemia |
| Moderate risk 0.2–11% | Cervical adenocarcinoma, breast cancer (infiltrating lobular subtype, stage IV), Ewing’s sarcoma, non-Hodgkin lymphoma, colon cancer | |
| Low risk <0.2% | Remaining malignancies | Osteosarcoma, rhabdomyosarcoma, synovial sarcoma, clear cell sarcoma, Burkitt’s lymphoma, juvenile myelomonocytic leukemia, central nervous system tumors |
| Malignancy | Proposed Markers | Potential Techniques | References |
|---|---|---|---|
| Overall | ABL class abnormalities | RT-qPCR | [25,27] |
| NUP98 gene fusion transcripts | RT-PCR | [56] | |
| WT1 mutations | PCR, qPCR | [57] | |
| RASSF1A hypermethylation | Quantitative MSP | [58] | |
| P53 mutations, transcript levels | NGS, RNA sequencing | [59,60] | |
| Notch aberrations | Sequencing | [61] | |
| CD19, CD20, CD22 | MFC, NGS, RT-qPCR | [62] | |
| GD2, B7H3 | MFC | [63] | |
| Acute lymphoblastic leukemia | Philadelphia chromosome-positive (Ph+; BCR-ABL1 fusion gene) | PCR, RT-PCR | [25,36] |
| (TCF3) E2A fusion genes | RT-qPCR;RT-ddPCR | [16,38] | |
| CD34, CD10, CD20, CD79a, CD3, TdT | IHC | [37] | |
| CD19, CD34, CD10-, negative for myeloid markers, CD45, HLA-DR2, CD10, CD19, CD22, CD33, CD38, CD2, cyCD3, CD5, CD7, CD33, CD45RA, CD123 | MFC | [26] | |
| Ig(H/K)/TCR targets | RT-PCR, RT-ddPCR | [16,25,27,38] | |
| RUNX1 (AML1)translocations, fusion genes | RT-PCR, FISH, RT-qPCR, PCR | [25,37,38,64] | |
| MLL (KMT2A) rearrangements, patient-specific on DNA | RT-qPCR, long-distance inverse PCR | [38,65] | |
| Intrachromosomal amplification of chromosome 21 (iAMP21) | FISH | [64] | |
| IKZF1 mutations | Sequencing | [66] | |
| SIL-TAL1 fusion gene | PCR | [67] | |
| Asparagine synthetase | RT-qPCR | [68] | |
| Chronic myeloid leukemia | BCR-ABL fusion gene | FISH, RT-PCR, PCR | [27,37,69] |
| MPO, glycophorin A, CD34, CD68, CD117, LCA/CD45, Factor VIII | IHC | [35,37] | |
| Acute promyelocytic leukemia | PML-RARα brc isoforms | RT-qPCR, FISH | [70] |
| Acute myeloid leukemia | RUNX1 (AML1) fusion genes | RT-PCR, RT-qPCR | [38,71,72] |
| NPM1 mutations | RT-qPCR | [38] | |
| FLT3fusion transcripts | RT-PCR | [25] | |
| BCR-ABL1 fusion genes | RT-PCR | [25] | |
| CD34, CD68, CD117, MPO, CD13, CD4 | IHC | [37] | |
| CD34, CD33, CD13, CD117, CD38, CD65, CD7, HLA-DR2, CD11c | MFC | [26] | |
| CBFB-MYH11 fusion genes | PCR, RT-qPCR | [37,71] | |
| NUP98 rearrangements | RT-qPCR, NGS | [56,73] | |
| FUS-ERG fusion gene | FISH, PCR, RNA sequencing | [74] | |
| KMT2A rearrangements | RT-qPCR, FISH, RT-PCR; long-distance inverse PCR | [65,75] | |
| RBM15-MKL1 (OTT-MAL) fusion gene/transcript | RT-PCR | [76] | |
| CEBPA mutations | PCR, sequencing | [77] | |
| UBTF mutations | NGS | [78] | |
| Acute myeloid leukemia–Down syndrome | GATA1 mutations, patient-specific on DNA | Sequencing | [79,80] |
| Anaplastic large cell lymphoma | ALK fusion genes, NPM1 mutations | RT-PCR, FISH, immunofluorescence | [78,81] |
| Juvenile myelomonocytic leukemia | CD3, CD4, CD68 | IHC | [37] |
| Lymphoma (Hodgkin and non-Hodgkin) | Immunoglobulin (Ig) or T-cell receptor (TCR) gene rearrangements | RT-qPCR | [38] |
| Multiplex PCR | [82] | ||
| 9p24 amplification/JAK2 | DNA copy number analysis, RT-qPCR | [83] | |
| 14q11/TRA/D | FISH | [84] | |
| Wilms tumor | WT1, WTX mutations | PCR, qPCR | [57] |
| CTNNB1 mutations | qPCR | [57] | |
| DROSHA/DGCR8 mutations | Sequencing (DNA, RNA) | [85] | |
| SIX1/SIX2 mutations | Sequencing (DNA, RNA) | [85] | |
| DICER1 mutations | Sequencing (DNA, RNA) | [85] | |
| DIS3L2 mutations | Sequencing (DNA, RNA) | [85] | |
| FBXW7 mutations | Chromosome copy number profiling, sequencing | [86] | |
| DIS3L2 mutations | PCR | [87] | |
| TP53 mutations | NGS | [88] | |
| Clear cell sarcoma of the kidney | BCOR internal tandem duplications | RNA sequencing, PCR, IHC, FISH | [88,89] |
| YWHAE-NUTM2 fusion transcript | RT-PCR | [90] | |
| TCF21 hypermethylation | Methylation-based methods, quantitative pyrosequencing methylation analysis | [90,91] | |
| Malignant rhabdoid tumor of the kidney | SMARCA4; SMARCB1 | IHC, WGS | [92,93] |
| Renal cell carcinoma | TFE3 (Xp11) translocations/fusion transcripts | FISH, WGS/NGS, IHC, RNA sequencing | [60,88,94,95] |
| TFEB translocations/fusion transcripts | RNA sequencing, FISH, IHC | [60,88,95] | |
| ALK rearrangements | IHC, FISH, RT-PCR, NGS | [96] | |
| Neuroblastoma | MYCN amplification/mutations | FISH, ddPCR, NGS | [88,97] |
| PHOX2B | IHC | [98] | |
| TH | RT-qPCR, multiplex RT-qPCR | [99] | |
| CHRNA3, DBH, GAP43, POSTN, PRRX1 and FMO3 | multiplex RT-qPCR | [100] | |
| MDM2 | RT-qPCR | [101] | |
| ATRX mutations | WGS, IHC | [102] | |
| ALK mutations | Sequencing, qPCR | [103] | |
| Synovial sarcoma | Bcl-2, SYT-SSX fusion gene | IHC, RT-qPCR | [34] |
| Clear cell sarcoma | Melanoma cocktail/S100 | IHC | [34] |
| EWS-ATF1 fusion gene | RT-qPCR | [34] | |
| BROC mutations | RT-qPCR | [104] | |
| YWHAE-NUTM2 fusion transcript | RT-qPCR | [104] | |
| Ewing Sarcoma | EWS fusion transcripts | RT-qPCR | [15,24,30,31,32,33,34] |
| t(X;22) EWS translocations | FISH | [34,88] | |
| CD99, INI1 | IHC | [34,88] | |
| Osteosarcoma | RB1 deletion | IHC, PCR, RT-PCR | [51] |
| Rhabdomyosarcoma | PAX3/7-FOXO1 fusion gene | RT-qPCR | [34,105] |
| MYOD1 | IHC, RT-qPCR | [34,105] | |
| MYOGENIN | IHC, RT-qPCR | [34,105] | |
| VGLL2 fusion transcripts | RT-PCR, RNA sequencing | [106] | |
| NTRK fusion transcripts | RT-PCR, RNA sequencing | [106] | |
| (B)RAF fusion transcripts | RT-PCR, RNA sequencing | [106] | |
| t(2;13) translocation alveolar | RT-PCR | [107] | |
| cfRRBS | shWGS | [108] | |
| Medulloblastoma | GFAP/NSE | IHC | [29] |
| Alterations in WNT and SHH pathways’ components | FISH, sequencing, methylation-based methods and combinations of them | [109] | |
| Astrocytoma | CD99/NSE GFAP | IHC, RT-ddPCR | [29] |
| Ependymoma | GFAP/NSE GFAP | IHC, RT-ddPCR | [29] |
| Germinoma | GFAP/NSE GFAP | IHC, RT-ddPCR | [29] |
| Glioblastoma | GFAP/NSE GFAP | IHC, RT-ddPCR | [29] |
| Primitive neuroectodermal tumor | GFAP/NSE GFAP | IHC, RT-ddPCR | [29] |
| Germ cell tumors | Chromosome 3p gain, miRNA | NGS, RT-qPCR | [110] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grubliauskaite, M.; van der Perk, M.E.M.; Bos, A.M.E.; Meijer, A.J.M.; Gudleviciene, Z.; van den Heuvel-Eibrink, M.M.; Rascon, J. Minimal Infiltrative Disease Identification in Cryopreserved Ovarian Tissue of Girls with Cancer for Future Use: A Systematic Review. Cancers 2023, 15, 4199. https://doi.org/10.3390/cancers15174199
Grubliauskaite M, van der Perk MEM, Bos AME, Meijer AJM, Gudleviciene Z, van den Heuvel-Eibrink MM, Rascon J. Minimal Infiltrative Disease Identification in Cryopreserved Ovarian Tissue of Girls with Cancer for Future Use: A Systematic Review. Cancers. 2023; 15(17):4199. https://doi.org/10.3390/cancers15174199
Chicago/Turabian StyleGrubliauskaite, Monika, M. E. Madeleine van der Perk, Annelies M. E. Bos, Annelot J. M. Meijer, Zivile Gudleviciene, Marry M. van den Heuvel-Eibrink, and Jelena Rascon. 2023. "Minimal Infiltrative Disease Identification in Cryopreserved Ovarian Tissue of Girls with Cancer for Future Use: A Systematic Review" Cancers 15, no. 17: 4199. https://doi.org/10.3390/cancers15174199
APA StyleGrubliauskaite, M., van der Perk, M. E. M., Bos, A. M. E., Meijer, A. J. M., Gudleviciene, Z., van den Heuvel-Eibrink, M. M., & Rascon, J. (2023). Minimal Infiltrative Disease Identification in Cryopreserved Ovarian Tissue of Girls with Cancer for Future Use: A Systematic Review. Cancers, 15(17), 4199. https://doi.org/10.3390/cancers15174199







