MicroRNA-1289 Functions as a Novel Tumor Suppressor in Oral Squamous Cell Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Cell Culture
2.2. Samples
2.3. Transfection with Human Mature miR Mimics and Small Interfering RNAs (siRNAs)
2.4. Cell Growth Assay
2.5. Total RNA Extraction
2.6. Microarray and Prediction of miR-1289 Target Genes
2.7. RT-qPCR
2.8. Luciferase Reporter Assay
2.9. Xenograft Model
2.10. Statistical Analysis
3. Results
3.1. Identification of TS-miR Candidates in Human OSCC Cells
3.2. Target Genes of miR-1289 in Human OSCC Cells
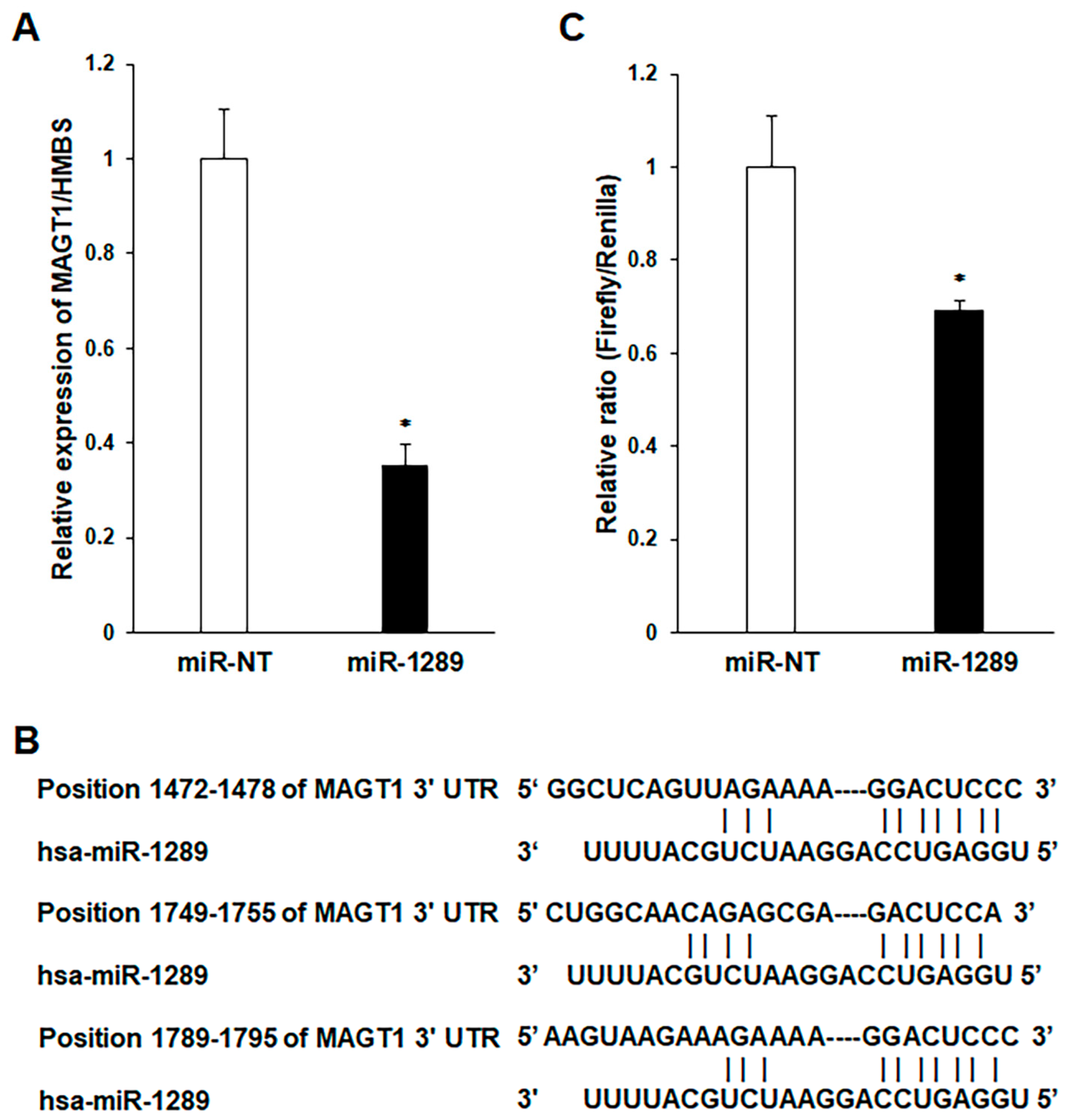
3.3. MAGT1 as a Direct Target Gene of miR-1289 in Human OSCC Cells
3.4. Effect of miR-1289 Mimics on the In Vivo Growth of Human OSCC Cells
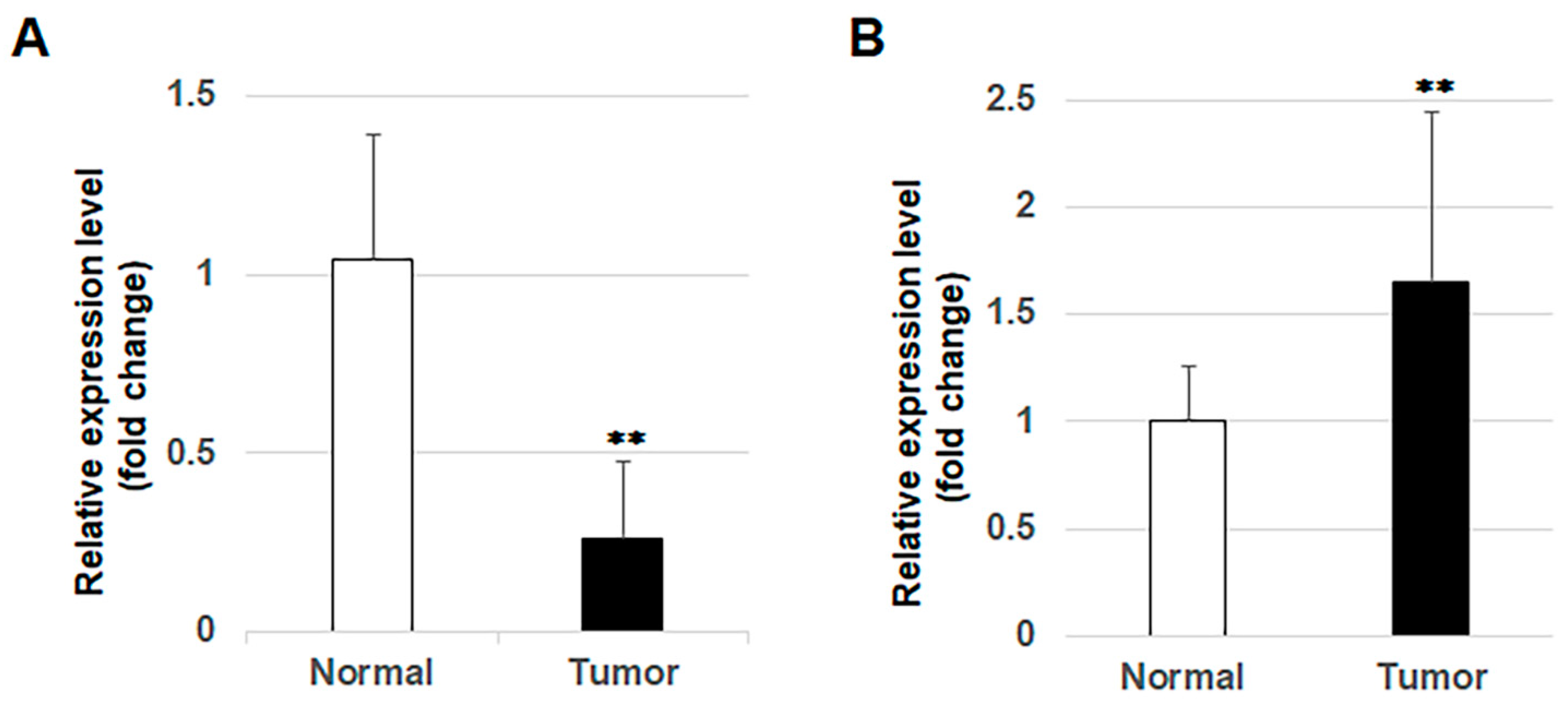
3.5. Expression of miR-1289 in OSCC Tissues
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Garzino-Demo, P.; Zavattero, E.; Franco, P.; Fasolis, M.; Tanteri, G.; Mettus, A.; Tosco, P.; Chiusa, L.; Airoldi, M.; Ostellino, O.; et al. Parameters and outcomes in 525 patients operated on for oral squamous cell carcinoma. J. Craniomaxillofac. Surg. 2016, 44, 1414–1421. [Google Scholar] [CrossRef]
- Agrawal, N.; Frederick, M.J.; Pickering, C.R.; Bettegowda, C.; Chang, K.; Li, R.J.; Fakhry, C.; Xie, T.X.; Zhang, J.; Wang, J.; et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011, 333, 1154–1157. [Google Scholar] [CrossRef] [PubMed]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef] [PubMed]
- Nakashiro, K.; Tanaka, H.; Goda, H.; Iwamoto, K.; Tokuzen, N.; Hara, S.; Onodera, J.; Fujimoto, I.; Hino, S.; Hamakawa, H. Identification of Akt1 as a potent therapeutic target for oral squamous cell carcinoma. Int. J. Oncol. 2015, 47, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Nakashiro, K.; Iwamoto, K.; Tokuzen, N.; Fujita, Y.; Shirakawa, R.; Oka, R.; Goda, H.; Hamakawa, H. Targeting Aurora kinase A suppresses the growth of human oral squamous cell carcinoma cells in vitro and in vivo. Oral Oncol. 2013, 49, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, K.; Nakashiro, K.; Tanaka, H.; Tokuzen, N.; Hamakawa, H. Ribonucleotide reductase M2 is a promising molecular target for the treatment of oral squamous cell carcinoma. Int. J. Oncol. 2015, 46, 1971–1977. [Google Scholar] [CrossRef]
- Tokuzen, N.; Nakashiro, K.; Tanaka, H.; Iwamoto, K.; Hamakawa, H. Therapeutic potential of targeting cell division cycle associated 5 for oral squamous cell carcinoma. Oncotarget 2016, 7, 2343–2353. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Garzon, R.; Calin, G.A.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Med. 2009, 60, 167–179. [Google Scholar] [CrossRef]
- Min, A.; Zhu, C.; Peng, S.; Rajthala, S.; Costea, D.E.; Sapkota, D. MicroRNAs as important players and biomarkers in oral carcinogenesis. Biomed. Res. Int. 2015, 2015, 186904. [Google Scholar] [CrossRef] [PubMed]
- Reis, P.P.; Tomenson, M.; Cervigne, N.K.; Machado, J.; Jurisica, I.; Pintilie, M.; Sukhai, M.A.; Perez-Ordonez, B.; Grénman, R.; Gilbert, R.W.; et al. Programmed cell death 4 loss increases tumor cell invasion and is regulated by miR-21 in oral squamous cell carcinoma. Mol. Cancer 2010, 9, 238. [Google Scholar] [CrossRef]
- Jung, H.M.; Phillips, B.L.; Patel, R.S.; Cohen, D.M.; Jakymiw, A.; Kong, W.W.; Cheng, J.Q.; Chan, E.K. Keratinization-associated miR-7 and miR-21 regulate tumor suppressor reversion-inducing cysteine-rich protein with Kazal motifs (RECK) in oral cancer. J. Biol. Chem. 2012, 287, 29261–29272. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, H.; Nakashiro, K.; Tokuzen, N.; Kuribayashi, N.; Goda, H.; Uchida, D. MicroRNA-361-3p is a potent therapeutic target for oral squamous cell carcinoma. Cancer Sci. 2020, 111, 1645–1651. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Gao, J.; Sun, Q.W.; Wang, C.X.; Deng, W.; Mao, G.Y.; Li, H.Q.; Guo, S.S.; Cheng, J.; Wu, Y.N.; et al. MiR-34a inhibits the proliferation, migration, and invasion of oral squamous cell carcinoma by directly targeting SATB2. J. Cell. Physiol. 2020, 235, 4856–4864. [Google Scholar] [CrossRef]
- Shao, Y.; Qu, Y.; Dang, S.; Yao, B.; Ji, M. MiR-145 inhibits oral squamous cell carcinoma (OSCC) cell growth by targeting c-Myc and Cdk6. Cancer Cell Int. 2013, 13, 51. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Yao, L.; Xiao, J.; Liu, D.; Ni, Z. MiR-206 functions as a tumor suppressor and directly targets K-Ras in human oral squamous cell carcinoma. Onco Targets Ther. 2014, 7, 1583–1591. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, S.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- Shintani, S.; Mihara, M.; Nakahara, Y.; Aida, T.; Tachikawa, T.; Hamakawa, H. Lymph node metastasis of oral cancer visualized in live tissue by green fluorescent protein expression. Oral Oncol. 2002, 38, 664–669. [Google Scholar] [CrossRef]
- Shintani, S.; Hamakawa, H.; Nakashiro, K.; Shirota, T.; Hatori, M.; Tanaka, M.; Kuroshita, Y.; Kurokawa, Y. Friend leukaemia insertion (Fli)-1 is a prediction marker candidate for radiotherapy resistant oral squamous cell carcinoma. Int. J. Oral Maxillofac. Surg. 2010, 39, 1115–1119. [Google Scholar] [CrossRef] [PubMed]
- Moriya, Y.; Nohata, N.; Kinoshita, T.; Mutallip, M.; Okamoto, T.; Yoshida, S.; Suzuki, M.; Yoshino, I.; Seki, N. Tumor suppressive microRNA-133a regulates novel molecular networks in lung squamous cell carcinoma. J. Hum. Genet. 2012, 57, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.M.; Liang, G.; Liu, C.-C.; Wolff, E.M.; Tsai, Y.C.; Ye, W.; Zhou, X.; Jones, P.A. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009, 69, 2623–2629. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, Y.; Nakada, C.; Noguchi, T.; Tanigawa, M.; Nguyen, L.T.; Uchida, T.; Hijiya, N.; Matsuura, K.; Fujioka, T.; Seto, M.; et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010, 70, 2339–2349. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guessous, F.; Zhang, Y.; Dipierro, C.; Kefas, B.; Johnson, E.; Marcinkiewicz, L.; Jiang, J.; Yang, Y.; Schmittgen, T.D.; et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009, 69, 7569–7576. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef]
- Kinoshita, T.; Hanazawa, T.; Nohata, N.; Kikkawa, N.; Enokida, H.; Yoshino, H.; Yamasaki, T.; Hidaka, H.; Nakagawa, M.; Okamoto, Y.; et al. Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion through targeting laminin-332 in head and neck squamous cell carcinoma. Oncotarget 2012, 3, 1386–1400. [Google Scholar] [CrossRef]
- Bolukbasi, M.F.; Mizrak, A.; Ozdener, G.B.; Madlener, S.; Ströbel, T.; Erkan, E.P.; Fan, J.B.; Breakefield, X.O.; Saydam, O. miR-1289 and “Zipcode”-like Sequence Enrich mRNAs in Microvesicles. Mol. Ther. Nucleic Acids 2012, 1, e10. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Noto, J.M.; Hammond, C.E.; Barth, J.L.; Argraves, W.S.; Backert, S.; Peek, R.M., Jr.; Smolka, A.J. Helicobacter pylori-induced posttranscriptional regulation of H-K-ATPase α-subunit gene expression by miRNA. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G606–G613. [Google Scholar] [CrossRef]
- Zhou, H.; Clapham, D.E. Mammalian MagT1 and TUSC3 are required for cellular magnesium uptake and vertebrate embryonic development. Proc. Natl. Acad. Sci. USA 2009, 106, 15750–15755. [Google Scholar] [CrossRef]
- Li, L.; Zhang, X.; Li, Y.; Xiao, B.; Pei, S.; Jiang, H.; Zhang, X. Transcription factor KLF16 activates MAGT1 to regulate the tumorigenesis and progression of breast cancer. Int. J. Mol. Med. 2022, 50, 115. [Google Scholar] [CrossRef]
- Bi, C.; Zhang, X.; Chen, Y.; Dong, Y.; Shi, Y.; Lei, Y.; Lv, D.; Cao, X.; Li, W.; Shi, H. MAGT1 is required for HeLa cell proliferation through regulating p21 expression, S-phase progress, and ERK/p38 MAPK MYC axis. Cell Cycle 2021, 20, 2233–2247. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, H.; Wei, D. Oncogenic magnesium transporter 1 upregulates programmed death-1-ligand 1 expression and contributes to growth and radioresistance of glioma cells through the ERK/MAPK signaling pathway. Bioengineered 2022, 13, 9575–9587. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, X.; Zhu, X.; Zhang, M.; Wang, Y.; Ma, M.; Lv, K. GINS1 promotes the proliferation and migration of glioma cells through USP15-mediated deubiquitination of TOP2A. iScience 2022, 25, 104952. [Google Scholar] [CrossRef]
- Hulin, J.A.; Tommasi, S.; Elliot, D.; Mangoni, A.A. Small molecule inhibition of DDAH1 significantly attenuates triple negative breast cancer cell vasculogenic mimicry in vitro. Biomed. Pharmacother. 2019, 111, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Song, D.; Yan, Y.; Huang, C.; Shen, C.; Lan, J.; Chen, Y.; Liu, A.; Wu, Q.; Sun, L.; et al. IL-6 regulates autophagy and chemotherapy resistance by promoting BECN1 phosphorylation. Nat. Commun. 2021, 12, 3651. [Google Scholar] [CrossRef] [PubMed]
- Van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and activity of microRNA-loaded mini-cells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017, 18, 1386–1396. [Google Scholar] [CrossRef]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions, and data contained in all publications are solely those of the individual author(s) and contributor(s), not of the MDPI and/or editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions, or products referred to in the content. |



| Gene Symbol | Gene Name | GFP-SAS (FC) 1 | Ca9-22 (FC) 1 | HSC2 (FC) 1 |
|---|---|---|---|---|
| SEPT8 | septin 8 | −3.56 | −6.18 | −3.90 |
| CCNE1 | cyclin E1 | −3.54 | −8.93 | −2.27 |
| FCF1 | FCF1 small subunit processome component homolog | −3.20 | −3.99 | −3.03 |
| SELK | selenoprotein K | −3.16 | −4.42 | −3.30 |
| DDAH1 | dimethylarginine dimethylaminohydrolase 1 | −2.48 | −2.85 | −5.87 |
| RMND5A | required for meiotic nuclear division 5 homolog A | −2.39 | −2.41 | −2.30 |
| MAGT1 | magnesium transporter 1 | −2.35 | −4.78 | −2.89 |
| GINS1 | GINS complex subunit 1 | −2.21 | −4.18 | −2.88 |
| PHC2 | polyhomeotic homolog 2 | −2.21 | −3.62 | −2.66 |
| BECN1 | beclin 1, autophagy related | −2.17 | −2.20 | −2.15 |
| PPP6C | protein phosphatase 6, catalytic subunit | −2.13 | −2.81 | −2.47 |
| EPCAM | epithelial cell adhesion molecule | −2.12 | −3.88 | −3.27 |
| GDPD5 | glycerophosphodiester phosphodiesterase domain containing 5 | −2.11 | −2.34 | −3.12 |
| SEC63 | SEC63 homolog | −2.08 | −2.37 | −2.17 |
| PLEKHB2 | pleckstrin homology domain containing, family B member 2 | −2.07 | −2.34 | −2.26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakashiro, K.-i.; Tokuzen, N.; Saika, M.; Shirai, H.; Kuribayashi, N.; Goda, H.; Uchida, D. MicroRNA-1289 Functions as a Novel Tumor Suppressor in Oral Squamous Cell Carcinoma. Cancers 2023, 15, 4138. https://doi.org/10.3390/cancers15164138
Nakashiro K-i, Tokuzen N, Saika M, Shirai H, Kuribayashi N, Goda H, Uchida D. MicroRNA-1289 Functions as a Novel Tumor Suppressor in Oral Squamous Cell Carcinoma. Cancers. 2023; 15(16):4138. https://doi.org/10.3390/cancers15164138
Chicago/Turabian StyleNakashiro, Koh-ichi, Norihiko Tokuzen, Masato Saika, Hiroyuki Shirai, Nobuyuki Kuribayashi, Hiroyuki Goda, and Daisuke Uchida. 2023. "MicroRNA-1289 Functions as a Novel Tumor Suppressor in Oral Squamous Cell Carcinoma" Cancers 15, no. 16: 4138. https://doi.org/10.3390/cancers15164138
APA StyleNakashiro, K.-i., Tokuzen, N., Saika, M., Shirai, H., Kuribayashi, N., Goda, H., & Uchida, D. (2023). MicroRNA-1289 Functions as a Novel Tumor Suppressor in Oral Squamous Cell Carcinoma. Cancers, 15(16), 4138. https://doi.org/10.3390/cancers15164138






