Single Nucleotide Polymorphisms in the Vitamin D Metabolic Pathway as Survival Biomarkers in Colorectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Ethic Statement
2.3. Study Population
2.4. Sociodemographic and Clinical Variables
2.5. Genetic Variables
2.5.1. DNA Isolation
2.5.2. Detection of Gene Polymorphisms
2.6. Survival Variables
2.7. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Influence of Clinical-Pathological Characteristics on Survival
3.2.1. Overall Survival
3.2.2. Progression-Free Survival
3.3. Genotypes and Haplotypes Distribution
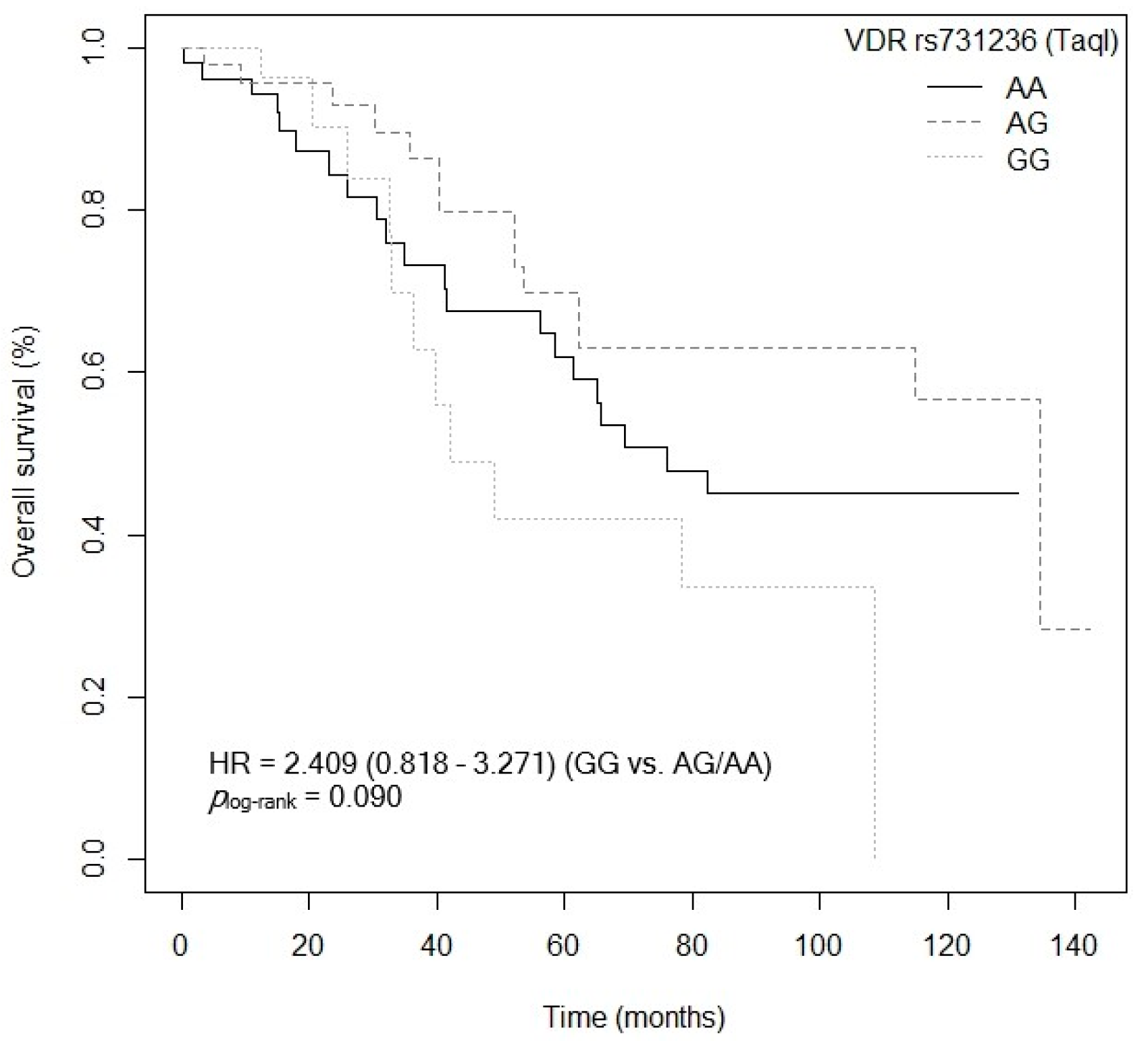
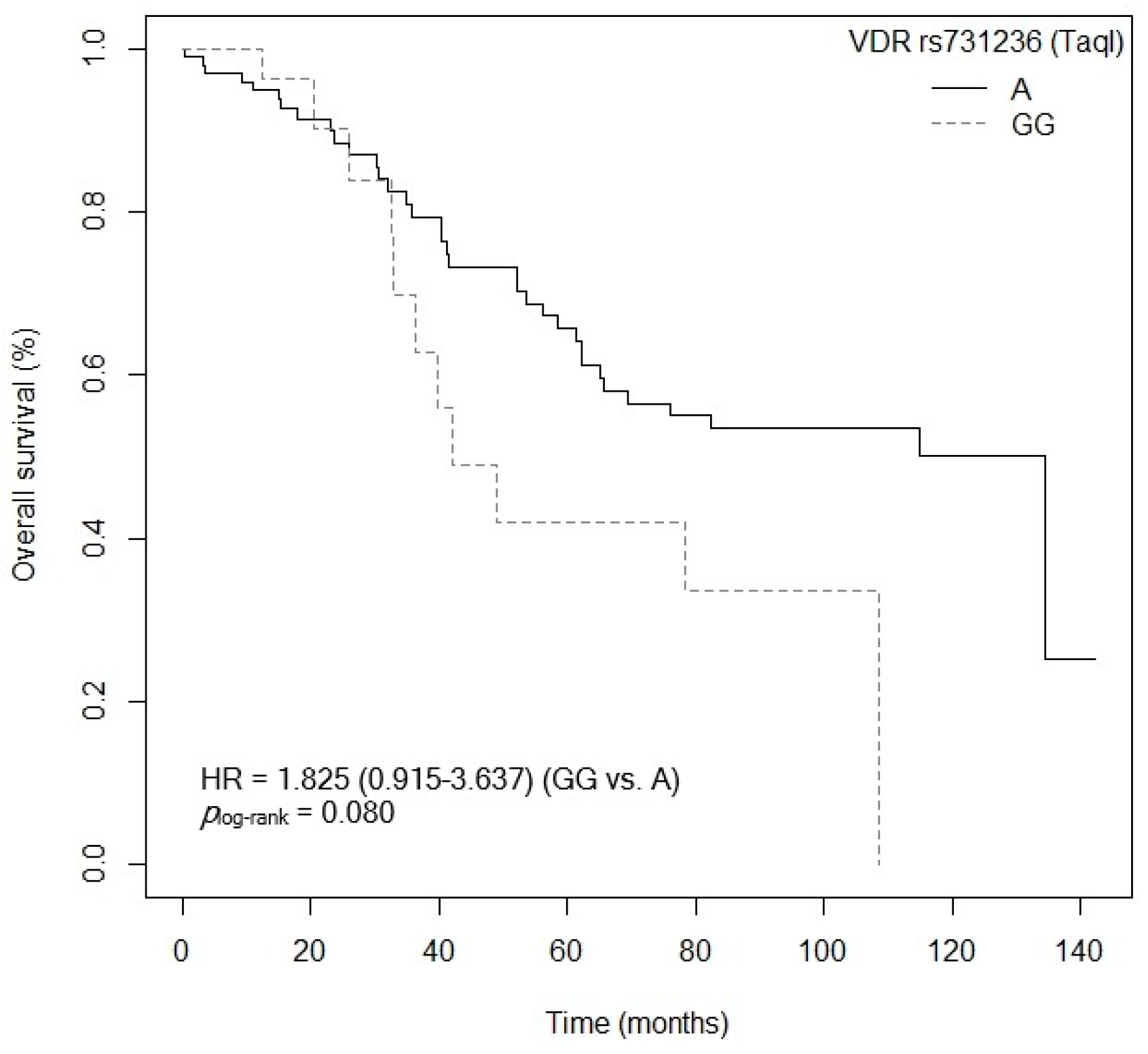
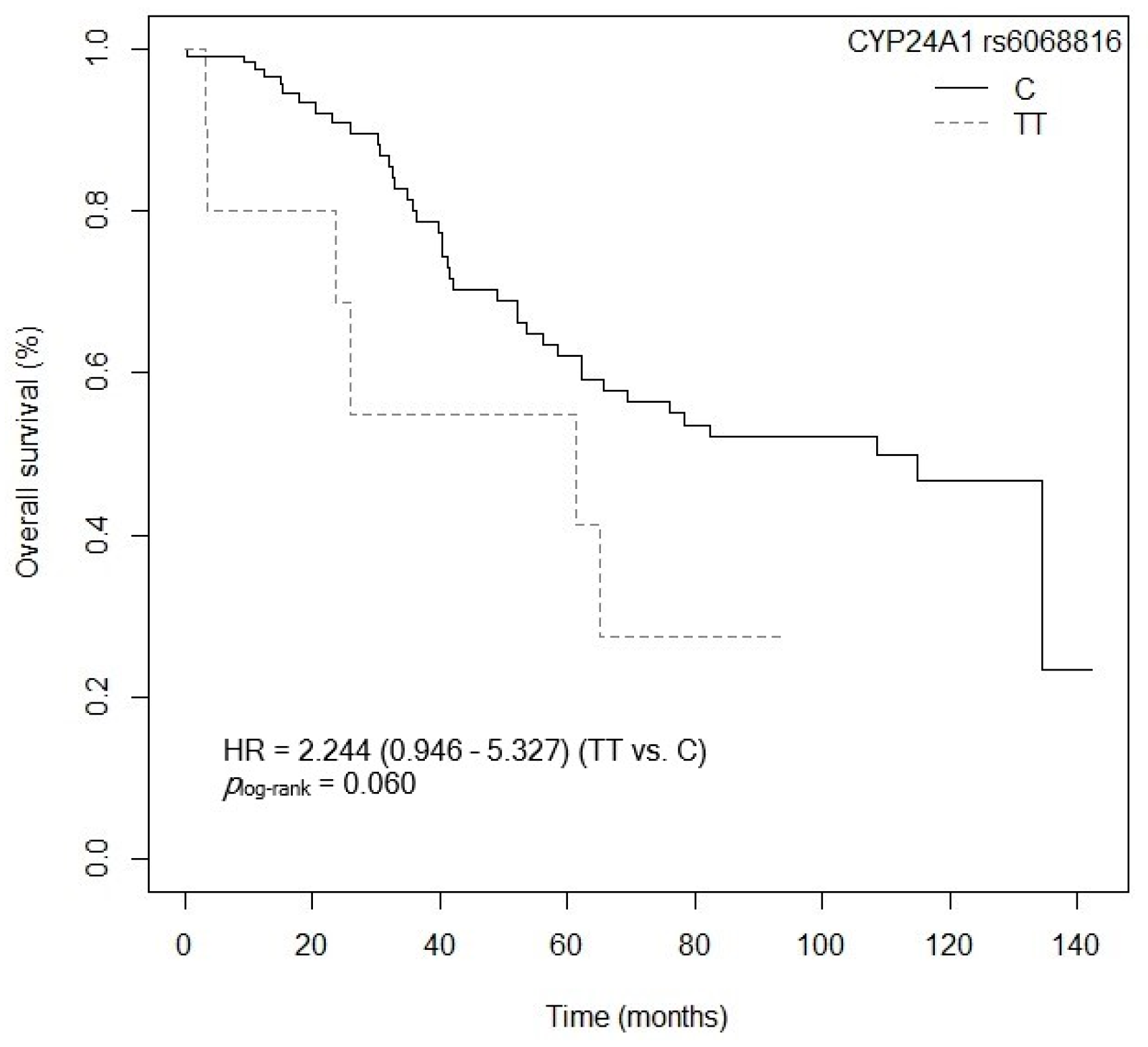
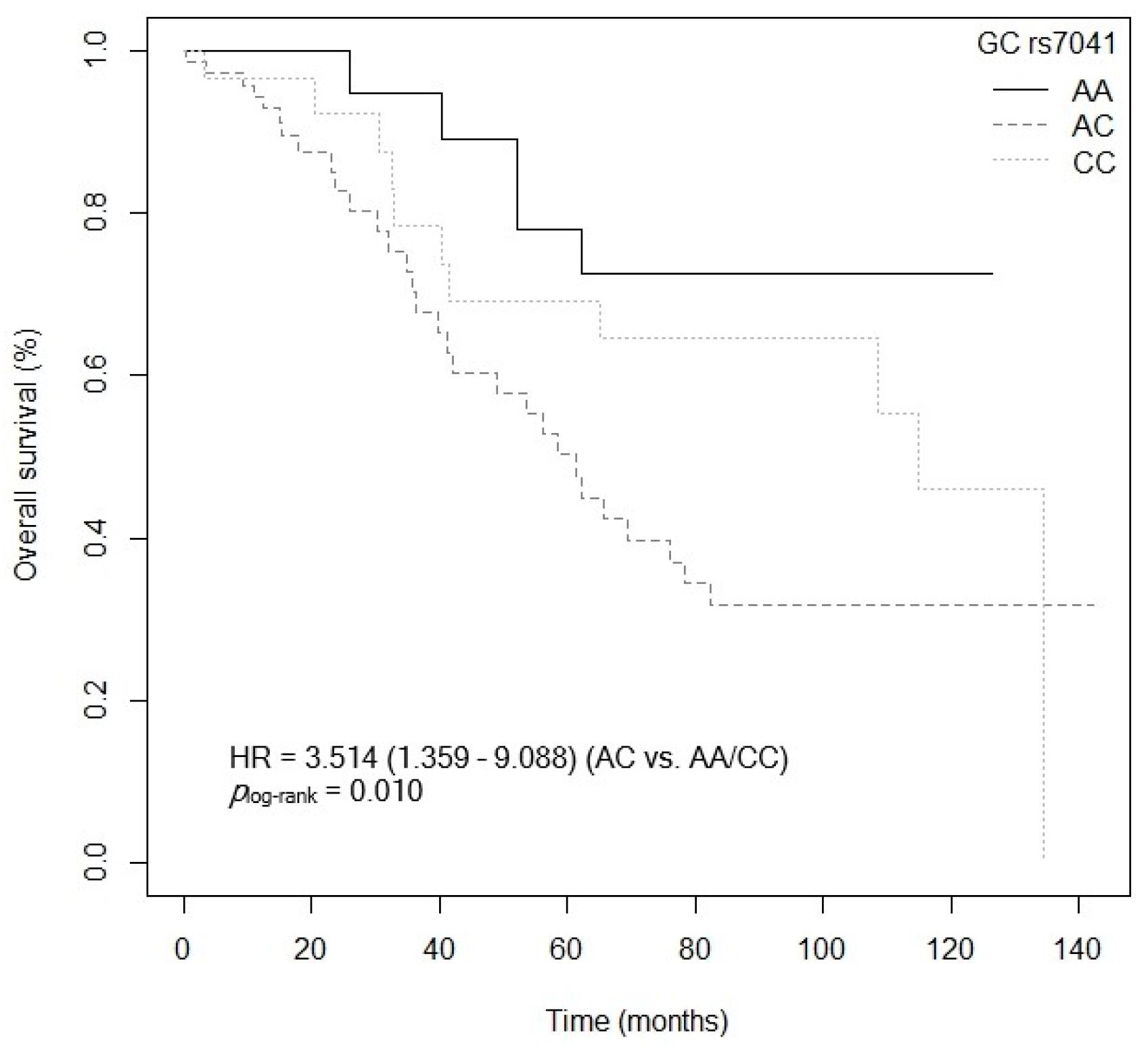
3.4. Influence of Gene Polymorphisms and Haplotypes on Survival
3.4.1. Overall Survival
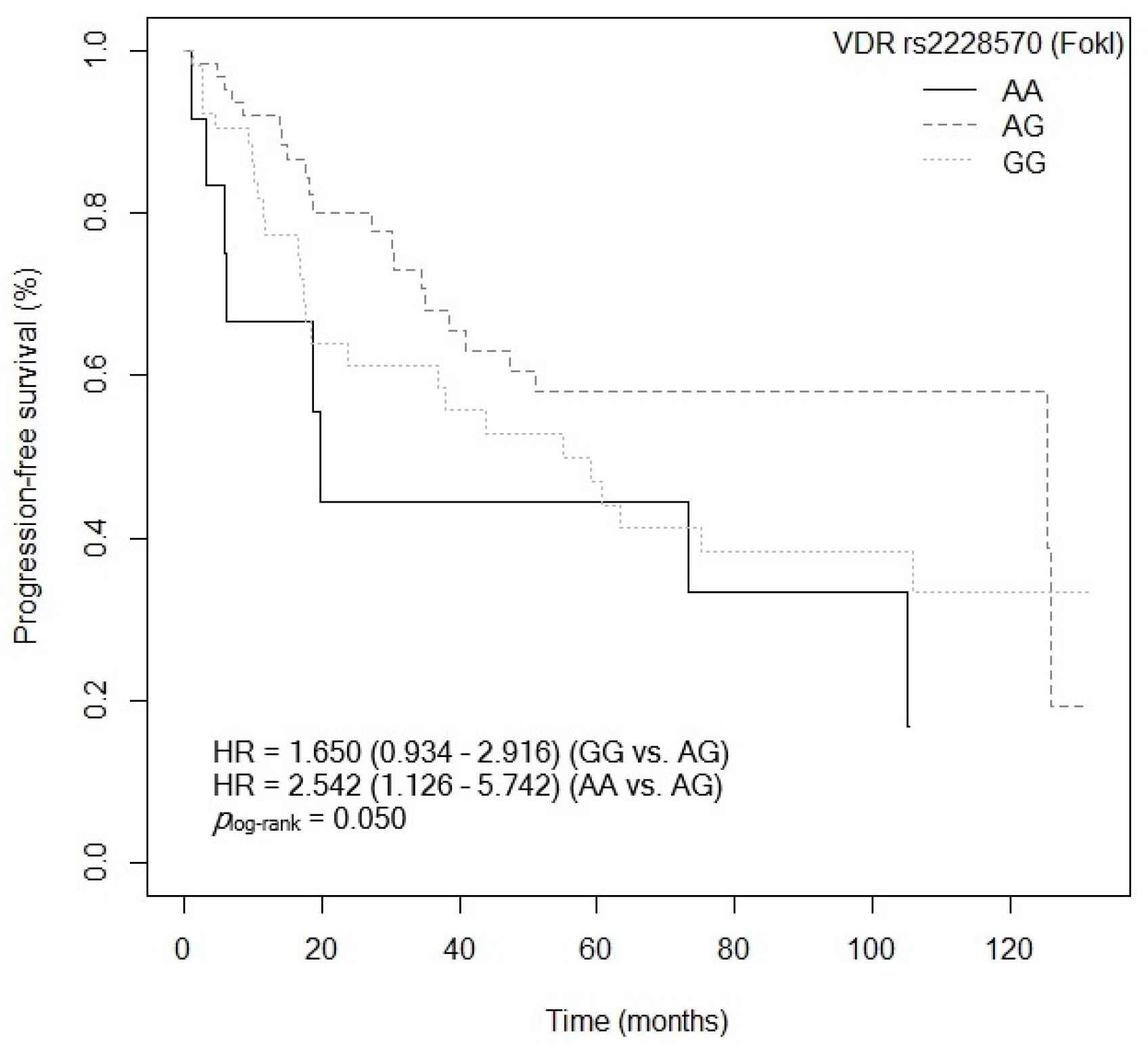
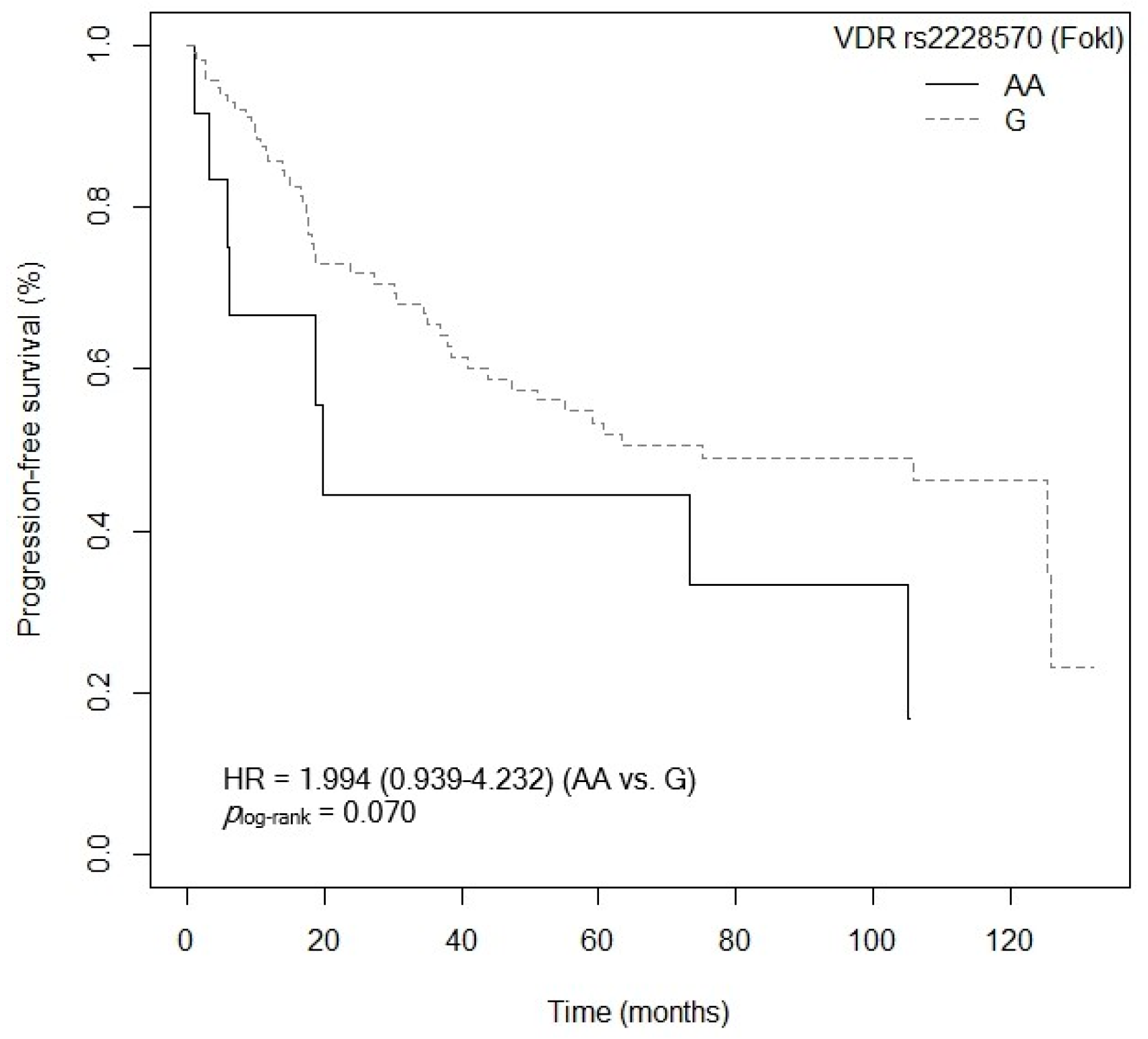
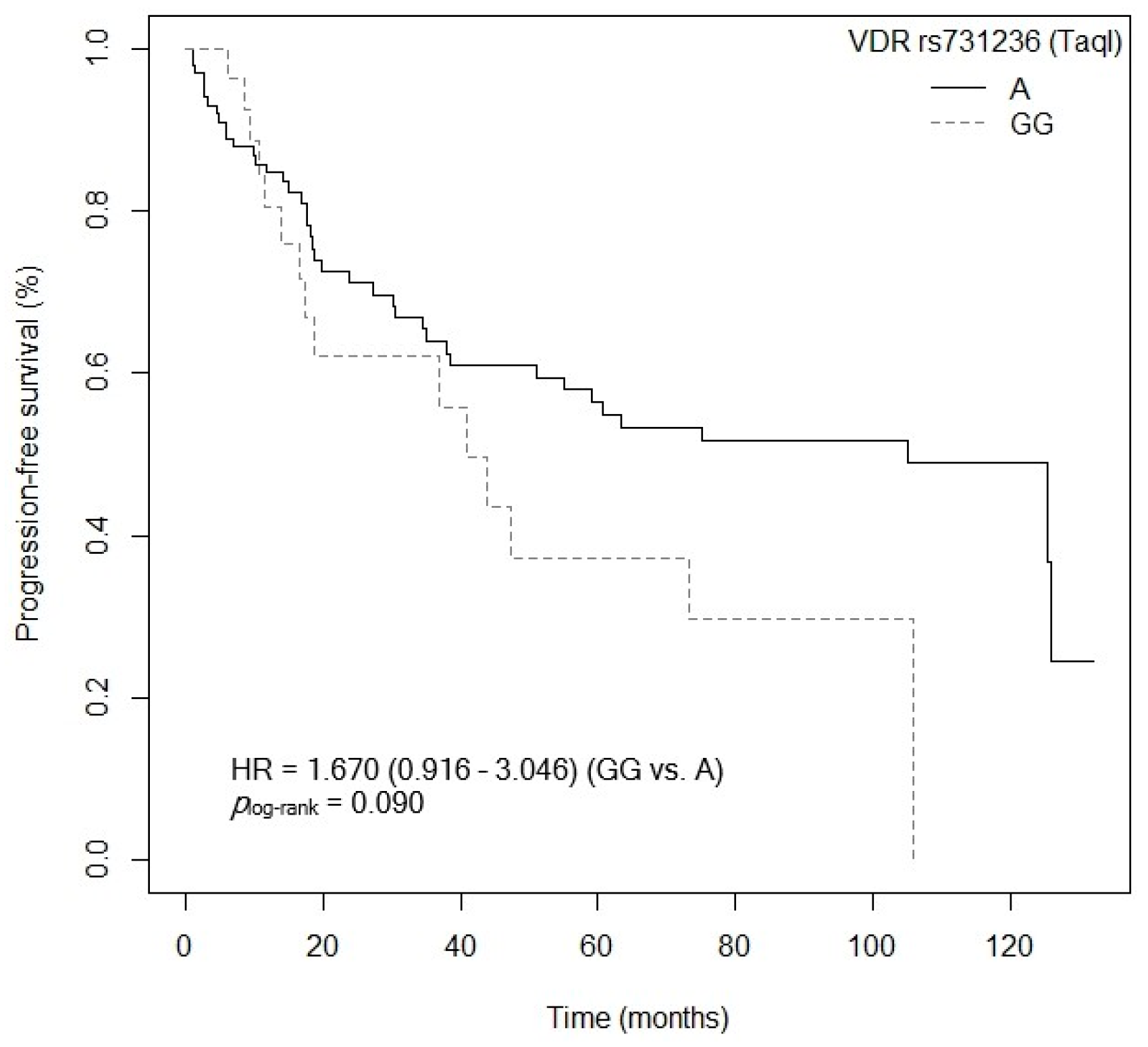
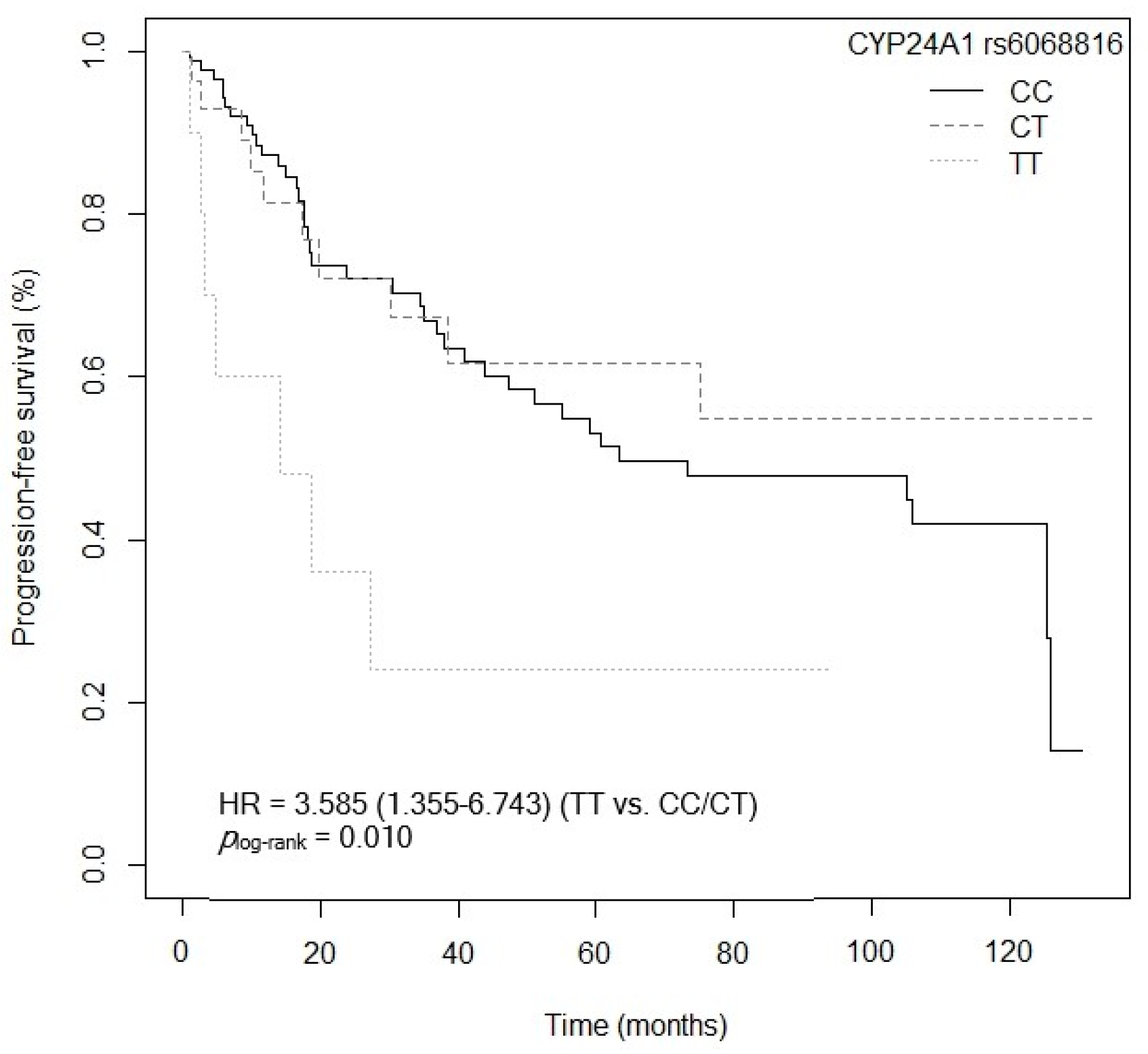
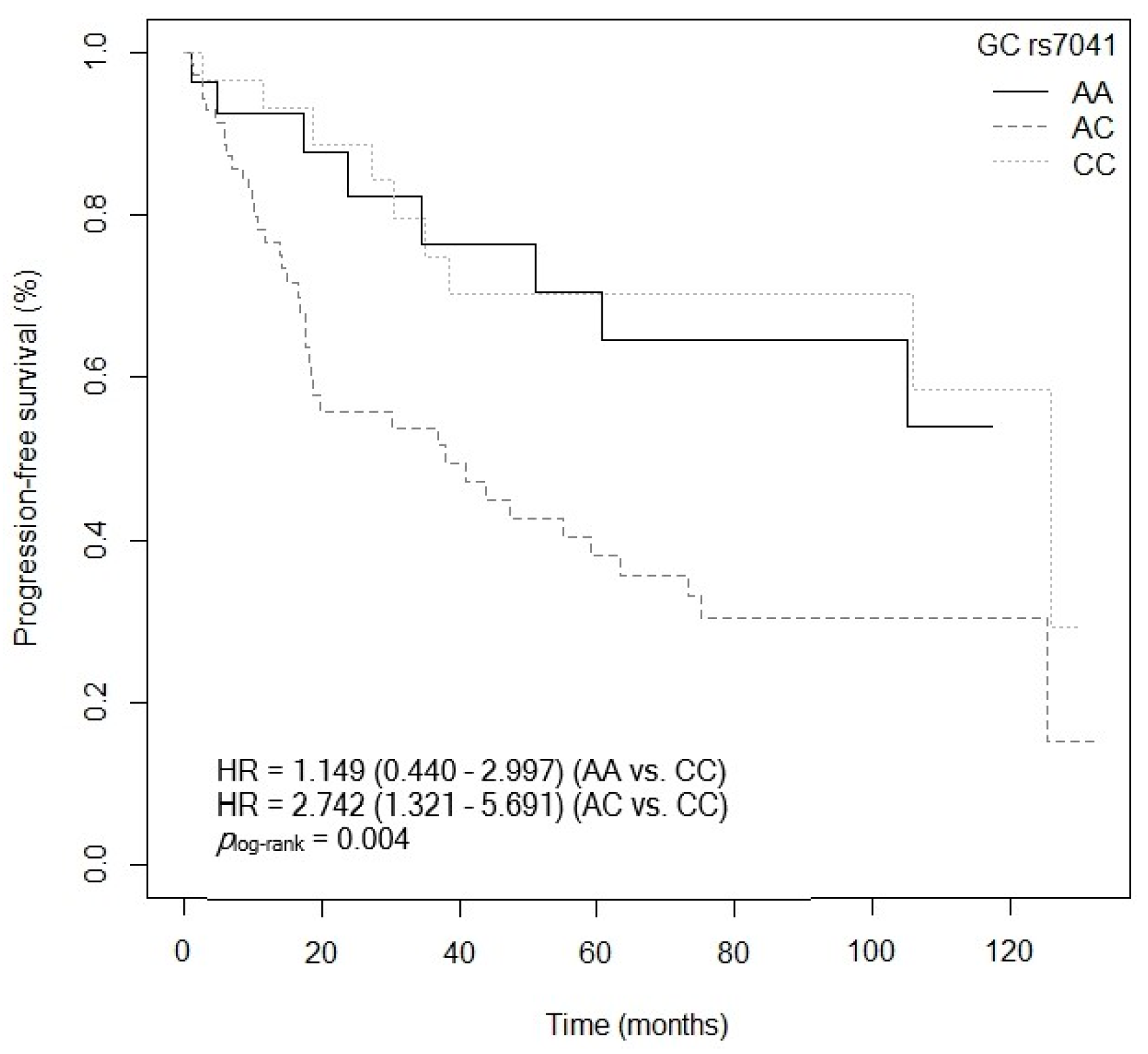
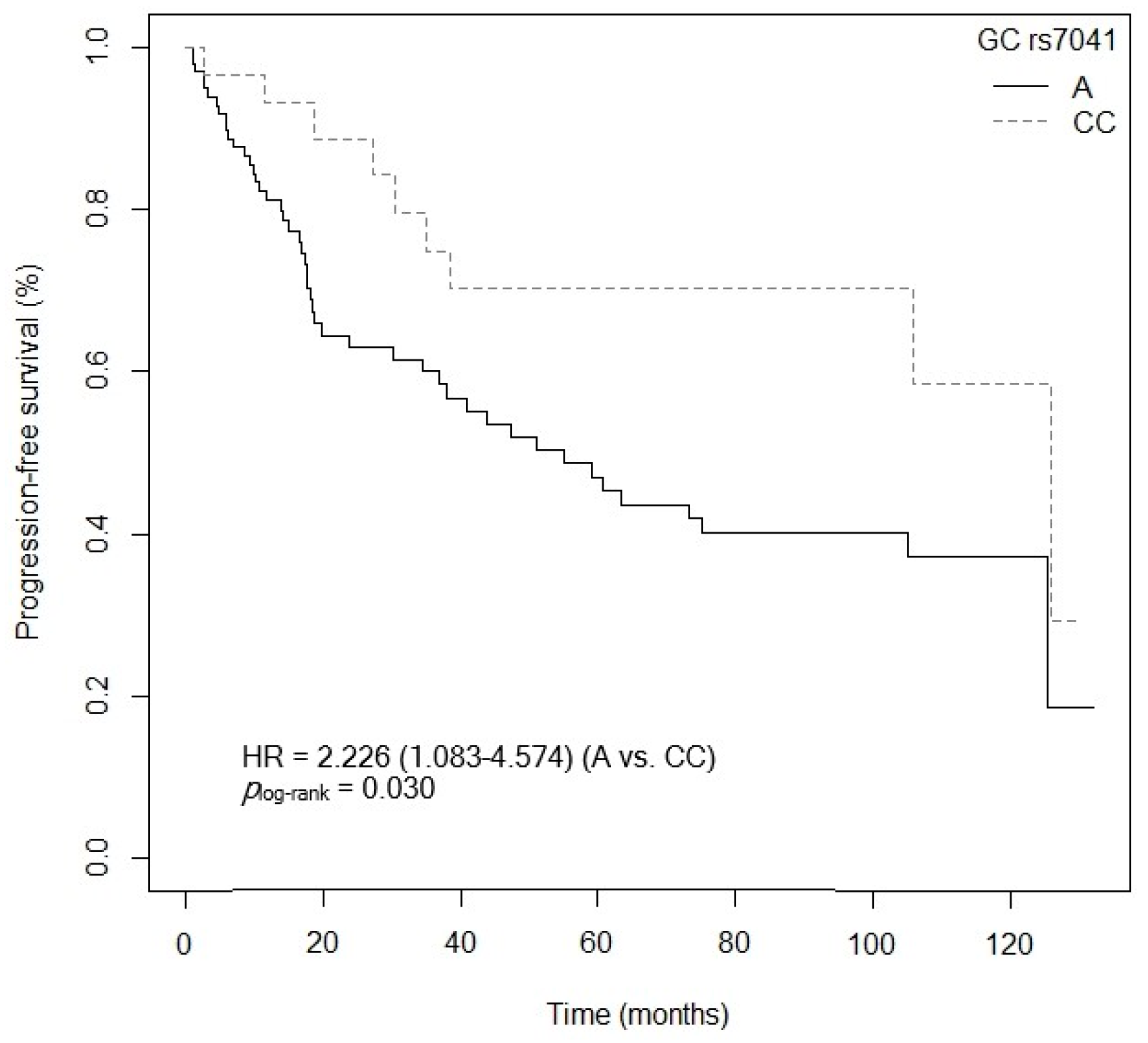
3.4.2. Progression-Free Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA A Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef] [PubMed]
- Meester, R.G.S.; Peterse, E.F.P.; Knudsen, A.B.; de Weerdt, A.C.; Chen, J.C.; Lietz, A.P.; Dwyer, A.; Ahnen, D.J.; Siegel, R.L.; Smith, R.A.; et al. Optimizing colorectal cancer screening by race and sex: Microsimulation analysis II to inform the American Cancer Society colorectal cancer screening guideline. Cancer 2018, 124, 2974–2985. [Google Scholar] [CrossRef] [PubMed]
- Simon, K.; Balchen, V. Colorectal cancer development and advances in screening. Clin. Interv. Aging 2016, 11, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Garrett, W.S.; Chan, A.T. Nutrients, Foods, and Colorectal Cancer Prevention. Gastroenterology 2015, 148, 1244–1260.e16. [Google Scholar] [CrossRef]
- Takeshige, N.; Yin, G.; Ohnaka, K.; Kono, S.; Ueki, T.; Tanaka, M.; Maehara, Y.; Okamura, T.; Ikejiri, K.; Maekawa, T.; et al. Associations between Vitamin D Receptor (VDR) Gene Polymorphisms and Colorectal Cancer Risk and Effect Modifications of Dietary Calcium and Vitamin D in a Japanese Population. Asian Pac. J. Cancer Prev. 2015, 16, 2019–2026. [Google Scholar] [CrossRef]
- Feldman, D.; Krishnan, A.V.; Swami, S.; Giovannucci, E.; Feldman, B.J. The role of vitamin D in reducing cancer risk and progression. Nat. Rev. Cancer 2014, 14, 342–357. [Google Scholar] [CrossRef]
- Mahendra, A.; Karishma; Choudhury, B.K.; Sharma, T.; Bansal, N.; Bansal, R.; Gupta, S. Vitamin D and gastrointestinal cancer. J. Lab. Physician 2018, 10, 1–5. [Google Scholar] [CrossRef]
- Mondul, A.M.; Weinstein, S.J.; Layne, T.M.; Albanes, D. Vitamin D and Cancer Risk and Mortality: State of the Science, Gaps, and Challenges. Epidemiol. Rev. 2017, 39, 28–48. [Google Scholar] [CrossRef]
- Nasiri, H.; Forouzandeh, M.; Rasaee, M.; Rahbarizadeh, F. Modified salting-out method: High-yield, high-quality genomic DNA extraction from whole blood using laundry detergent. J. Clin. Lab. Anal. 2005, 19, 229–232. [Google Scholar] [CrossRef]
- Christakos, S.; Dhawan, P.; Verstuyf, A.; Verlinden, L.; Carmeliet, G. Vitamin D: Metabolism, Molecular Mechanism of Action, and Pleiotropic Effects. Physiol. Rev. 2016, 96, 365–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, D.; Liu, J.; Feng, M.; Wu, X. A novel CpG-methylation-based nomogram predicts survival in colorectal cancer. Epigenetics 2020, 15, 1213–1227. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Yang, J.; Jiang, Y.; Song, R.; Lu, Q. Vitamin D Receptor BsmI Polymorphism and Colorectal Cancer Risk: An Updated Analysis. Asian Pac. J. Cancer Prev. 2014, 15, 4801–4807. [Google Scholar] [CrossRef]
- Vaughan-Shaw, P.G.; O’Sullivan, F.; Farrington, S.M.; Theodoratou, E.; Campbell, H.; Dunlop, M.G.; Zgaga, L. The impact of vitamin D pathway genetic variation and circulating 25-hydroxyvitamin D on cancer outcome: Systematic review and meta-analysis. Br. J. Cancer 2017, 116, 1092–1110. [Google Scholar] [CrossRef]
- Ferrer-Mayorga, G.; Larriba, M.J.; Crespo, P.; Muñoz, A. Mechanisms of action of vitamin D in colon cancer. J. Steroid Biochem. Mol. Biol. 2019, 185, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zgaga, L.; Theodoratou, E.; Farrington, S.M.; Din, F.V.N.; Ooi, L.Y.; Glodzik, D.; Johnston, S.; Tenesa, A.; Campbell, H.; Dunlop, M.G. Plasma Vitamin D Concentration Influences Survival Outcome After a Diagnosis of Colorectal Cancer. J. Clin. Oncol. 2014, 32, 2430–2439. [Google Scholar] [CrossRef]
- Thakkinstian, A.; McEvoy, M.; Minelli, C.; Gibson, P.; Hancox, B.; Duffy, D.; Thompson, J.; Hall, I.; Kaufman, J.; Leung, T.-F.; et al. Systematic Review and Meta-Analysis of the Association between β2-Adrenoceptor Polymorphisms and Asthma: A HuGE Review. Am. J. Epidemiol. 2005, 162, 201–211. [Google Scholar] [CrossRef]
- Weiss, J.M.; Gupta, S.; Burke, C.A.; Axell, L.; Chen, L.-M.; Chung, D.C.; Clayback, K.M.; Dallas, S.; Felder, S.; Gbolahan, O.; et al. NCCN Guidelines® Insights: Genetic/Familial High-Risk Assessment: Colorectal.; Version 1.2021: Featured Updates to the NCCN Guidelines. J. Natl. Compr. Cancer Netw. 2021, 19, 1122–1132. [Google Scholar] [CrossRef]
- Ochoa Mangado, E.; Madoz-Gúrpide, A.; Vicente Muelas, N. Diagnóstico y tratamiento de la dependencia de alcohol. Med. Segur. Trab. 2009, 55, 26–40. [Google Scholar] [CrossRef][Green Version]
- Edge, S.B.; Byrd, D.R.; Compton, C.C.; Fritz, A.G.; Greene, F.L.; Trotti, A. AJCC Cancer Staging Manual, 7th ed.; Springer: New York, NY, USA, 2010; pp. 347–376. ISBN 978-0-387-88440-0. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; ISBN 3-900051-07-0. Available online: http://www.R-project.org/ (accessed on 25 April 2023).
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Tregouet, D.-A.; Tiret, L. Cox proportional hazards survival regression in haplotype-based association analysis using the Stochastic-EM algorithm. Eur. J. Hum. Genet. 2004, 12, 971–974. [Google Scholar] [CrossRef] [PubMed]
- Pineda Lancheros, L.E.; Pérez Ramírez, C.; Sánchez Martín, A.; Gálvez Navas, J.M.; Martínez Martínez, F.; Ramírez Tortosa, M.C.; Jiménez Morales, A. Impact of Genetic Polymorphisms on the Metabolic Pathway of Vitamin D and Survival in Non-Small Cell Lung Cancer. Nutrients 2021, 13, 3783. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Timofeeva, M.; Zhang, X.; Xu, W.; Li, X.; Din, F.V.N.; Svinti, V.; Farrington, S.M.; Campbell, H.; Dunlop, M.G.; et al. Colorectal cancer risk variants rs10161980 and rs7495132 are associated with cancer survival outcome by a recessive mode of inheritance. Int. J. Cancer 2021, 148, 2774–2778. [Google Scholar] [CrossRef]
- Song, N.; Kim, K.; Shin, A.; Park, J.W.; Chang, H.J.; Shi, J.; Cai, Q.; Kim, D.Y.; Zheng, W.; Oh, J.H. Colorectal cancer susceptibility loci and influence on survival. Genes Chromosom. Cancer 2018, 57, 630–637. [Google Scholar] [CrossRef]
- Dou, R.; Ng, K.; Giovannucci, E.L.; Manson, J.E.; Qian, Z.R.; Ogino, S. Vitamin D and colorectal cancer: Molecular, epidemiological and clinical evidence. Br. J. Nutr. 2016, 115, 1643–1660. [Google Scholar] [CrossRef]
- Jeon, S.-M.; Shin, E.-A. Exploring vitamin D metabolism and function in cancer. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef]
- Latacz, M.; Rozmus, D.; Fiedorowicz, E.; Snarska, J.; Jarmołowska, B.; Kordulewska, N.; Savelkoul, H.; Cieślińska, A. Vitamin D Receptor (VDR) Gene Polymorphism in Patients Diagnosed with Colorectal Cancer. Nutrients 2021, 13, 200. [Google Scholar] [CrossRef]
- Zmuda, J.M.; Cauley, J.A.; Ferrell, R.E. Molecular Epidemiology of Vitamin D Receptor Gene Variants. Epidemiol. Rev. 2000, 22, 203–217. [Google Scholar] [CrossRef]
- Perna, L.; Butterbach, K.; Haug, U.; Schöttker, B.; Müller, H.; Arndt, V.; Holleczek, B.; Burwinkel, B.; Brenner, H. Vitamin D Receptor Genotype rs731236 (Taq1) and Breast Cancer Prognosis. Cancer Epidemiol. Biomark. Prev. 2013, 22, 437–442. [Google Scholar] [CrossRef]
- Berger, M.D.; Stintzing, S.; Heinemann, V.; Cao, S.; Yang, D.; Sunakawa, Y.; Matsusaka, S.; Ning, Y.; Okazaki, S.; Miyamoto, Y.; et al. A Polymorphism within the Vitamin D Transporter Gene Predicts Outcome in Metastatic Colorectal Cancer Patients Treated with FOLFIRI/Bevacizumab or FOLFIRI/Cetuximab. Clin. Cancer Res. 2018, 24, 784–793. [Google Scholar] [CrossRef] [PubMed]
- Fedirko, V.; Riboli, E.; Tjønneland, A.; Ferrari, P.; Olsen, A.; Bueno-De-Mesquita, H.B.; van Duijnhoven, F.J.; Norat, T.; Jansen, E.H.; Dahm, C.C.; et al. Prediagnostic 25-Hydroxyvitamin D, VDR and CASR Polymorphisms, and Survival in Patients with Colorectal Cancer in Western European Populations. Cancer Epidemiol. Biomark. Prev. 2012, 21, 582–593. [Google Scholar] [CrossRef] [PubMed]
- Heist, R.S.; Zhou, W.; Wang, Z.; Liu, G.; Neuberg, D.; Su, L.; Asomaning, K.; Hollis, B.W.; Lynch, T.J.; Wain, J.C.; et al. Circulating 25-Hydroxyvitamin D, VDR Polymorphisms, and Survival in Advanced Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2008, 26, 5596–5602. [Google Scholar] [CrossRef]
- Akiba, T.; Morikawa, T.; Odaka, M.; Nakada, T.; Kamiya, N.; Yamashita, M.; Yabe, M.; Inagaki, T.; Asano, H.; Mori, S.; et al. Vitamin D Supplementation and Survival of Patients with Non–small Cell Lung Cancer: A Randomized, Double-Blind, Placebo-Controlled Trial. Clin. Cancer Res. 2018, 24, 4089–4097. [Google Scholar] [CrossRef]
- Chakraborty, M.; Arora, M.; Ramteke, A.; Yadav, V.; Naaz, H.; Muntakhab, M.; Tripathi, P.; Chandran, K.N. FokI polymorphism of Vitamin D receptor gene and deficiency of serum Vitamin D increases the risk of breast cancer in North Indian women. Endocrine 2023, 81, 168–174. [Google Scholar] [CrossRef]
- Nam, S.E.; Jin, T.; Park, K.S.; Saindane, M.; Noh, W.C.; Yoo, Y.B.; Park, W.S.; Yun, I.J. Vitamin D receptor (VDR) mRNA overexpression is associated with poor prognosis in breast carcinoma. Ann. Surg. Treat. Res. 2022, 103, 183–194. [Google Scholar] [CrossRef]
- Messaritakis, I.; Koulouridi, A.; Boukla, E.; Sfakianaki, M.; Vogiatzoglou, K.; Karagianni, M.; Gouvas, N.; Tsiaoussis, J.; Xynos, E.; Athanasakis, E.; et al. Investigation of Microbial Translocation, TLR and VDR Gene Polymorphisms, and Recurrence Risk in Stage III Colorectal Cancer Patients. Cancers 2022, 14, 4407. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, H.; Zhang, Z.; Qin, L.; Li, B. Association of the CYP24A1-rs2296241 polymorphism of the vitamin D catabolism enzyme with hormone-related cancer risk: A meta-analysis. OncoTargets Ther. 2015, 8, 1175–1183. [Google Scholar] [CrossRef]
- Chen, X.-Q.; Mao, J.-Y.; Li, W.-B.; Li, J.; Yang, H.; Qian, J.-M.; Li, J.-N. Association between CYP24A1 polymorphisms and the risk of colonic polyps and colon cancer in a Chinese population. World J. Gastroenterol. 2017, 23, 5179–5186. [Google Scholar] [CrossRef]
- Yi, C.; Huang, C.; Wang, H.; Wang, C.; Dong, L.; Gu, X.; Feng, X.; Chen, B. Association study between CYP24A1 gene polymorphisms and cancer risk. Pathol.-Res. Pract. 2020, 216, 152735. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Long, Z.; Yu, Y.; Zhu, L.; Tian, J.; Li, S.; Li, J.; Yu, H.; Chi, Q.; Piao, D.; et al. Dietary factors and polymorphisms in vitamin D metabolism genes: The risk and prognosis of colorectal cancer in northeast China. Sci. Rep. 2017, 7, 8827. [Google Scholar] [CrossRef]
- Zeng, R.; Li, H.; Jia, L.; Lee, S.H.; Jiang, R.; Zhang, Y.; Hu, X.; Ye, T.; Wang, X.; Yan, X.; et al. Association of CYP24A1 with survival and drug resistance in clinical cancer patients: A meta-analysis. BMC Cancer 2022, 22, 1317. [Google Scholar] [CrossRef]
- Chen, G.; Kim, S.H.; King, A.N.; Zhao, L.; Simpson, R.U.; Christensen, P.J.; Wang, Z.; Thomas, D.G.; Giordano, T.J.; Lin, L.; et al. CYP24A1 Is an Independent Prognostic Marker of Survival in Patients with Lung Adenocarcinoma. Clin. Cancer Res. 2011, 17, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Xu, F.; Qu, J.; Wang, Y.; Gao, M.; Yu, H.; Qian, B. Genetic polymorphisms in the vitamin D pathway in relation to lung cancer risk and survival. Oncotarget 2015, 6, 2573–2582. [Google Scholar] [CrossRef] [PubMed]
- Latacz, M.; Snarska, J.; Kostyra, E.; Fiedorowicz, E.; Savelkoul, H.F.J.; Grzybowski, R.; Cieślińska, A. Single Nucleotide Polymorphisms in 25-Hydroxyvitamin D3 1-Alpha-Hydroxylase (CYP27B1) Gene: The Risk of Malignant Tumors and Other Chronic Diseases. Nutrients 2020, 12, 801. [Google Scholar] [CrossRef]
- Ramagopalan, S.V.; Heger, A.; Berlanga, A.J.; Maugeri, N.J.; Lincoln, M.R.; Burrell, A.; Handunnetthi, L.; Handel, A.E.; Disanto, G.; Orton, S.-M.; et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: Associations with disease and evolution. Genome Res. 2010, 20, 1352–1360. [Google Scholar] [CrossRef] [PubMed]
- Dong, A.N.; Tan, B.H.; Pan, Y.; Ong, C.E. The CYP2R1 Enzyme: Structure, Function, Enzymatic Properties and Genetic Polymorphism. J. Pharm. Pharm. Sci. 2021, 24, 94–112. [Google Scholar] [CrossRef]
- Nissen, J.; Rasmussen, L.B.; Ravn-Haren, G.; Andersen, E.W.; Hansen, B.; Andersen, R.; Mejborn, H.; Madsen, K.H.; Vogel, U. Common Variants in CYP2R1 and GC Genes Predict Vitamin D Concentrations in Healthy Danish Children and Adults. PLoS ONE 2014, 9, e89907. [Google Scholar] [CrossRef]
- Slater, N.A.; Rager, M.L.; Havrda, D.E.; Harralson, A.F. Genetic Variation in CYP2R1 and GC Genes Associated with Vitamin D Deficiency Status. J. Pharm. Pract. 2017, 30, 31–36. [Google Scholar] [CrossRef]
- Rozmus, D.; Ciesielska, A.; Płomiński, J.; Grzybowski, R.; Fiedorowicz, E.; Kordulewska, N.; Savelkoul, H.; Kostyra, E.; Cieślińska, A. Vitamin D Binding Protein (VDBP) and Its Gene Polymorphisms—The Risk of Malignant Tumors and Other Diseases. Int. J. Mol. Sci. 2020, 21, 7822. [Google Scholar] [CrossRef]
- Pibiri, F.; Kittles, R.A.; Sandler, R.S.; Keku, T.O.; Kupfer, S.S.; Xicola, R.M.; Llor, X.; Ellis, N.A. Genetic variation in vitamin D-related genes and risk of colorectal cancer in African Americans. Cancer Causes Control 2014, 25, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Chun, R.F. New perspectives on the vitamin D binding protein. Cell Biochem. Funct. 2012, 30, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, S.J.; Mondul, A.M.; Layne, T.M.; Yu, K.; Huang, J.; Stolzenberg-Solomon, R.Z.; Ziegler, R.G.; Purdue, M.P.; Huang, W.-Y.; Abnet, C.C.; et al. Prediagnostic Serum Vitamin D, Vitamin D Binding Protein Isoforms, and Cancer Survival. JNCI Cancer Spectr. 2022, 6, pkac019. [Google Scholar] [CrossRef] [PubMed]






| Gene | dbSNP ID | Location | Alleles | Assay ID |
|---|---|---|---|---|
| VDR (12q13.11) | rs1544410 (BsmI) | Intron 8 | C > T | C___8716062_20 |
| rs11568820 (Cdx2) | Intron 1 | C > T | C___2880808_10 | |
| rs2228570 (FokI) | Exon 2 | A > G | C__12060045_20 | |
| rs7975232 (ApaI) | Intron 8 | C > A | C__28977635_10 | |
| rs731236 (TaqI) | Exon 9 | A > G | C___2404008_10 | |
| CYP27B1 (12q14.1) | rs4646536 | Intron 6 | A > G | C__25623453_10 |
| rs3782130 | 5′ Promoter | G > C | ANGZRHH 1 | |
| rs10877012 | 5′ UTR | G > T | C__26237740_10 | |
| rs703842 | 3′ UTR | A > G | AN9HX2K 1 | |
| CYP24A1 (20q13.2) | rs6068816 | Exon 6 | C > T | C__25620091_20 |
| rs4809957 | 3′ UTR | A > G | C___3120981_20 | |
| CYP2R1 (11p15.2) | rs10741657 | 5′ UTR | A > G | C___2958430_10 |
| GC (4q13.3) | rs7041 | Exon 11 | A > C | C___3133594_30 |
| Variable | Baseline | ||
|---|---|---|---|
| N | % | Mean ± Standard Deviation | |
| Sex | 127 | 100 | |
| Female | 42 | 33.07 | - |
| Male | 85 | 66.93 | - |
| Family history of cancer | |||
| Yes | 64 | 50.39 | - |
| No | 63 | 49.61 | - |
| Family history of CRC | |||
| Yes | 27 | 21.26 | - |
| No | 100 | 78.74 | - |
| Smoking status | |||
| Smokers | 27 | 21.26 | - |
| Ex-smokers | 44 | 34.65 | - |
| Nonsmokers | 56 | 44.09 | - |
| Drinking status | |||
| Drinkers | 24 | 18.90 | - |
| Ex-drinkers | 7 | 5.51 | - |
| Nondrinkers | 96 | 75.59 | - |
| Body mass index (BMI) | |||
| <24 | 27 | 21.26 | - |
| >24 | 100 | 78.74 | - |
| Type of CRC | |||
| Colon | 89 | 70.08 | - |
| Rectum | 38 | 29.92 | - |
| Age of diagnosis | 127 | - | 62.52 ± 10.55 |
| ECOG score | |||
| 0–1 | 116 | 91.34 | - |
| 2.4 | 11 | 8.66 | - |
| Histopathology | |||
| ADC | 107 | 84.25 | - |
| Mucinous ADC | 20 | 15.75 | - |
| Tumor size | 127 | - | 4.40 (3.00–5.50) |
| Lymph node involvement | |||
| Yes | 92 | 72.44 | - |
| No | 35 | 27.56 | - |
| Number of lymph node involvement | |||
| None | 35 | 27.56 | - |
| 1 | 61 | 48.03 | - |
| 2 | 31 | 24.41 | - |
| Histological grade | |||
| Grade 1 | 18 | 14.17 | - |
| Grade 2 | 94 | 74.02 | - |
| Grade 3 | 15 | 11.81 | - |
| Metastasis | |||
| Yes | 36 | 28.35 | - |
| No | 91 | 71.65 | - |
| Stage | |||
| I, II, or IIIA | 71 | 55.91 | - |
| IIIB, IV, or IVB | 56 | 44.09 | - |
| TNM stage | |||
| I/II | 11 | 8.66 | - |
| III/IV | 116 | 91.34 | - |
| Primary tumor resection | |||
| Yes | 119 | 93.70 | - |
| No | 8 | 6.30 | - |
| Adjuvant chemotherapy | |||
| Fluoropyrimidines | 35 | 27.56 | - |
| Fluoropyrimidines + Oxiplatin | 70 | 55.12 | - |
| Fluoropyrimidines + Biological drugs | 19 | 14.96 | - |
| None | 3 | 2.36 | - |
| Survival | |||
| PFS | 127 | - | 73.3 (41.0-NR) |
| OS | 127 | - | 82.3 (62.1-NR) |
| Overall Survival | ||
|---|---|---|
| Independent Variables | HR (CI95%) | p-Value |
| Family history of CRC (No) | 1.442 (0.268–1.792) | 0.449 |
| Stage (IIIB, IV, or IVB) | 1.130 (0.311–4.104) | 0.852 |
| Adjuvant chemotherapy (Fluoropyrimidines) | 1.455 (0.595–3.558) | 0.411 |
| Adjuvant chemotherapy (Fluoropyrimidines + biologics drugs) | 4.248 (1.655–10.904) | 0.003 |
| Adjuvant chemotherapy (None) | 5.148 (0.507–52.332) | 0.166 |
| Lymph node involvement (Yes) | 1.503 (0.751–3.006) | 0.249 |
| ECOG score (2–4) | 1.342 (0.505–3.568) | 0.555 |
| Metastasis (yes) | 1.360 (0.335–5.548) | 0.664 |
| Age of CRC diagnosis | 1.067 (1.029–1.107) | <0.001 |
| CYP24A1 rs6068816 (TT vs. C) | 6.237 (2.297–16.936) | <0.001 |
| GC rs7041 (C vs. AA) | 4.939 (1.578–15.457) | 0.006 |
| VDR rs7975232 (ApaI) (AA vs. C) | 1.974 (1.045–3.731) | 0.036 |
| Overall Survival | ||
|---|---|---|
| Independent Variables | HR (CI95%) | p-Value |
| Age of CRC diagnosis | 1.055 (1.023–1.090) | 0.001 |
| ECOG score (2–4) | 1.029 (0.412–2.577) | 0.950 |
| Lymph node involvement (Yes) | 1.994 (0.982–4.049) | 0.056 |
| Metastasis (yes) | 3.142 (1.096–9.004) | 0.033 |
| Stage (IIIB, IV, or IVB) | 1.268 (0.452–3.556) | 0.651 |
| Primary tumor resection (No) | 1.271 (0.311–1.992) | 0.613 |
| Adjuvant chemotherapy (Fluoropyrimidines) | 2.306 (1.059–5.019) | 0.035 |
| Adjuvant chemotherapy (Fluoropyrimidines + biologics drugs) | 1.861 (0.823–4.208) | 0.135 |
| Adjuvant chemotherapy (None) | 1.870 (0.231–15.143) | 0.557 |
| CYP24A1 rs6068816 (TT vs. C) | 6.095 (2.413–15.398) | <0.001 |
| GC rs7041 (A vs. CC) | 1.543 (1.143–1.935) | 0.268 |
| VDR rs2228570 (FokI) (G vs. AA) | 1.003 (0.412–2.420) | 0.999 |
| VDR rs7975232 (ApaI) (AA vs. C) | 1.692 (1.402–2.092) | 0.066 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Durán, C.; Márquez-Pete, N.; Gálvez-Navas, J.M.; Cura, Y.; Rojo-Tolosa, S.; Pineda-Lancheros, L.E.; Ramírez-Tortosa, M.; García-Collado, C.; Maldonado-Montoro, M.d.M.; Villar-del-Moral, J.M.; et al. Single Nucleotide Polymorphisms in the Vitamin D Metabolic Pathway as Survival Biomarkers in Colorectal Cancer. Cancers 2023, 15, 4077. https://doi.org/10.3390/cancers15164077
Pérez-Durán C, Márquez-Pete N, Gálvez-Navas JM, Cura Y, Rojo-Tolosa S, Pineda-Lancheros LE, Ramírez-Tortosa M, García-Collado C, Maldonado-Montoro MdM, Villar-del-Moral JM, et al. Single Nucleotide Polymorphisms in the Vitamin D Metabolic Pathway as Survival Biomarkers in Colorectal Cancer. Cancers. 2023; 15(16):4077. https://doi.org/10.3390/cancers15164077
Chicago/Turabian StylePérez-Durán, Carmen, Noelia Márquez-Pete, José María Gálvez-Navas, Yasmin Cura, Susana Rojo-Tolosa, Laura Elena Pineda-Lancheros, MCarmen Ramírez-Tortosa, Carlos García-Collado, María del Mar Maldonado-Montoro, Jesús María Villar-del-Moral, and et al. 2023. "Single Nucleotide Polymorphisms in the Vitamin D Metabolic Pathway as Survival Biomarkers in Colorectal Cancer" Cancers 15, no. 16: 4077. https://doi.org/10.3390/cancers15164077
APA StylePérez-Durán, C., Márquez-Pete, N., Gálvez-Navas, J. M., Cura, Y., Rojo-Tolosa, S., Pineda-Lancheros, L. E., Ramírez-Tortosa, M., García-Collado, C., Maldonado-Montoro, M. d. M., Villar-del-Moral, J. M., Pérez-Ramírez, C., & Jiménez-Morales, A. (2023). Single Nucleotide Polymorphisms in the Vitamin D Metabolic Pathway as Survival Biomarkers in Colorectal Cancer. Cancers, 15(16), 4077. https://doi.org/10.3390/cancers15164077








