Simple Summary
Pemetrexed, a multi-target anti-folate agent, is the treatment choice for advanced or metastatic thymoma at the second or later line setting. Thymoma patients occasionally show an exceptionally durable and deep response to pemetrexed treatment. Recent studies using next-generation sequencing have identified some genomic aberrations in such patients, but the mechanism underlying their sensitivity to pemetrexed remains unclear. This study explores certain somatic single-nucleotide variants or copy number variations (CNVs) in exceptional responders to pemetrexed treatment. To elucidate any genomic changes, we performed whole-exome sequencing in patients with advanced thymomas treated with pemetrexed. We found no differences between the exceptional and typical responders, but the highest number of whole arm gain or the loss of chromosomal CNVs was observed in an exceptional responder to pemetrexed. Our study provides additional genomic findings on thymomas and chemosensitivity.
Abstract
Background: Pemetrexed is used for the chemotherapy of advanced thymoma. Exceptional responses of thymoma to pemetrexed treatment are not frequently observed. The underlying genetic mechanism of the exceptional responses remains unclear. We used whole-exome sequencing to explore the specific genomic aberrations that lead to an extreme and durable response. Methods: Whole-exome sequencing using NovaSeq6000 (150 bp paired-end sequencing) was performed on nine formalin-fixed paraffin-embedded tissues from patients with advanced thymomas treated with pemetrexed (two exceptional responders and seven typical responders). Results: We identified 284 somatic single-nucleotide variants (SNVs; 272 missense, 8 missense/splice-site, 3 stop-gain, and 1 stop-gain/splice-site), 34 insertions and deletions (Indels; 33 frameshift and one splice region), and 21 copy number variations (CNVs; 15 gains and six losses). No difference in the number of SNVs variants and distribution of deleterious Indels was observed between the exceptional and typical responders. Interestingly, arm-level chromosomal CNVs (15 gains and six losses) were detected in four patients, including an exceptional responder. The highest number of arm-level CNVs was observed in an exceptional responder. Conclusion: Exceptional responders to pemetrexed for metastatic thymomas may be characterized by arm-level CNVs. Further, whole-genome and RNA sequencing studies should be performed.
1. Introduction
Thymomas are rare anterior mediastinal neoplasms that arise from thymic epithelial cells and are classified according to the World Health Organization (WHO) by histopathological classification (subtypes A, AB, B1, B2, B3, and carcinoma) [1]. Surgical resection cures early-stage thymomas, whereas systemic chemotherapy is required for metastatic or recurrent thymomas. Although anthracycline-based chemotherapy is commonly used as first-line treatment for advanced thymomas [2,3]; however, no standard chemotherapy regimen has been established.
Pemetrexed is an approved multi-target anti-folate agent for the treatment of malignant pleural mesothelioma and non-small cell lung carcinoma [4]. A single-arm phase II trial [5] showed that pemetrexed is also useful for the treatment of heavily pretreated thymomas. Pemetrexed rarely produces an exceptional response in advanced thymoma, i.e., a profound and durable reduction in tumor size. In a small subset of patients with solid tumors, molecular-targeted treatments lead to extreme responses [6,7,8,9,10]. In previous studies, genomic alterations on the mammalian target of rapamycin (mTOR) pathway activation conferred exceptional response to everolimus or pazopanib [7] in various tumors [6]. Additionally, patients with lung adenocarcinoma exhibiting KRAS mutation and EGFR amplification showed an exceptional response to erlotinib therapy [8]. Another study identified very few extreme responders to anticancer therapy among patients with advanced breast cancer [9]. Moreover, patients with low-grade ovarian cancer showed an exceptional clinical response to ibrutinib when guided by organoid drug testing [10]. In these cases, certain genomic aberrations may confer exceptional sensitivity to target therapies [7,9].
Recently, next-generation sequencing (NGS) studies have revealed new genomic mutations in several tumors, including the lung, breast, and colon [11,12,13]. Additionally, case studies that performed NGS found genomic aberrations in thymomas, such as GTF2I [14,15], ASXL2, DNMT3A [16], KRAS, HRAS [17], CDKN2A/B [18], and TP53 [18], as well as chromosomal abnormalities [19,20]. However, the molecular basis of pemetrexed sensitivity in thymomas remains unclear [21,22].
However, to date, no predictive markers for pemetrexed treatment are available. Previous studies reported that mRNA levels of TPX2, CPA3, EZH2, MCM2, and TOP2A [23], a folylpoly-γ-glutamate synthase single nucleotide polymorphism [24], may be potential predictive biomarkers of pemetrexed sensitivity in non-small cell lung cancer. On the other hand, elevated thymidylate synthase expression levels, including [25] BMI1 [26], AMPK [27], and ABCC5 [28], may confer acquired resistance to pemetrexed or predict poor therapeutic sensitivity of pemetrexed chemotherapy. The useful predictive molecular markers for pemetrexed-based treatment have been expected.
Furthermore, genomic mutations associated with oncogenic drivers, particularly drug sensitivity, have not been identified in thymomas by targeted sequencing.
In the present study, we performed the whole-exome sequencing of patients with thymoma, including those who showed an exceptional response to pemetrexed treatment. The purpose of this study was to identify genomic alterations that could predict an exceptional treatment response to pemetrexed.
2. Materials and Methods
2.1. Patients and Tissue Collection
We retrospectively reviewed the medical records of 12 Japanese patients with advanced thymoma treated with pemetrexed monotherapy between August 2011 and April 2020 at the Department of Thoracic Oncology, National Cancer Center Hospital, Tokyo, Japan. Formalin-fixed paraffin-embedded (FFPE) thymoma samples were collected from six hospitals in Japan. The histopathological analysis was performed by expert pathologists in accordance with the 2015 WHO classification.
2.2. Assessment and Tumor Response
Pemetrexed was administered at 500 mg/m2 every 21 days. Clinical efficacy was assessed every 2–3 months. The tumor response was evaluated using The Response Criteria in Solid Tumors (version 1.1) [29]. The objective response rate (ORR) was defined as the proportion of patients who achieved a complete response (CR) or partial response (PR). The disease control rate (DCR) was defined as proportion of the patients who achieved CR, PR, or stable disease.
2.3. Definition of Exceptional Response to Pemetrexed
Based on previous studies [7,9], a CR or PR to pemetrexed monotherapy for 15 months until disease progression or death (equivalent to progression-free survival [PFS]) was considered to indicate an exceptional response.
2.4. DNA Extraction
Genomic DNA was extracted from FFPE specimens from advanced thymomas using the Maxwell RSC DNA FFPE Kit (Promega, Madison, WI, USA). A DNA library was prepared using the SureSelect Human All Exon Kit v6 (Agilent Technologies Inc., Santa Clara, CA, USA) in accordance with the manufacturer’s guidelines. The DNA quality and quantity were evaluated using a 2100 Bioanalyzer DNA 1000 kit (Agilent Technologies Inc.). The total DNA (0.2 µg) met the quality control criteria and was determined to be acceptable for analysis. Genomic DNA was fragmented, purified, end-repaired, adenylated on the 3′ ends, ligated to indexed pair-end adaptors, purified again, and amplified by PCR. The genomic DNA was then cleaved into fragments ranging from approximately 236 to 367 bp, and sequencing adapters were attached to these fragments. We subsequently performed whole-exome sequencing using NovaSeq6000 (150 bp paired-end sequencing) (Illumina, San Diego, CA, USA).
2.5. Sequencing Analysis
The obtained FASTQ files were aligned to the hg38 human reference using Burrows–Wheeler Aligner (version 0.7.17) (Figure S1). The output binary alignment map files were then processed using the MarkDuplicates tool in Picard (version 2.18.2). For somatic analysis, the base-recalibrated tumors were analyzed using GATK (version 4.0.5.1) to call somatic variants at chromosome positions covered in the target bed (Agilent SureSelect V6). The resulting somatic variant call files were hard-filtered at a read depth of ≥20. Realignment and somatic variant calling were performed using Mutect2. Genetic single-nucleotide variants (SNVs) and insertions and deletions (Indels) were annotated using SnpEff (version 4.3).
To retain high-confidence nonsynonymous coding variants, we applied the following criteria for the additional filtration of the initial call set: (1) filter depth > 20, (2) quality score > 100, and (3) the removal of common germline single-nucleotide polymorphisms using a public database (dbSNP 138). Detected SNVs were searched using information from publicly available databases, including the Genome Aggregation Database, 1000G-integration, ExAC, and ExAC non-TCGA, to obtain a <0.01 minor allele frequency.
SNVs were identified as potentially pathogenic if they were identified by at least two of the following methods: SIFT, LRT, MutationTaster, MutationAssessor, FATHMM, PROVEAN, MetaSVM, MetaLR, MCAP, and fathmm-MKL (coding).
2.6. Copy Number Variation (CNV) Analysis
Briefly, the CNVs were analyzed, and heatmap plots were drawn using CNVkit (version 0.9.6) [30]. Data were entered into the CNVkit, and copy number calls were generated for each sample based on the cutoff value for deletions and duplications specified in the settings. The identified CNVs were interpreted according to standard methods, and the data were normalized according to the average depth. The median CNV was used as the cutoff for classifying CNVs in our cohort. The gene copy number gain and loss were indicated by all exons that had a copy number > 0.3 and <−0.3 log2, respectively.
2.7. Statistical Analysis
Statistical analysis was performed using the EZR program (version 1.37; Saitama Medical Center, Jichi Medical University, Saitama, Japan) [31]. Overall survival (OS) curves were drawn using the Kaplan–Meier method. p < 0.05 was considered statistically significant. p values and hazard ratios were calculated using the log-rank test.
3. Results
3.1. Patient Characteristics and Sample Collection
Figure 1 shows the CONSORT diagram. We identified 38 patients with advanced thymoma who visited the participating institutes; of these patients, 12 (three exceptional responders and nine typical responders) were included in the study. One exceptional responder and one typical responder were excluded from the analysis because they had no or insufficient tumor specimens available for sequencing. Therefore, the final analysis included two exceptional responders (patients 3 and 8) and seven typical responders (Figure 2). Of the nine thymoma patients, one (11%), two (22%), four (44%), and two (22%) had B1, B2, B3, and B2 + B3 WHO subtype thymomas, respectively (Table 1). The patients included two males and seven females, with a median age of 51 years (range: 36–77 years). Three (33%) patients met the stage III Masaoka criteria at diagnosis, whereas six (66%) met the stage IV criteria.
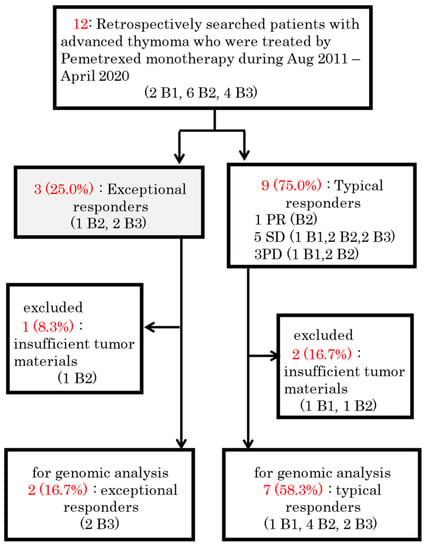
Figure 1.
Schematic diagram of the study protocol. PR = partial response. SD = stable disease. PD = progressive disease.
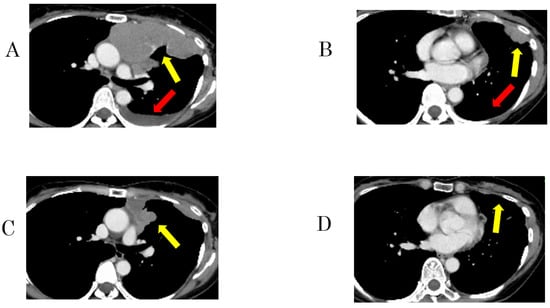
Figure 2.
Representative imaging of a 51-year-old woman with type B3 thymoma showing an exceptional response to pemetrexed monotherapy (patient 8). (A,B) Enhanced computed tomography (CT) revealed massive pleural dissemination and pleural effusion in the left hemithorax before pemetrexed treatment. (C,D) After 7 months of pemetrexed treatment, CT showed shrinkage of the massive lesions. Images of pleural dissemination (yellow arrows) and pleural effusion (red arrows).

Table 1.
Clinical characteristics of 9 Thymoma patients treated by pemetrexed monotherapy.
All patients had an Eastern Cooperative Oncology Group performance status of 0–1. Of the responders, 20 (67%) had received pemetrexed as a first-line treatment, whereas 7 (78%) had previously received anthracycline-containing regimens, 1 (11%) had received paclitaxel, and 1 (11%) was chemotherapy-naïve.
3.2. Response and Survival Analysis
After a median follow-up period of 18.0 months (95% confidence interval [CI] = 12.6–29.1), six patients experienced recurrence, and two died. The patients underwent a median of 10 (range: 2–33) pemetrexed treatment cycles. Of the nine patients, PR and stable disease were achieved in two and five, respectively, for a response rate of 22.2% (95% CI = 3.8–60.0) and a DCR of 77.8% (95% CI = 21.2–86.3). The median PFS of all patients was 16.0 months [95% CI = 1.0 − not reached (NR)]. The median OS was NR (95% CI = 12 − NR). The median PFS of exceptional responders was longer than that of typical responders (22.5 vs. 3.1 months; p = 0.37) (Figure 3A,B).
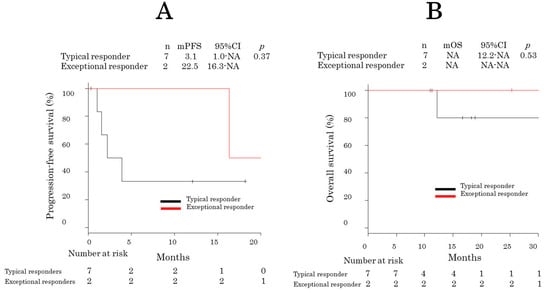
Figure 3.
Kaplan–Meier progression-free survival curves of exceptional and typical responders. Kaplan–Meier progression-free survival (A) and overall survival (B) curves.
3.3. Whole-Exome Sequencing
Whole-exome sequencing was performed on the nine FFPE samples from the nine thymoma patients. We used an analysis pipeline that involved rigorous quality control (QC) and filtering. To identify the mutations, we obtained an average sequencing at a depth of 153.4× (range: 39.7–205×) in tumor DNA. Whole-exome sequencing was not performed for patient 7 because of low coverage depth.
3.4. Identification of SNVs and Indels
In total, 284 somatic SNVs were identified in the tumor genomes (Table S1). A median of 35.1 variants per sample was identified in the samples (Figure 4A). There was no difference in variants between exceptional and typical responders.
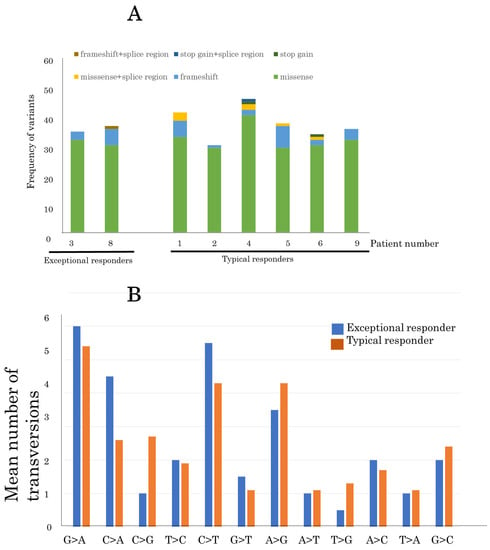
Figure 4.
Comparison of single-nucleotide variants between exceptional and typical responders (A) Comparison of the frequency of variants in advanced thymoma patients between exceptional and typical responders to pemetrexed. (B) The mean number of somatic transversion mutations in nine advanced thymomas from exceptional and typical responders.
With regard to transversions, G>A substitutions were the most common mutation in all samples (50/271, 18.5%), indicating formalin fixation artifacts (Figure 4B). The second most common mutation was C>T substitution (41/271, 15.1%). The mutation pattern was similar between exceptional and typical responders. Furthermore, 17 SNVs were identified in genes that were predicted to be potentially pathogenic by at least one of the above-mentioned methods (Table 2).

Table 2.
List of SNVs that were predicted to be deletorious.
Next, we filtered out the 282 Indels. The remaining 34 mutations included frameshift mutations (Table 3). Of the Indels, variations in GTF3C1, SCN3A, and RPL5 were identified as potentially deleterious if they were detected more than twice using computational filtered methods. No difference in the distribution of the deleterious Indels was identified between exceptional and typical responders to pemetrexed. Unlike previous studies, aberrations were not detected in GTF2I, ASXL2, DNMT3A, and KRAS/HRAS.

Table 3.
Validated Indels detected in patients with advanced thymoma treated by Pemetrexed.
3.5. Identification of CNVs
Of the nine samples, two were excluded (patients 3 and 7) from the CNV analysis to reduce the number of false positives arising due to a low gene coverage depth (average read depth < 100×). Consequently, data from one exceptional responder and six typical responders were analyzed.
According to previous studies, CNVs that covered > 25% [32] to 33% of the chromosome arm [32] were considered arm-level CNVs; these were detected in 57.1% (4/7) of thymoma patients (with a total of 15 copy number gains and six copy number losses) (Table 4 and Table S2, Figure S2).

Table 4.
The distribution of cromosomal arm-level CNVs per patient with advanced thymoma treated by Pemetrexed.
The most common chromosomal gain was 1q (4/7, 57.1%), followed by 5p (3/7, 42.9%), 9p (2/7, 28.6%), and 3p (1/7, 14.3%) (Table 5). The most common whole-arm gain was chromosome 7 (1/7, 14.3%), followed by chromosome 14 (1/7, 14.3%). The most common chromosomal loss was the whole arm of chromosome 6 (2/7, 28.6%), followed by 2p (1/7, 14.3%) and 3p (1/7, 14.3%). The whole-arm chromosome gain was seen most commonly on chromosomes 7 (2/7, 28.6%), 5 (1/7, 14.3%), and 14 (1/7, 14.3%). Whole-arm chromosome loss was seen most commonly on chromosomes 6 (2/7, 28/6%) and 13 (1/7, 14.3%).

Table 5.
Frequency of CNVs per chromosomes among 7 advanced thymoma patients treated by Pemetrexed.
A hierarchical clustering algorithm generated four distinct clusters (Figure 5).
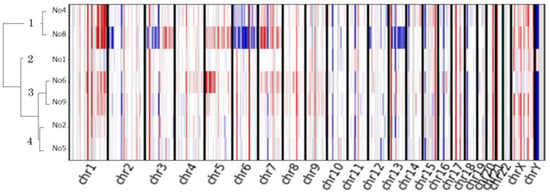
Figure 5.
Copy number analysis of nine advanced thymomas. Unsupervised hierarchical clustering heatmap for the nine thymoma patients treated with pemetrexed; copy number gains are depicted in red, and losses are in blue.
Cluster 1 (patients 4 and 8) was characterized by multiple chromosomal arm-level CNVs, whereas cluster 3 (patients 6 and 9) showed arm-level chromosome gain. An exceptional responder had the most arm-level CNVs (patient 8).
4. Discussion
In this study, we performed the whole-exome sequencing of tissues from patients with advanced thymoma to explore the genomic variants related to exceptional responses of pemetrexed monotherapy. We identified certain Indels that were potentially deleterious, including SCN3A, GTF3C1, and RPL5. However, none of the genomic alterations were candidate driver oncogenes, and none were specific to exceptional responders. No novel driver oncogene was identified in advanced thymomas. Interestingly, arm-level chromosomal CNVs were detected in some patients with advanced thymomas treated with pemetrexed.
Cluster analysis revealed that cluster 1, which was detected in an exceptional responder (patient 8), was associated with remarkably increased arm-level chromosomal CNVs, suggesting that arm-level chromosomal CNVs might be related to the response to pemetrexed. A previous study showed that the MYC chromosomal copy number was related to the clinical response to paclitaxel and the in vitro antitumor effect of mTORC1/2 inhibition [33]. Another retrospective study showed that chromosomal 1q gain may have a detrimental impact on the prognosis of multiple myeloma when treated with bortezomib-based regimens [34]. However, that study included only one exceptional responder.
Our results also suggest that arm-level chromosomal CNVs are present mainly in type B2 or B3 thymomas with a chromosomal gain of 1q, 5p, 9p, or 3p; with chromosomal loss of 2p or 3p; with a whole-arm chromosomal gain of 7, 5, or 14; or with whole-arm chromosomal loss of 6 or 13. Previous studies have found that chromosomal arm-level CNVs were detectable mainly in the thymoma subtypes B2 and B3 (Table S3) [17,20,35,36]. These studies showed that some type B3 thymomas were characterized by chromosomal 1q gain and chromosomal 6 loss [17,20]. Another study revealed the loss of 13q in type B3 thymomas [20]. Petrini et al. [35] reported a case of B3 thymoma with the gain of chromosomes 1q, 5, and 7 and the loss of chromosomes 3p, 6, 13, and part of chromosome 11q. A comprehensive analysis of The Cancer Genome Atlas (TCGA) showed that CNVs are rare in type A and AB thymomas, whereas arm-level CNVs are frequently detected in type B2 and B3 thymomas [37]. These results suggest that chromosomal arm-level CNVs might be related to the aggressiveness of thymomas. These results suggest that chromosomal arm-level CNVs might be related to the aggressiveness of thymomas. A recent study showed that arm-level CNVs can be used to screen early-stage colorectal cancer [38] or lung cancer [39]. However, the clinical significance of arm-level CNVs as a screening tool for early thymomas remains unclear. Although arm-level CNVs might play a role in thymoma oncogenesis, this is difficult to confirm because of the rarity of thymomas.
In this study, chromosomal 1q gain and whole chromosome 6 loss were detected in advanced thymomas. In other tumors, chromosome 1q gain was identified in multiple myeloma [34,40], ependymoma [41], Wilms tumor [42], and New Zealand virus-negative Merkel cell carcinomas [43]. Whole chromosome 6 loss has been detected in acute lymphoblastic lymphoma [44]. The gain of whole chromosome 9, the loss of whole chromosome 22, and the loss of the Y chromosome were detected in polymorphous low-grade adenocarcinoma of the head and neck [45]. The loss of whole chromosome 7 or loss of 7q and 5q were the most frequent primary abnormalities significantly related to myelodysplasia [46].
With respect to the prognosis or malignant grade, chromosomal 1q gain predict a poor prognosis of hepatocellular carcinoma [47]. Additionally, the aggressiveness of diffuse leptomeningeal glioneuronal tumors might be related to chromosomal 1q gain [48]. Another study showed that the loss of whole chromosome 3, the loss of 6q, and the gain of 8q were significantly associated with poor overall survival in patients with metastatic primary uveal melanoma [49]. The co-occurrence of chromosomal 1p loss and 1q gain was associated with a poor prognosis in patients with low-grade serous ovarian carcinoma [50]. The gain of whole chromosome 7 showed a more aggressive clinical nature in grade II and III gliomas [51]. However, no study to date has clarified whether chromosomal gain or loss has any effect on the prognosis or malignancy of thymomas.
TCGA suggested that patients with thymoma combined with myasthenia gravis (MG) have more chromosomal arm-level CNVs [37]. Another study suggested that the loss of whole chromosome 6, including the HLA locus, might play a role in paraneoplastic autoimmunity in patients with type B3 thymoma [20]. In our study, only patient 2 had concomitant MG; this patient had chromosomal arm-level CNVs. However, the sample size of the present study was insufficient to evaluate the relationship between MG and arm-level CNVs.
Certain potentially harmful SNVs were identified in the present study. Among these, a STAT6 missense variant was reported as a potential cause of primary atopic disorders [52]. Additionally, the ERBB2 missense mutations D769Y and D742N were associated with acquired resistance to tyrosine kinase inhibitors [53]. The missense V419L variant in TGFBR2 associated with Loeys–Dietz syndrome was also related to impaired TGF-β signaling [54]. Patients with pathogenic missense mutations in LZTR1 exhibited a characteristic Noonan syndrome [55]. A missense mutation in NROB1 was associated with isolated mineralocorticoid deficiency [56]. A missense mutation in HERC2 was associated with intellectual disability, autism, and gait disturbance [57]. PCDHA9 has been identified as a potential candidate gene for Hirschsprung’s disease [58]. A homozygous missense mutation in STIL causes holoprosencephaly and microcephaly [59]. In one study, genetic aberrations in FBXW7 were detected in 55% of patients with primary uveal melanoma [60]. A recent study showed that EPHA5 mutations, along with other molecular alterations in the DNA damage response pathway and a favorable antitumor immune signature, could contribute to exceptional responses [61]. These deleterious SNVs do not seem to be related to the oncogenesis of thymoma or sensitivity to pemetrexed.
This study identified the potentially deleterious Indels SCN3A, GTF3C1, and RPL5. These mutations might have played oncogenic roles in the advanced thymomas in our study. A previous study showed that SCN3A, which encodes the voltage-gated sodium channel subunit Nav1.3, caused severe epilepsy and disordered cortical development [62]. GTF3C1 is part of the GTF3 family, which is related to the expansion of different types of cancers [63]. 1p22 deletion encompassing the RPL5 gene caused Diamond–Blackfan anemia [64]. However, the same deleterious Indels were not observed in other cases.
Furthermore, these deleterious SNVs and Indels were not detected in patients with advanced thymomas or exceptional sensitivity to pemetrexed. Although certain SNVs or Indels appeared to be harmful, no deleterious SNVs or Indels were found to be candidates for oncogenic driver mutations in relation to drug sensitivity and pemetrexed in patients with advanced thymomas.
Regarding pemetrexed, two previous studies performed the whole-exome sequencing of non-small cell carcinoma patients with exceptional responses to pemetrexed and carboplatin [61,65]. However, the molecular aberrations identified in the previous studies did not match those identified in the present one. Therefore, the underlying genetic mechanism of exceptional response to pemetrexed might differ between non-small cell carcinoma and thymomas. In particular, an exceptional responder (patient 8) had a whole-arm gain and loss of chromosome 3 that contained the CAND2 genes. CAND2 is a translationally upregulated gene that encodes a muscle-specific protein and is dependent on the activity of mTOR [66]. CAND2 has also been identified as a novel obesity susceptibility gene [67]. In the present study, we were unable to identify a relationship between CAND2 and its sensitivity to pemetrexed. Furthermore, this was seen in only one patient. Further studies are required to validate our results. GTF2I mutations, which were frequently found in the comprehensive genomic profiling study of thymomas [14], were not detected in the present study. GTF2I mutations were exclusively detected in early-stage thymoma [14,37]. However, our study did not include patients with thymoma subtypes A or AB.
Our study had some limitations. First, the number and size of the tumor samples were limited, which prevented us from drawing definitive conclusions. In particular, only two exceptional responders were included, one of whom was excluded from the analysis of CNVs due to a small amount of genomic data. No further conclusions could be drawn from this single case. Second, sequencing is difficult to perform on archived FFPE samples, and our RNA sequencing, including enrichment analysis using the Gene Ontology/Kyoto Encyclopedia of Genes and Genomes analysis of the CNVs, failed because the RNA of almost all samples was fragmented. Therefore, the fusion genes of thymomas could not be explored. Future studies should collect frozen tissues of advanced thymoma patients, including exceptional responders, to allow for comprehensive analysis of genomic aberrations, including mutations, CNVs, and structural genomic variations by whole-genome or RNA sequencing.
In conclusion, this is the first study to perform the whole-exome sequencing of patients with advanced thymomas treated with pemetrexed. Although we failed to identify the molecular mechanisms of exceptional response to pemetrexed, arm-level CNVs could be related to the genetic mechanism underlying extreme sensitivity to pemetrexed treatment or the deterioration of the thymoma. Further studies are required to confirm our results.
5. Conclusions
In the present study, none of the evaluated genomic alterations were specific to exceptional responders to pemetrexed treatment, and no new driver gene was identified in advanced thymomas. However, arm-level chromosomal CNVs were detected in four of seven patients. Because this was a retrospective study of a small sample, the results will not affect the treatment choice for thymomas. Further, whole-genome and RNA sequencing studies should be performed.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15164018/s1, Figure S1: The bioinformatics analysis pipeline used in this study. Single-nucleotide variants (SNVs) and insertion and deletions (Indels) were analyzed using GATK. CNVkit was used for calling copy number variations (CNVs). Figure S2: Scatter plots of copy number variations in thymomas. Table S1: Validated SNVs detected by whole exome sequencing. Table S2: Predicted copy number in 7 FFPE advanced thymomas. Table S3: Reports of chromosomal arm-level CNVs detected in advanced thymomas. The authors confirm that all Tables and Figures are original and that they own the copyright.
Author Contributions
T.T. and Y.G. contributed to the writing—original draft. M.H. contributed to the formal analysis. K.M., Y.S., Y.M., Y.O. (Yusuke Okuma), T.Y., H.H., N.Y. and Y.O. (Yuichiro Ohe) contributed to the writing—review and editing. N.M., Y.Y. and S.W. contributed to the data curation. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study protocol was approved by the Institutional Review Board of the National Cancer Center on 19 March 2020 (no.: 2019-123) and was performed in accordance with the Declaration of Helsinki.
Informed Consent Statement
Written informed consent was obtained from all patients prior to their inclusion in this study.
Data Availability Statement
The datasets presented in this article are not readily available due to ethical concerns regarding patient privacy. Requests to access the datasets should be directed to the corresponding author.
Acknowledgments
We thank Tokyo University Hospital, Keio University Hospital, Hachinohe City Hospital, Yokohama Municipal Hospital, and Jikei University Hospital for lending the FFPE samples.
Conflicts of Interest
Yuichiro Ohe received an honoraria (lecture fees) from Eli Lilly Japan K.K. and received research grants from Eli Lilly Japan K.K.; Hidehito Horinouchi received an honoraria (lecture fees) from Eli Lilly Japan K.K.; Yasushi Goto received an honoraria (lecture fees) from Eli Lilly Japan K.K.; Noboru Yamamoto received research grants from Eli Lilly Japan K.K.; Yusuke Okuma received an honoraria (lecture fees) from Eli Lilly Japan K.K.; Yuki Shinno received a honoraria (lecture fees) from Eli Lilly; Tomohiro Tanaka, Ken Masuda, Yuji Matsumoto, Tatsuya Yoshida, Noriko Motoi, Yasushi Yatabe, Shunichi Watanabe, and Masayuki Horie have no conflict of interest.
References
- Travis, W.D.; Brambilla, E.; Burke, A.P.; Marx, A.; Nicholson, A.G. Pathology and Genetics: Tumors of the Lung, Pleura, Thymus and Heart, 4th ed.; IRCA Press: Gallarate, Italy, 2015; Volume 7. [Google Scholar]
- Hirai, F.; Toyozawa, R.; Nosaki, K.; Seto, T. Are Anthracycline-Based Regimens Truly Indicated To Be the Standard Chemotherapy Regimen for Thymic Carcinoma? J. Thorac. Oncol. 2016, 11, 115–121. [Google Scholar] [CrossRef][Green Version]
- Loehrer, P.J.; Chen, M.; Kim, K.; Aisner, S.C.; Einhorn, L.H.; Livingston, R.; Johnson, D. Cisplatin, doxorubicin, and cyclophosphamide plus thoracic radiation therapy for limited-stage unresectable thymoma: An intergroup trial. J. Clin. Oncol. 1997, 15, 3093–3099. [Google Scholar] [CrossRef]
- Villela, L.R.; Stanford, B.L.; Shah, S.R. Pemetrexed, a novel antifolate therapeutic alternative for cancer chemotherapy. Pharmacotherapy 2006, 26, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Gbolahan, O.B.; Porter, R.F.; Salter, J.T.; Yiannoutsos, C.; Burns, M.; Chiorean, E.G.; Loehrer, P.J. A Phase II Study of Pemetrexed in Patients with Recurrent Thymoma and Thymic Carcinoma. J. Thorac. Oncol. 2018, 13, 1940–1948. [Google Scholar] [CrossRef]
- Lim, S.M.; Park, H.S.; Kim, S.; Ali, S.M.; Greenbowe, J.R.; Yang, I.S.; Kwon, N.J.; Lee, J.L.; Ryu, M.H.; Ahn, J.H.; et al. Next-generation sequencing reveals somatic mutations that confer exceptional response to everolimus. Oncotarget 2016, 7, 10547–10556. [Google Scholar] [CrossRef]
- Wagle, N.; Grabiner, B.C.; Van Allen, E.M.; Hodis, E.; Jacobus, S.; Supko, J.G.; Stewart, M.; Choueiri, T.K.; Gandhi, L.; Cleary, J.M.; et al. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer Discov. 2014, 4, 546–553. [Google Scholar] [CrossRef]
- Krejci, J.; Pesek, M.; Grossmann, P.; Krejci, M.; Ricar, J.; Benesova, L.; Minarik, M. Extraordinary response to erlotinib therapy in a patient with lung adenocarcinoma exhibiting KRAS mutation and EGFR amplification. Cancer Genom. Proteom. 2011, 8, 135–138. [Google Scholar]
- Lim, S.M.; Kim, E.; Jung, K.H.; Kim, S.; Koo, J.S.; Kim, S.I.; Park, S.; Park, H.S.; Park, B.W.; Cho, Y.U.; et al. Genomic landscape of extraordinary responses in metastatic breast cancer. Commun. Biol. 2021, 4, 449. [Google Scholar] [CrossRef]
- Gray, H.J.; Chatterjee, P.; Rosati, R.; Appleyard, L.R.; Durenberger, G.J.; Diaz, R.L.; Swan, H.A.; Peretti, D.; Pollastro, M.; Ainge, T.; et al. Extraordinary clinical response to ibrutinib in low-grade ovarian cancer guided by organoid drug testing. NPJ Precis. Oncol. 2023, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Jiang, Z.; Liu, J.; Haverty, P.M.; Guan, Y.; Stinson, J.; Yue, P.; Zhang, Y.; Pant, K.P.; Bhatt, D.; et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature 2010, 465, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.J.; Ding, L.; Shen, D.; Luo, J.; Suman, V.J.; Wallis, J.W.; Van Tine, B.A.; Hoog, J.; Goiffon, R.J.; Goldstein, T.C.; et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 2012, 486, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Network, C.G.A. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Petrini, I.; Meltzer, P.S.; Kim, I.K.; Lucchi, M.; Park, K.S.; Fontanini, G.; Gao, J.; Zucali, P.A.; Calabrese, F.; Favaretto, A.; et al. A specific missense mutation in GTF2I occurs at high frequency in thymic epithelial tumors. Nat. Genet. 2014, 46, 844–849. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Lei, Y.; Wu, X.; Huang, Y.; Rao, H.; Zhang, Y.; Wang, F. GTF2I mutation frequently occurs in more indolent thymic epithelial tumors and predicts better prognosis. Lung Cancer 2017, 110, 48–52. [Google Scholar] [CrossRef]
- Belani, R.; Oliveira, G.; Erikson, G.A.; Ra, S.; Schechter, M.S.; Lee, J.K.; Shipman, W.J.; Haaser, S.M.; Torkamani, A. ASXL1 and DNMT3A mutation in a cytogenetically normal B3 thymoma. Oncogenesis 2014, 3, e111. [Google Scholar] [CrossRef] [PubMed]
- Girard, N.; Shen, R.; Guo, T.; Zakowski, M.F.; Heguy, A.; Riely, G.J.; Huang, J.; Lau, C.; Lash, A.E.; Ladanyi, M.; et al. Comprehensive genomic analysis reveals clinically relevant molecular distinctions between thymic carcinomas and thymomas. Clin. Cancer Res. 2009, 15, 6790–6799. [Google Scholar] [CrossRef]
- Girard, N.; Basse, C.; Schrock, A.; Ramkissoon, S.; Killian, K.; Ross, J.S. Comprehensive Genomic Profiling of 274 Thymic Epithelial Tumors Unveils Oncogenic Pathways and Predictive Biomarkers. Oncologist 2022, 27, 919–929. [Google Scholar] [CrossRef]
- Inoue, M.; Marx, A.; Zettl, A.; Ströbel, P.; Müller-Hermelink, H.K.; Starostik, P. Chromosome 6 suffers frequent and multiple aberrations in thymoma. Am. J. Pathol. 2002, 161, 1507–1513. [Google Scholar] [CrossRef]
- Zettl, A.; Ströbel, P.; Wagner, K.; Katzenberger, T.; Ott, G.; Rosenwald, A.; Peters, K.; Krein, A.; Semik, M.; Müller-Hermelink, H.K.; et al. Recurrent genetic aberrations in thymoma and thymic carcinoma. Am. J. Pathol. 2000, 157, 257–266. [Google Scholar] [CrossRef]
- Qian, X.; Song, Z. Efficacy of pemetrexed-based regimen in relapsed advanced thymic epithelial tumors and its association with thymidylate synthetase level. OncoTargets Ther. 2016, 9, 4527–4531. [Google Scholar] [CrossRef]
- Bai, Y.; Ma, X.; Zhang, Y.H. Systematic review and meta-analysis of the predictive power of MTHFR polymorphisms for pemetrexed drug efficacy and toxicity in non-small cell lung cancer patients. J. Chemother. 2021, 34, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Visser, S.; Hou, J.; Bezemer, K.; de Vogel, L.L.; Hegmans, J.P.J.J.; Stricker, B.H.; Philipsen, S.; Aerts, J.G.J.V. Prediction of response to pemetrexed in non-small-cell lung cancer with immunohistochemical phenotyping based on gene expression profiles. BMC Cancer 2019, 19, 440. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Oguri, T.; Kunii, E.; Sone, K.; Uemura, T.; Takakuwa, O.; Maeno, K.; Kanemitsu, Y.; Ohkubo, H.; Takemura, M.; et al. A folylpoly-γ-glutamate synthase single nucleotide polymorphism associated with response to pemetrexed treatment combined with platinum for non-small cell lung cancer. Lung Cancer 2016, 102, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Shan, F.; Liu, Y.L.; Wang, Q.; Shi, Y.L. Thymidylate synthase predicts poor response to pemetrexed chemotherapy in patients with advanced breast cancer. Oncol. Lett. 2018, 16, 3274–3280. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.T.; Chien, P.J.; Chen, S.H.; Sheu, G.T.; Jan, M.S.; Wang, B.Y.; Chang, W.W. BMI1-Mediated Pemetrexed Resistance in Non-Small Cell Lung Cancer Cells Is Associated with Increased SP1 Activation and Cancer Stemness. Cancers 2020, 12, 2069. [Google Scholar] [CrossRef]
- Qin, Y.; Sekine, I.; Hanazono, M.; Morinaga, T.; Fan, M.; Takiguchi, Y.; Tada, Y.; Shingyoji, M.; Yamaguchi, N.; Tagawa, M. AMPK activation induced in pemetrexed-treated cells is associated with development of drug resistance independently of target enzyme expression. Mol. Oncol. 2019, 13, 1419–1432. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Z.; Gao, S.; Wu, K.; Bai, F.; Zhang, Q.; Wang, H.; Ye, Q.; Xu, F.; Sun, H.; et al. Correction to: Human drug efflux transporter ABCC5 confers acquired resistance to pemetrexed in breast cancer. Cancer Cell. Int. 2021, 21, 183. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Talevich, E.; Shain, A.H.; Botton, T.; Bastian, B.C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput. Biol. 2016, 12, e1004873. [Google Scholar] [CrossRef]
- Kanda, Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013, 48, 452–458. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Martins, F.C.; Couturier, D.L.; de Santiago, I.; Sauer, C.M.; Vias, M.; Angelova, M.; Sanders, D.; Piskorz, A.; Hall, J.; Hosking, K.; et al. Clonal somatic copy number altered driver events inform drug sensitivity in high-grade serous ovarian cancer. Nat. Commun. 2022, 13, 6360. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Han, X.; Zheng, G.; Yang, Y.; Li, Y.; Zhang, E.; Yang, L.; Dong, M.; He, D.; He, J.; et al. The adverse impact of a gain in chromosome 1q on the prognosis of multiple myeloma treated with bortezomib-based regimens: A retrospective single-center study in China. Front. Oncol. 2022, 12, 1084683. [Google Scholar] [CrossRef]
- Petrini, I.; Rajan, A.; Pham, T.; Voeller, D.; Davis, S.; Gao, J.; Wang, Y.; Giaccone, G. Whole genome and transcriptome sequencing of a B3 thymoma. PLoS ONE 2013, 8, e60572. [Google Scholar] [CrossRef] [PubMed]
- Petrini, I.; Meltzer, P.S.; Zucali, P.A.; Luo, J.; Lee, C.; Santoro, A.; Lee, H.S.; Killian, K.J.; Wang, Y.; Tsokos, M.; et al. Copy number aberrations of BCL2 and CDKN2A/B identified by array-CGH in thymic epithelial tumors. Cell. Death Dis. 2012, 3, e351. [Google Scholar] [CrossRef]
- Radovich, M.; Pickering, C.R.; Felau, I.; Ha, G.; Zhang, H.; Jo, H.; Hoadley, K.A.; Anur, P.; Zhang, J.; McLellan, M.; et al. The Integrated Genomic Landscape of Thymic Epithelial Tumors. Cancer Cell. 2018, 33, 244–258.210. [Google Scholar] [CrossRef]
- Xu, J.F.; Kang, Q.; Ma, X.Y.; Pan, Y.M.; Yang, L.; Jin, P.; Wang, X.; Li, C.G.; Chen, X.C.; Wu, C.; et al. A Novel Method to Detect Early Colorectal Cancer Based on Chromosome Copy Number Variation in Plasma. Cell. Physiol. Biochem. 2018, 45, 1444–1454. [Google Scholar] [CrossRef]
- Yu, D.; Liu, Z.; Su, C.; Han, Y.; Duan, X.; Zhang, R.; Liu, X.; Yang, Y.; Xu, S. Copy number variation in plasma as a tool for lung cancer prediction using Extreme Gradient Boosting (XGBoost) classifier. Thorac. Cancer 2020, 11, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.M.; Barwick, B.G.; Joseph, N.; Heffner, L.T.; Hofmeister, C.C.; Bernal, L.; Dhodapkar, M.V.; Gupta, V.A.; Jaye, D.L.; Wu, J.; et al. Gain of Chromosome 1q is associated with early progression in multiple myeloma patients treated with lenalidomide, bortezomib, and dexamethasone. Blood Cancer J. 2019, 9, 94. [Google Scholar] [CrossRef] [PubMed]
- Rajeshwari, M.; Sharma, M.C.; Kakkar, A.; Nambirajan, A.; Suri, V.; Sarkar, C.; Singh, M.; Saran, R.K.; Gupta, R.K. Evaluation of chromosome 1q gain in intracranial ependymomas. J. Neurooncol. 2016, 127, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Gratias, E.J.; Jennings, L.J.; Anderson, J.R.; Dome, J.S.; Grundy, P.; Perlman, E.J. Gain of 1q is associated with inferior event-free and overall survival in patients with favorable histology Wilms tumor: A report from the Children’s Oncology Group. Cancer 2013, 119, 3887–3894. [Google Scholar] [CrossRef] [PubMed]
- Robb, T.J.; Ward, Z.; Houseman, P.; Woodhouse, B.; Patel, R.; Fitzgerald, S.; Tsai, P.; Lawrence, B.; Parker, K.; Print, C.G.; et al. Chromosomal Aberrations Accumulate during Metastasis of Virus-Negative Merkel Cell Carcinoma. J. Investig. Dermatol. 2023, 143, 1168–1177.e1162. [Google Scholar] [CrossRef]
- McEvoy, C.R.; Morley, A.A.; Firgaira, F.A. Evidence for whole chromosome 6 loss and duplication of the remaining chromosome in acute lymphoblastic leukemia. Genes. Chromosomes Cancer 2003, 37, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Persson, F.; Fehr, A.; Sundelin, K.; Schulte, B.; Löning, T.; Stenman, G. Studies of genomic imbalances and the MYB-NFIB gene fusion in polymorphous low-grade adenocarcinoma of the head and neck. Int. J. Oncol. 2012, 40, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Pedersen-Bjergaard, J.; Pedersen, M.; Roulston, D.; Philip, P. Different genetic pathways in leukemogenesis for patients presenting with therapy-related myelodysplasia and therapy-related acute myeloid leukemia. Blood 1995, 86, 3542–3552. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, K.; Wang, X.; Liu, Y.; Guo, D.; Bian, Z.; Su, L.; Liu, K.; Gu, X.; Guo, X.; et al. Multiple-level copy number variations in cell-free DNA for prognostic prediction of HCC with radical treatments. Cancer Sci. 2021, 112, 4772–4784. [Google Scholar] [CrossRef]
- Chiang, J.; Moreira, D.C.; Li, X.; Furtado, L.V. Prognostic significance of chromosome arm 1q gain and methylation class in molecularly defined diffuse leptomeningeal glioneuronal tumor. Acta Neuropathol. 2022, 144, 1185–1187. [Google Scholar] [CrossRef]
- Aalto, Y.; Eriksson, L.; Seregard, S.; Larsson, O.; Knuutila, S. Concomitant loss of chromosome 3 and whole arm losses and gains of chromosome 1, 6, or 8 in metastasizing primary uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2001, 42, 313–317. [Google Scholar]
- Thomson, J.P.; Hollis, R.L.; van Baal, J.; Ilenkovan, N.; Churchman, M.; van de Vijver, K.; Dijk, F.; Meynert, A.M.; Bartos, C.; Rye, T.; et al. Whole exome sequencing of low grade serous ovarian carcinoma identifies genomic events associated with clinical outcome. Gynecol. Oncol. 2023, 174, 157–166. [Google Scholar] [CrossRef]
- Hattori, N.; Hirose, Y.; Sasaki, H.; Nakae, S.; Hayashi, S.; Ohba, S.; Adachi, K.; Hayashi, T.; Nishiyama, Y.; Hasegawa, M.; et al. World Health Organization grade II-III astrocytomas consist of genetically distinct tumor lineages. Cancer Sci. 2016, 107, 1159–1164. [Google Scholar] [CrossRef]
- Takeuchi, I.; Yanagi, K.; Takada, S.; Uchiyama, T.; Igarashi, A.; Motomura, K.; Hayashi, Y.; Nagano, N.; Matsuoka, R.; Sugiyama, H.; et al. STAT6 gain-of-function variant exacerbates multiple allergic symptoms. J. Allergy Clin. Immunol. 2023, 151, 1402–1409.e1406. [Google Scholar] [CrossRef]
- Zhou, Y.; Jiang, W.; Yang, N.; Zhang, Y. Concurrent ERBB2 missense mutations D769Y and D742 N Are Novel Acquired Mechanism of Gefitinib Resistance but Responds to Gefitinib plus Pyrotinib. Lung Cancer 2020, 144, 90–91. [Google Scholar] [CrossRef] [PubMed]
- Cousin, M.A.; Zimmermann, M.T.; Mathison, A.J.; Blackburn, P.R.; Boczek, N.J.; Oliver, G.R.; Lomberk, G.A.; Urrutia, R.A.; Deyle, D.R.; Klee, E.W. Functional validation reveals the novel missense V419L variant in. Cold Spring Harb. Mol. Case Stud. 2017, 3, a001727. [Google Scholar] [CrossRef] [PubMed]
- Güemes, M.; Martín-Rivada, Á.; Ortiz-Cabrera, N.V.; Martos-Moreno, G.; Pozo-Román, J.; Argente, J. LZTR1: Genotype Expansion in Noonan Syndrome. Horm. Res. Paediatr. 2019, 92, 269–275. [Google Scholar] [CrossRef]
- Verrijn Stuart, A.A.; Ozisik, G.; de Vroede, M.A.; Giltay, J.C.; Sinke, R.J.; Peterson, T.J.; Harris, R.M.; Weiss, J.; Jameson, J.L. An amino-terminal DAX1 (NROB1) missense mutation associated with isolated mineralocorticoid deficiency. J. Clin. Endocrinol. Metab. 2007, 92, 755–761. [Google Scholar] [CrossRef]
- Puffenberger, E.G.; Jinks, R.N.; Wang, H.; Xin, B.; Fiorentini, C.; Sherman, E.A.; Degrazio, D.; Shaw, C.; Sougnez, C.; Cibulskis, K.; et al. A homozygous missense mutation in HERC2 associated with global developmental delay and autism spectrum disorder. Hum. Mutat. 2012, 33, 1639–1646. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Zhang, H.; Su, Y.; Wen, Z.; Zhu, Z.; Chen, G.; Peng, L.; Du, C.; Xie, H.; Li, H.; et al. Identification of two novel PCDHA9 mutations associated with Hirschsprung’s disease. Gene 2018, 658, 96–104. [Google Scholar] [CrossRef]
- Mouden, C.; de Tayrac, M.; Dubourg, C.; Rose, S.; Carré, W.; Hamdi-Rozé, H.; Babron, M.C.; Akloul, L.; Héron-Longe, B.; Odent, S.; et al. Homozygous STIL mutation causes holoprosencephaly and microcephaly in two siblings. PLoS ONE 2015, 10, e0117418. [Google Scholar] [CrossRef]
- Kowalik, A.; Karpinski, P.; Markiewicz, A.; Orlowska-Heitzman, J.; Romanowska-Dixon, B.; Donizy, P.; Hoang, M.P. Molecular profiling of primary uveal melanoma: Results of a Polish cohort. Melanoma Res. 2023, 33, 104–115. [Google Scholar] [CrossRef]
- Bilusic, M.; Girardi, D.; Zhou, Y.; Jung, K.; Pei, J.; Slifker, M.; Chen, Q.; Meerzaman, D.; Alpaugh, K.; Young, D.; et al. Molecular Profiling of Exceptional Responders to Cancer Therapy. Oncologist 2021, 26, 186–195. [Google Scholar] [CrossRef]
- Zaman, T.; Helbig, K.L.; Clatot, J.; Thompson, C.H.; Kang, S.K.; Stouffs, K.; Jansen, A.E.; Verstraete, L.; Jacquinet, A.; Parrini, E.; et al. SCN3A-Related Neurodevelopmental Disorder: A Spectrum of Epilepsy and Brain Malformation. Ann. Neurol. 2020, 88, 348–362. [Google Scholar] [CrossRef] [PubMed]
- Anuraga, G.; Tang, W.C.; Phan, N.N.; Ta, H.D.K.; Liu, Y.H.; Wu, Y.F.; Lee, K.H.; Wang, C.Y. Comprehensive Analysis of Prognostic and Genetic Signatures for General Transcription Factor III (GTF3) in Clinical Colorectal Cancer Patients Using Bioinformatics Approaches. Curr. Issues Mol. Biol. 2021, 43, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, M.; Hamanoue, S.; Goto, H.; Saito, T.; Nagai, J.I.; Masuno, M.; Umeda, Y.; Kurosawa, K. Diamond-Blackfan anemia caused by chromosome 1p22 deletion encompassing. Hum. Genome Var. 2019, 6, 36. [Google Scholar] [CrossRef]
- Wheeler, D.A.; Takebe, N.; Hinoue, T.; Hoadley, K.A.; Cardenas, M.F.; Hamilton, A.M.; Laird, P.W.; Wang, L.; Johnson, A.; Dewal, N.; et al. Molecular Features of Cancers Exhibiting Exceptional Responses to Treatment. Cancer Cell. 2021, 39, 38–53.e37. [Google Scholar] [CrossRef] [PubMed]
- Górska, A.A.; Sandmann, C.; Riechert, E.; Hofmann, C.; Malovrh, E.; Varma, E.; Kmietczyk, V.; Ölschläger, J.; Jürgensen, L.; Kamuf-Schenk, V.; et al. Muscle-specific Cand2 is translationally upregulated by mTORC1 and promotes adverse cardiac remodeling. EMBO Rep. 2021, 22, e52170. [Google Scholar] [CrossRef]
- Dong, S.S.; Yao, S.; Chen, Y.X.; Guo, Y.; Zhang, Y.J.; Niu, H.M.; Hao, R.H.; Shen, H.; Tian, Q.; Deng, H.W.; et al. Detecting epistasis within chromatin regulatory circuitry reveals CAND2 as a novel susceptibility gene for obesity. Int. J. Obes. 2019, 43, 450–456. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).