1. Introduction
The cellular mechanisms leading to cancer in an individual are heterogeneous, nuanced, and not well understood. It is well appreciated that cancer is a disease of aberrant signaling, and the state of a cancer cell can be described in terms of abnormally functioning cellular signaling pathways. Precision oncology depends on the ability to identify the abnormal cellular signaling pathways causing a patient’s cancer, so that patient-specific effective treatments can be prescribed—including targeting multiple abnormal pathways during a treatment regime. Aberrant signaling in cancer cells usually results from somatic genomic alterations (SGAs) that perturb the function of signaling proteins. Although large-scale cancer genomic data are available, such as those from The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC), it remains a very difficult and unsolved task to reliably infer how the SGAs in a cancer cell cause aberrations in cellular signaling pathways based on the genomic data of a tumor. One challenge is that the majority of the genomic alterations observed in a tumor are non-consequential (
passenger genomic alterations) with respect to cancer development and only a few are
driver genomic alterations, i.e., genomic alterations that cause cancer. Furthermore, even if the driver genomic alterations of a tumor are known, it remains challenging to infer how aberrant signals of perturbed proteins affect the cellular system of cancer cells, because the states of signaling proteins (or pathways) in the signaling system are not measured (latent). This requires one to study the causal relationships among latent (i.e., hidden or unobserved) variables, which represent the state of individual signaling proteins, protein complexes, or certain biological processes within a cell, in addition to the observed variables in order to understand the disease mechanisms of an individual tumor and identify drug targets. Most causal discovery algorithms have been developed to find the causal structure and the parameterization of the causal structure relative to the
observed variables of a dataset [
1,
2,
3,
4,
5,
6]. Only a small number of causal discovery algorithms also find the latent causal structure [
7,
8,
9,
10,
11]. Interestingly, Xie et al. [
11] and Huang et al. [
10] developed algorithms to learn hierarchical structures among latent variables based on compositional statistical structures. However, due to the large search space, these algorithms are not suitable for handling high-dimension data, which we deal with in our study.
Deep learning represents a group of machine learning strategies, based on neural networks, that learn a function mapping inputs to outputs. The signals of input variables are processed and transformed with many hidden layers of latent variables (i.e., hidden nodes) [
12,
13,
14]. These hidden layers learn hierarchical or compositional statistical structures, meaning that different hidden layers capture structures of different degrees of complexity [
9,
15,
16,
17]. Researchers have previously shown that deep learning models can represent the hierarchical organization of signaling molecules in a cell [
18,
19,
20,
21,
22], with latent variables as natural representations of unobserved activation states of signaling molecules (e.g., membrane receptors or transcription factors). However, deep learning models have not been broadly used as tools to infer
causal relationships in a computational biology setting, partly due to deep learning’s “black box” nature.
Relevant to the work presented in this paper, here, we briefly describe some of the studies that used neural-network-based approaches to discover gene regulatory networks (GRNs). A study published in 1999 by Weaver and Stormo [
23] modeled the relationships in gene regulatory networks as coefficients in weight matrices (using the familiar concepts of weighted sums and an activation function in their model). However, they only tested their algorithm on simulated time-series data, as large gene expression datasets were not yet available and performing these calculations on large numbers of data was quite challenging at the time. Similarly, Vohradsky [
24] and Keedwell et al. [
25] also interpreted the weights of a quasi-recurrent neural network as relationships in gene regulatory networks. Again, simulated time-series data were used in these studies with reasonable results. A more recent study published in 2015 [
26] used a linear classifier with one input “layer” and one output “layer” (which they called a neural network) to infer regulatory relationships (as represented by the weights of the weight matrix in the linear classifier) among genes in lung adenocarcinoma gene expression data. However, they did not evaluate their learned regulatory network, did not use DNA mutation data (as we do in this work), and did not use neural networks. There have also been studies that used genetic algorithms (evolving a weight matrix of regulatory pathways) to infer gene regulatory networks [
27]. Like the studies above, this work was ahead of its time, and they were only able to test their algorithm on simulated expression data. Improving upon Ando and Iba [
27], Keedwell and Narayanan [
28] used the weights representing a single-layer artificial neural network (ANN) (trained with gradient descent) to represent regulatory relationships among genes in gene expression data. Interestingly, they also used a genetic algorithm for a type of feature selection to make the task more tractable. They achieved good results on simulated temporal data and also tested their method on real temporal expression data (112 genes over nine time points) but did not have ground truth for comparison. In another study, Narayanan et al. [
29] used single-layer ANNs to evaluate real, non-temporal gene expression data in a classification setting (i.e., they did not recover GRNs). However, the high dimensionality of the problem was again a major limiting factor.
Many of the studies discussed above used the weights of a neural network to represent relationships in gene regulatory networks. However, it does not appear that these studies attempted to use a neural network to identify latent causal structures at different hierarchical levels (i.e., cellular signaling system), as we do in this work. In general, the above studies used very basic versions of neural network (without regularization limiting the magnitude of the weights and thereby the complexity of the learned function) and, due to computing constraints and the absence of large genomic datasets, were unable to train their networks on high-dimension data. Also, most of the methods above require temporal data, as opposed to the static genomic data that we utilize here. Overall, none of the studies discussed above used a deep neural network (DNN) to predict expression data from genomic alteration data and then recover causal relationships in the weights of a DNN, as we do in this paper.
More recent work from our group used deep learning to simulate cellular signaling systems that were shared by human and rat cells [
18] and to recover components of the yeast cellular signaling system, including transcription factors [
19]. These studies utilized unsupervised learning methods, in contrast to the supervised methods used in this study. Also, the previous studies by our group did not attempt to find causal relationships representing the cellular signaling system.
In a recent work [
9], we developed a deep learning algorithm, named redundant-input neural network (RINN), to learn causal relationships among latent variables from data inspired by cellular signaling pathways. The RINN solves the problem where a set of input variables
cause the change in another set of output variables, and this causal interaction is mediated by a set of an unknown number of latent variables. The constraint of inputs causing outputs is necessary to interpret the latent structure as causal relationships (see [
9] for more details regarding the causal assumptions of the RINN). A key innovation of the RINN model is that it is a partially transparent model, allowing input variables to directly interact with all latent variables in its hierarchy. This allows the RINN to constrain (in conjunction with
regularization) an input variable to be connected to a set of latent variables that can sufficiently encode the impact of the input variable on the output variables. In Young et al. [
9], we showed that the RINN outperformed other algorithms, including neural-network-based algorithms and a causal discovery algorithm known as DM (Detect MIMIC (Multiple Indicators, Multiple Input Causes)), in identifying latent causal structures in various types of simulated data.
In the current study, we took advantage of the partially transparent nature of the RINN model and used the model to learn a representation of the cancer cellular signaling system. In this setting, we interpreted the cellular signaling system as a hierarchical causal model of interactions among the activation states of proteins or protein complexes within a cell. Based on the assumption that the somatic genome alterations (SGAs) that drive the development of a cancer often influence gene expression, we trained the RINN on a large number of tumors from TCGA using tumor SGAs as inputs to predict cancer differentially expressed genes (DEGs) (outputs). We then evaluated the latent structure learned with the hidden layers of the RINN in an attempt to learn components of the cancer cellular signaling system. We show that the model is capable of detecting the shared functional impact of SGAs affecting members of a common pathway. We also show that the RINN can capture cancer signaling pathway relationships within its hidden variables.
3. Discussion
In this study, we show that deep learning models, RINNs and DNNs, can capture the statistical relationships between genomic alterations and transcriptomic events in tumor cells with reasonably high accuracy, despite the small number of training cases relative to the high dimensionality of the data. Our findings further indicate that a regularized deep learning model with redundant inputs (i.e., RINN) can capture cancer signaling pathway relationships within its hidden variables and weights. The RINN models correctly captured much of the functional similarity among SGAs that perturb a common signaling pathway, as reflected by the SGAs’ similar interactions with the hidden nodes of the RINN models (i.e., cosine similarity of SGA weight signatures). This shows that SGAs in the same pathway share similar interactions (in terms of connection and weights) with a set of latent variables. These are very encouraging results for eventually using a future version of the RINN to find signaling pathways robustly. Many of the most well-known cancer driver genes (EGFR, TP53, CDKN2A, APC, and PIK3CA) were found to have dense SGA weight signatures and weights with larger values relative to the other genes we analyzed, reinforcing the importance of these genes in driving cancer gene expression and the validity of our models. Our results indicate that an RINN consistently employs certain hidden nodes to represent the shared functional impact of SGAs perturbing a common pathway, although different instantiations of the RINN could use totally different hidden nodes. The ability of an RINN to explicitly connect SGAs and hidden nodes throughout the latent hierarchy essentially makes the RINN a partially transparent deep learning model, so that one can interpret which hidden nodes encode and transmit the signals (i.e., functional impact) of SGAs in cancer cells. Finally, we show that RINNs are capable of capturing some causal relationships (given our interpretation of the hidden nodes) among the signaling proteins perturbed by SGAs. All these results indicate that by allowing SGAs to directly interact with hidden nodes in a deep learning model, the RINN model provides constraints, information, and flexibility to enable certain hidden nodes to encode the impact of specific SGAs.
Overall, both RINNs and DNNs are capable of capturing statistical relationships between SGAs and DEGs. However, latent variables in a DNN model (except those directly connected to SGAs) are less interpretable because the latent variable information deeper in the network is more convoluted. In a DNN model, all SGAs have to interact with the first layer of hidden variables, and their information is then propagated through the whole hierarchy of the model. In such a model, it is difficult to pinpoint how the signal of each SGA is propagated. Hidden layers in a DNN are alternate representations of all the information in the input necessary to calculate the output, whereas each hidden layer of an RINN does not need to capture all the information in the input necessary to predict the output because there are multiple chances to learn what is needed from the input (i.e., redundant inputs). This difference gives RINNs more freedom in how to choose to use the information in the input. The redundant inputs of an RINN represent an attempt to deconvolute the signal of each SGA by giving the model more freedom to take advantage of the hierarchical structure and choose latent variables at the right level of granularity to encode the signal of an SGA, e.g., early in the network in the first hidden layer or in later layers in the network. This approach is biologically sensible because different SGAs do affect proteins at different levels in the hierarchy of the cellular signaling system. It is expected that an SGA perturbing a transcription factor (e.g., STAT3) impacts a relatively small number of genes in comparison to an SGA that perturbs at a high level in the signaling system (e.g., EGFR). Refined granularity enables RINNs to search for the "optimal" structure in order to encode the signaling between SGAs and DEGs while satisfying our sparsity constraints, leading to RINNs with three to four relatively sparsely connected layers of hidden variables; whereas DNNs tend to use two layers of relatively densely connected latent variables.
A DNN cannot capture the same causal relationships that an RINN can. By nature of its architecture and design, a DNN can only capture
direct causal relationships (i.e., edges starting from an observed variable) between the input and the first hidden layer—DNNs cannot capture direct causal relationships between the input and any other hidden layers. This means that a DNN cannot be used to generate causal graphs like the ones shown in
Figure 6. In addition, a DNN cannot capture causal relationships among
input SGAs as described in the Inferring Causal Relationships Using RINNs section, meaning that one cannot infer the causal relationships among SGAs with a DNN. This is because DNNs do not have edges between hidden nodes in the same layer (or redundant inputs). Let us consider
and
as the paths, with
KEAP1 and
NFE2L2 as the source nodes, respectively, in a DNN. In a DNN, to determine that there is a dependency between
KEAP1 and
NFE2L2, eventually, these two paths would need to collide on a hidden node. When these two paths collide on a hidden node, the number of edges in each path will be the same, meaning that the direction of the causal relationship is ambiguous. This limitation of DNNs can be remedied by adding redundant inputs (i.e., RINN). Using the RINN architecture allows us to infer order to the causal relationships among SGAs; this design difference and the extension of causal interpretability are what sets RINNs apart from DNNs.
It is intriguing to further examine whether the hierarchy of hidden nodes can capture causal relationships among the signals encoded by SGA-affected proteins. We have shown that many of the pathway relationships and some known causal relationships were present in the hierarchy reflected by the weight matrices of our trained models. However, we also noticed that in our RINN models, an SGA was often connected to a large number of hidden nodes, which were in turn connected to a large number of other hidden nodes—meaning that the current RINN model learns relatively dense causal graphs. While one can infer the relationships between the signal perturbed by distinct SGAs of a pathway, our current model cannot directly output a causal network that looks like those commonly shown in the literature. We plan to develop the RINN into an algorithm that is able to find more easily interpretable cellular signaling pathways when trained on SGA and DEG data. The following algorithm modifications will potentially lead to better results in the future: (1) incorporating differential regularization of the weights, (2) using constrained and parallelized versions of evolutionary algorithms to optimize the weights and avoid the need to threshold weights, and (3) training an autoencoder with a bottleneck layer to encourage hidden nodes to more easily represent biological entities and then using these weights (and architecture) to initialize an RINN.
In order to interpret the weights of a neural network as causal relationships among biological entities, we assume that the causal relationships among biological entities can be approximated with a linear function combined with a simple nonlinear function (e.g.,
), where all variables have scalar values and
represents a simple nonlinear function such as ReLU or softplus. This is a necessary assumption in order to interpret all nonzero hidden nodes as biological entities; however, it could also be the case that some hidden nodes are not biological entities but rather some intermediate calculation required to compute a relationship among biological entities that cannot be modeled with
. Given the high density of the models learned with TCGA data, it is possible that the relationships among some biological entities cannot be modeled with
, suggesting that more complex activation functions are needed or that biological entities may be present in every other hidden layer. It would be interesting to explore using more complex activation functions and specifically using an unregularized one in a hidden-layer neural network as an activation function for each hidden node in an RINN. This setup would account for even quite complex relationships among biological entities captured as latent variables. See [
9] for additional discussion of this topic.
A cellular signaling system is a complex information-encoding/-processing machine that processes signals arising from extrinsic environmental changes or perturbations (genetic or pharmacological) affecting the intrinsic state of a cell. The relationships of cellular signals are hierarchical and nonlinear by nature, and deep learning models are particularly suitable for modeling such a system [
18,
19,
20,
21,
22]. However, conventional deep learning models behave like “black boxes”, such that it is almost impossible to determine what signal a hidden node encodes, with few exceptions in image analysis, where human-interpretable image patterns can be represented with hidden nodes [
15,
16,
17]. Here, we took advantage of our knowledge of cancer biology that SGAs causally influence the transcriptomic programs of cells, and we adopted a new approach that allows SGAs to directly interact with hidden nodes in an RINN. We conjecture that this approach forces hidden nodes to explicitly and thus more effectively encode the impact of SGAs on transcriptomic systems. This hypothesis is supported by the discoveries of this paper that SGAs in a common pathway share similar connection patterns to hidden nodes and that there are hidden nodes that are connected to multiple members of a pathway in different instances of the model. Essentially, our approach also allows certain hidden nodes to be “labeled” and “partially interpretable”. An interpretable deep learning model provides a unique opportunity to study how cellular signals are encoded and perturbed under pathological conditions. Understanding and representing the state of a cellular system further opens directions for translational applications of such information, such as predicting the drug sensitivity of cancer cells based on the states of their signaling systems. To our knowledge, this is the first time that a partially interpretable deep learning model has been developed and applied to study cancer signaling, and we anticipate this approach laying a foundation for developing future explainable deep learning models in this domain.
4. Experimental Procedures
4.1. Data
The data used in this paper were originally downloaded from TCGA [
31,
32]. RNA Seq, mutation, and copy number variation (CNV) data over multiple cancer types were used to generate two binary datasets. A binary differentially expressed gene (DEG) dataset was created by comparing the expression value of a gene in a tumor against the distribution of the expression values of the gene across normal samples from the same tissue of origin. A gene was deemed a DEG in a tumor if its value was outside the 2.5% percentile on either side of the normal sample distribution; then, that gene’s value was set to 1. Otherwise, the gene’s value was set to 0. A somatic genome alteration (SGA) dataset was created using mutation and CNV data. A gene was deemed to be perturbed by an SGA event if it hosted a non-synonymous mutation, small insert/deletion, or somatic copy number alteration (deletion or amplification). If perturbed, the value in the tumor for that gene was set to 1, otherwise the value was set to 0.
We applied the tumor-specific causal inference (TCI) algorithm [
32,
33] to these two matrices to identify the SGAs that causally influence gene expression in tumors and the union of their target DEGs. TCI is an algorithm that finds causal relationships between SGAs and DEGs in each individual tumor, without determining how the signal from the SGA is propagated in the cellular signaling system [
32,
33]. We identified 372 SGAs that were deemed driver SGAs, as well as 5259 DEGs that were deemed target DEGs of the 372 SGAs using TCI. Overall, combining data from 5097 tumors led to two data matrices (with dimensions of 5097
and 5097
) as inputs and outputs for the RINN, where SGAs (inputs) were used to predict DEGs (outputs).
4.2. Deep Learning Strategies: RINN and DNN
In this study, we used two deep learning strategies: RINN and DNN. A DNN is a conventional supervised feed-forward deep neural network. A DNN learns a function mapping inputs (
x) to outputs (
y) according to
where
represent the weight matrices between layers of a neural network,
is some nonlinear function (i.e., an activation function such as ReLU, softplus, or sigmoid), · represents vector–matrix multiplication, and
is the predicted output. This function represents our predicted value for
and, in other words, represents a vector–matrix multiplication followed by the application of a nonlinear function repeated multiple times. An iterative procedure (stochastic gradient descent) is used to slowly change the values of all
to bring
closer and closer to
(hopefully). The left side of
Figure 1, without the redundant-input nodes and corresponding redundant-input weights, represents a DNN. DNNs also have bias vectors added to each layer (i.e., to each
, where
is the previous layer’s output), which are omitted in the above equation for clarity. For a more detailed explanation of DNNs, please see [
13].
We introduced the RINN for latent causal structure discovery in [
9]. An RINN is similar to a DNN with a modification of the architecture. An RINN not only has the input fully connected to the first hidden layer but also has copies of the input fully connected to each additional hidden layer (
Figure 1). This structure allows an RINN to learn direct causal relationships between an input SGA and any hidden node in the RINN. Forward propagation with an RINN is performed as it is with a DNN with multiple vector–matrix multiplications and nonlinear functions, except that an RINN has hidden layers concatenated to copies of the input (
Figure 1). Each hidden layer of an RINN with redundant inputs is calculated according to
where
represents the previous layer’s output vector,
is the vector input to the neural network,
represents concatenation into a single vector,
represent the weight matrix between hidden and redundant nodes in layer
and hidden layer
i,
is a nonlinear function (e.g., ReLU), · represents vector–matrix multiplication, and
represents the bias vector for layer
i. In contrast to an RINN, a plain DNN calculates each hidden layer as
The backpropagation of errors and stochastic gradient descent with RINNs are the same as with DNNs but with additional weights to be optimized. For much more detailed information about the RINN, we recommend that you see our paper where we introduce it [
9]. In addition to the architecture modification, all RINNs in this study importantly included
regularization of the weights as part of the objective function. The other component of the objective function was the binary cross-entropy error. All DNNs also used
regularization of weights plus binary cross-entropy error as the objective function to be optimized. All RINNs used in this study had eight hidden layers (with varying sizes of the hidden layers), as we hypothesized that cancer cellular signaling pathways do not have more than eight levels of hierarchy. However, this assumption can be easily updated if found to be false.
4.3. Model Selection
We hypothesized that since the RINN mimics the signaling processes through which the SGAs of a tumor exert their impact on gene expression (DEGs), the better an RINN model captures the relationships between SGAs and DEGs, the more closely the RINN can represent the signaling processes of cancer cells, i.e., the causal network connecting SGAs and DEGs. We performed an extensive amount of model selection to search for the structures and parameterization that best balanced loss and sparsity or, in other words, the models that fitted the data well. To this end, we performed model selection over 23,000 different sets of hyperparameters (23,000 for RINNs and 23,000 for DNNs).
Model selection was performed using 3 folds of 10-fold cross-validation. Using 3 folds of 10-fold cross-validation gave us multiple validation datasets so we avoided chance overfitting, which can occur with a single validation set. This setup also allowed us to train the models on 90% of the data (and validate them on 10%) for each split of the data, which is important, considering the small number of instances relative to the number of output DEGs that we were trying predict. Importantly, using 3 folds of 10-fold cross-validation takes significantly less time than training on all 10 folds. The running time was especially important in this study because of the large number of hyperparameter sets to be evaluated. All metrics recorded in this study (i.e., AUROC, cross-entropy error, etc.) represented the mean across the three validation sets.
The hyperparameters used in this study included learning rate, regularization rate, number of training epochs, activation function, size of hidden layers, number of hidden layers (DNN only; in RINNs, it was set to eight), and batch size. We used a combined random and grid search approach [
34,
35] to find the best sets of hyperparameters with the main objective of finding the optimal balance between sparsity and cross-entropy error, as explained in the next section and in [
9].
4.4. Ranking Models Based on Balance between Sparsity and Prediction Error
The model selection in this study was more complex than standard DNN model selection, as we needed to find a balance between the sparsity of the model (i.e., sets of weight matrices for each trained network) and prediction error. In contrast, many DNNs are trained by simply finding the model with the lowest prediction error on a hold-out dataset. As was shown in previous work by our group [
9], RINN models trained on simulated data with relatively high sparsity and low cross-entropy error were able to recover much of the latent causal structure contained within the data. We followed the same procedure as in our previous work [
9] for selecting the best models (i.e., the models with the highest chance of containing correct causal structures). In brief, we plotted prediction error versus sparsity and measured the Euclidean distance from the origin to a set of hyperparameters, i.e., a unique, trained neural network (blue circles in the model selection figure). This distance was measured according to
where
L is the validation set’s cross-entropy loss for neural network
x;
m is the number of weight matrices in neural network
x;
and
are the numbers of rows and columns in matrix
i, respectively; and
w is a scalar weight. The sets of hyperparameters were ranked according to the shortest distance from the origin (the smallest
); then, we retrained the models on all data for further analysis of the learned weights. Please see [
9] for a more detailed explanation.
4.5. AUROC and Other Metrics
Area under the receiver operating characteristics (AUROC) values were calculated for each DEG and then averaged over the three validation sets, leading to 5259 AUROC values for each classifier when plotted as a histogram of AUROC values.
k-Nearest neighbors (
kNN) was performed using sklearn’s KNeighborsClassifier [
36]). Both Euclidean and Jaccard distance metrics were evaluated, and the best
k values were 21 and 45, respectively. Other distance metrics were evaluated with results worse than those obtained using the Jaccard distance metric. For random control, we sampled predictions from a uniform distribution over the interval
for each of the validation sets. Then, we used these predictions to calculate the AUROC values for each validation set. As with the other AUROC calculations, we took the mean over the three validation sets.
When displaying a single value for a classifier’s AUROC, the AUROC values were calculated as described above; then, the mean over all DEGs was calculated. The same procedure was followed for calculating cross-entropy error, area under the precision–recall (AUPR) curve, and the sum of the absolute values of all weights. The same random predictions described above were also used to calculate cross-entropy error and AUPR.
4.6. Evaluation of Learned Relationships among SGAs
Throughout this work, the results from [
30] were used as ground truth for comparison purposes. Specifically,
Figure S1 and
Figure 2 were used as ground-truth causal relationships that we hypothesized the RINN may have been able to find.
Figure S1 is reproduced in
Supporting Information, with a minor formatting modification for convenience (
Figure S1). Of the 372 genes in our SGA dataset, there were 35 genes that overlapped with the genes in
Figure S1. Therefore, the causal relationships that we could find were limited to the relationships among these 35 genes (
Table 6).
4.7. SGA Weight Signature
To better understand how an RINN learns to connect SGAs to hidden nodes, we only analyzed the weights going from SGA nodes to hidden nodes. To accomplish this, we generated an “SGA weight signature” for each SGA, which is the concatenation, into a single vector, of all weights for a single SGA going from that SGA to all hidden nodes in all hidden layers. This can be visualized as the concatenation of the blue weights in
Figure 1 into one long vector for each SGA. The end result was an SGA weight-signature matrix of SGAs by hidden nodes. The weight signatures for a DNN were generated using only a single weight matrix, the weight matrix between the inputs and the first hidden layer (i.e.,
), as these are the only weights in a DNN that are specific to individual SGAs. Cosine similarity between SGA weight signatures was measured using the sklearn function
cosine_similarity [
36]. Hierarchical clustering using cosine similarity and average linkage was performed on the SGA weight signatures using the seaborn python module function
clustermap.
Cosine-similarity community detection figures were generated using Gephi [
37] and the cosine similarity between SGA weight signatures. Edges represented the highest or three highest cosine similarity for a given SGA. After generating a graph where nodes were SGAs and edges were the highest cosine similarity values, we ran the
Modularity function in Gephi to perform community detection.
Modularity runs an algorithm based on [
38,
39]. Next, we partitioned and colored the graph based on the community.
4.8. Visualizing an RINN as a Causal Graph
Let
, where
is a causal directed acyclic graph with vertices
V and edges
E for neural network
i and set of SGAs
j. For this work,
V represents SGAs and hidden nodes, and
E represents directed weighted edges with weight values corresponding to the weights of an RINN. The edges are directed from SGAs (input) to DEGs (output), as we know from biology that SGAs cause changes in expression. Please see [
9] for further explanation of interpreting an RINN in a causal framework. If we simply interpreted any nonzero weight in a trained RINN as an edge in
, there would be hundreds of thousands to millions of edges in the causal graph (depending on the size of the hidden layers), as
regularization encourages weight values toward zero, but weights are often not actually zero. This means that some thresholding of the weights is required [
9].
Even after selecting models based on the best balance between sparsity and error (the smallest
), our weight matrices were still very dense in terms of what could be readily visually interpreted (even after rounding weights to zero decimals). Therefore, a threshold weight value was needed to limit weight visualizations to only the largest (and, we suspect, most causally important) weights. To accomplish this, we first limited the weights to be visualized to only those that were descendants (i.e., downstream) of any of the 35 SGAs from
Figure S1. Next, for each of the top ten RINN models (the ten shortest
), we found the absolute-value weight threshold, which led to 300 edges in total, including all SGA-to-hidden and hidden-to-hidden edges. This threshold varied slightly from model to model, ranging from 0.55 to 0.71. This threshold was chosen as it seemed to give a biologically reasonable density of edges, allowed us to recover some of the relationships in
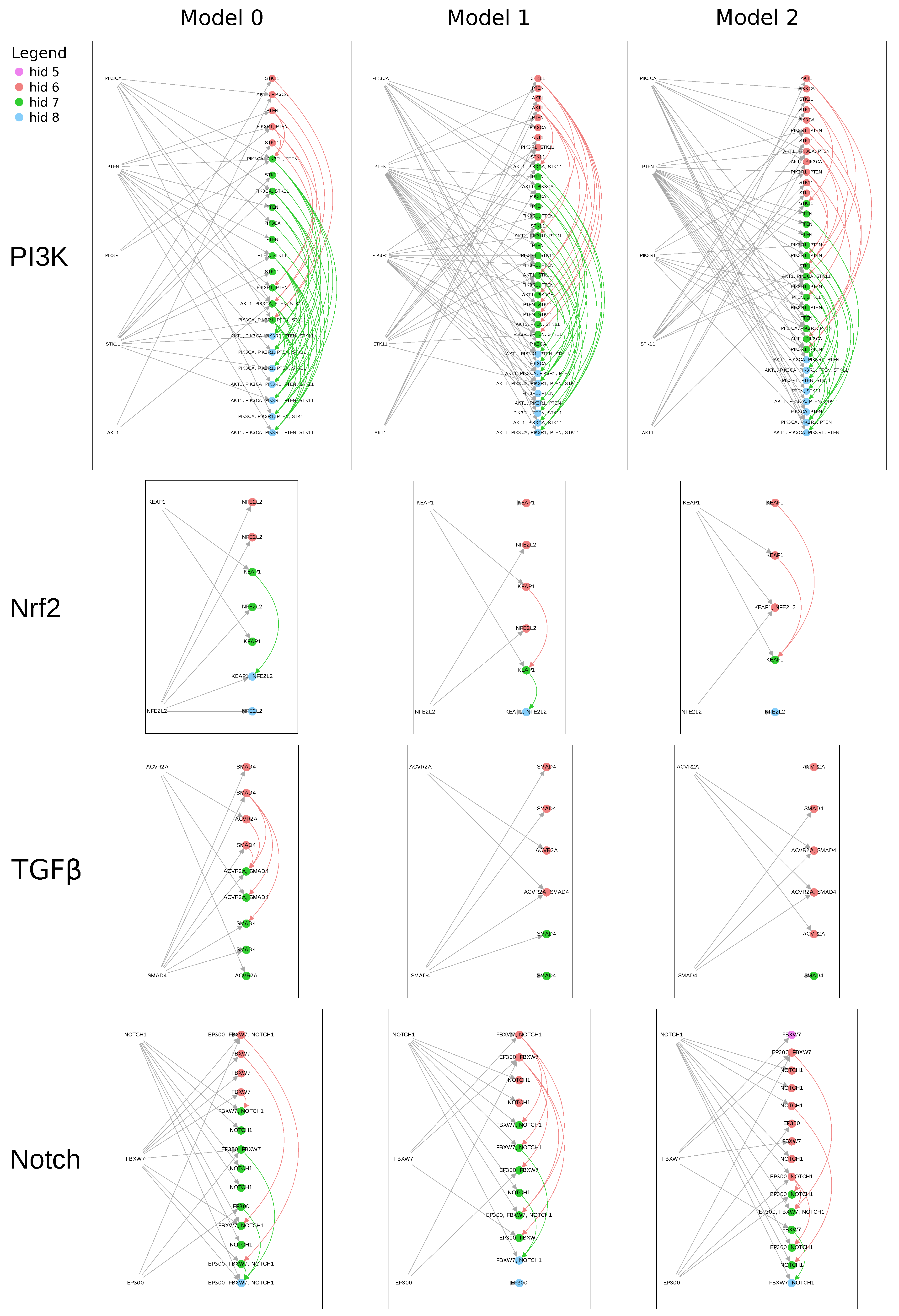
Figure S1, and was low enough to allow at least some of the causal paths to proceed from the input all the way to the output.
After finding a threshold for each model, any weight whose absolute value was greater than the threshold was added as an edge to causal graph for that model. The causal graphs were then plotted as modified bipartite graphs with SGAs on one side and all hidden nodes on the other. Hidden-to-hidden edges were included as arcing edges on the outside of the bipartite graph. We labeled the hidden nodes using a recursive algorithm that found all ancestor or upstream SGAs (i.e., on a path to that hidden node) for a given hidden node and graph .
4.9. Finding Hidden Nodes Encoding Similar Information with Respect to SGAs
To determine if hidden nodes with similar connectivity patterns with respect to the input SGAs were shared across the best models, we needed a method to map the hidden nodes to some meaningful label. To accomplish this, we used the same recursive algorithm (described at the end of the previous section) to map each hidden node in causal graph
to the set of SGAs that were ancestors of that hidden node. For this mapping, only the SGAs in set
j could be used to label a hidden node, as these were the only SGAs in
. We performed this mapping using graphs generated with set
j to only the SGAs in the individual pathways from
Figure S1 (e.g.,
for the PI3K pathway). For each model in the best ten models and each pathway in
Figure S1, we mapped hidden nodes to a set of SGAs. Next, we compared the labeled hidden nodes across models and determined the number of models that shared identically labeled hidden nodes.
We compared the number of models that shared specific hidden nodes with random controls. Random controls were performed in the same way as described above for the experimental results, except that
j was not set to the SGAs in one of the pathways in
Figure S1—rather
j was randomly selected from the set of SGAs including all 372 SGAs minus the 35 SGAs in
Figure S1. Then, ten
, one for each of the top ten RINN models, were generated using the random SGAs. The number of models with shared hidden nodes was recorded. This procedure was repeated 30 times for each possible number of SGAs in
j. For example, the PI3K pathway (in
Figure S1) has five SGAs in it that were also in our SGA dataset. To perform random control for PI3K, we performed 30 replicates of randomly selecting five SGAs (from the set of SGAs described above) and then recorded the mean number of models sharing an
n-SGA labeled hidden node, where
n is the number of SGAs that a hidden node was mapped to using our recursive algorithm for finding ancestors.