Hematopoietic Stem Cell (HSC)-Independent Progenitors Are Susceptible to Mll-Af9-Induced Leukemic Transformation
Abstract
Simple Summary
Abstract
1. Introduction
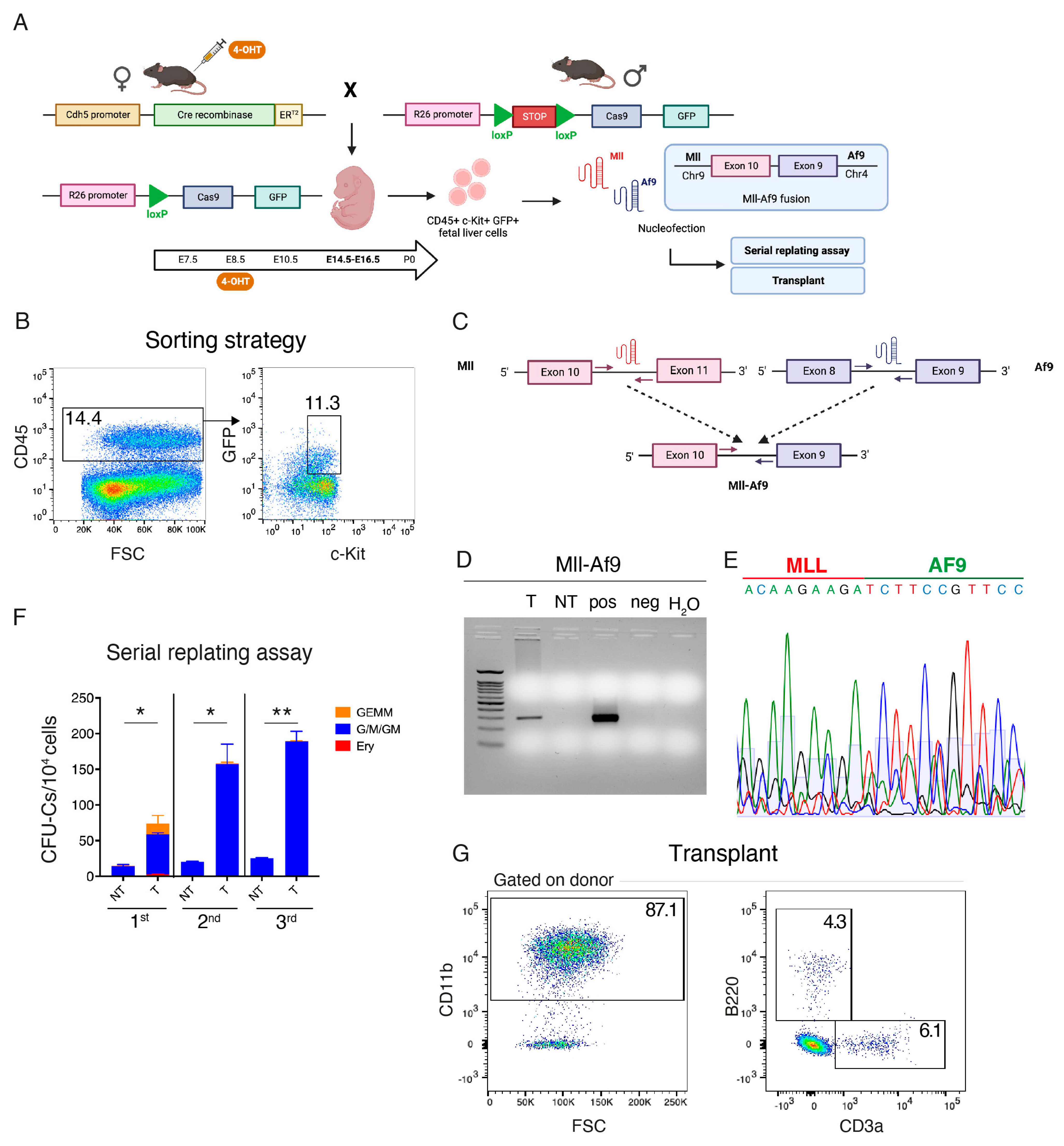
2. Materials and Methods
2.1. Mouse Strains
2.2. Polymerase Chain Reaction (PCR) for Mll-Af9 Detection and Sequencing
2.3. Flow Cytometry and Hematological Analysis
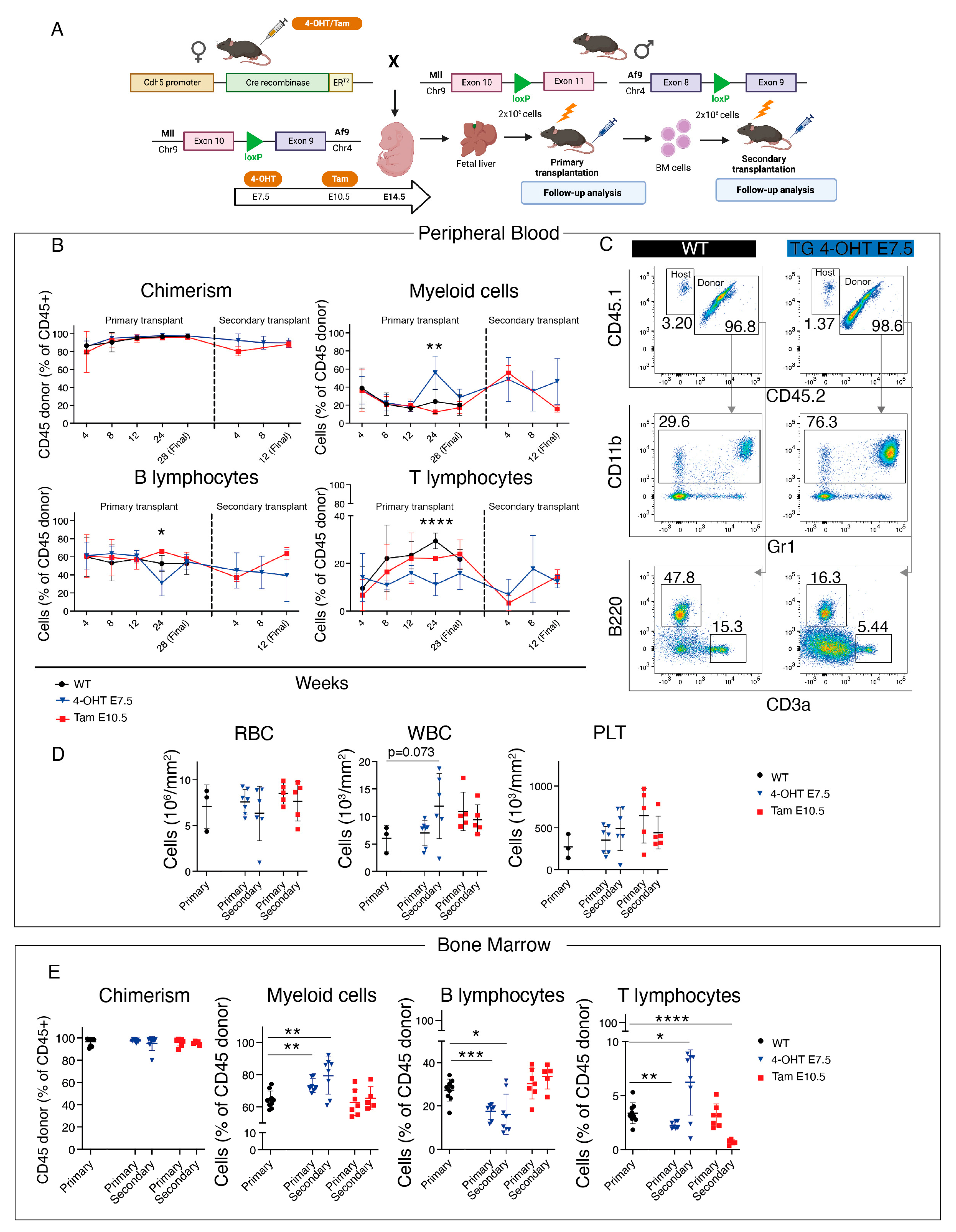
2.4. Primary and Secondary Transplants
2.5. Histological Analysis
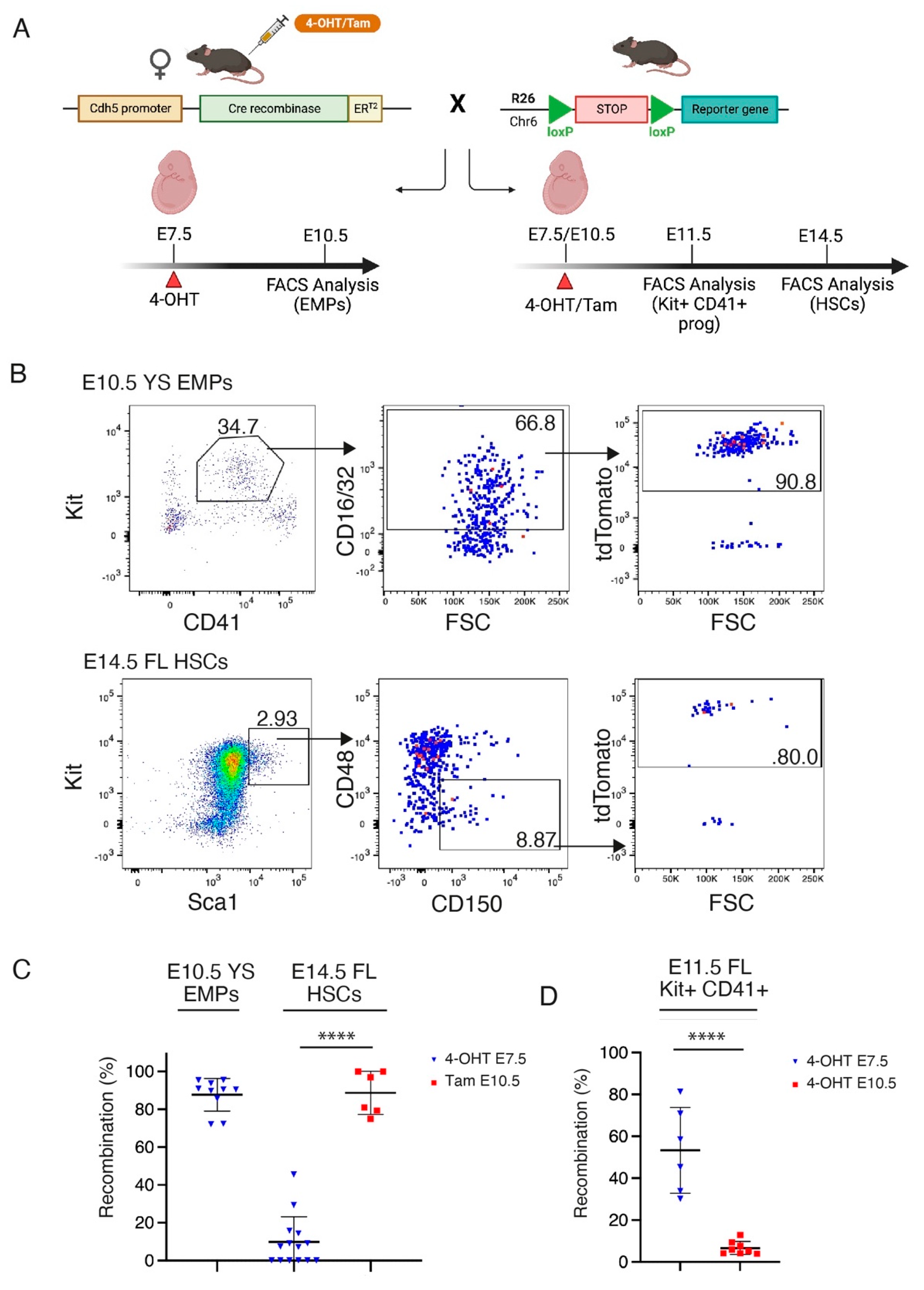
2.6. Fluorescence-Activated Cell Sorting (FACS) for the Isolation of HSC-Independent Hematopoietic Progenitors
2.7. Single Guide RNAs (sgRNA) Design and Nucleofection
2.8. CFU-C Assays
2.9. Cell Cultures
2.10. Statistical Analysis
3. Results
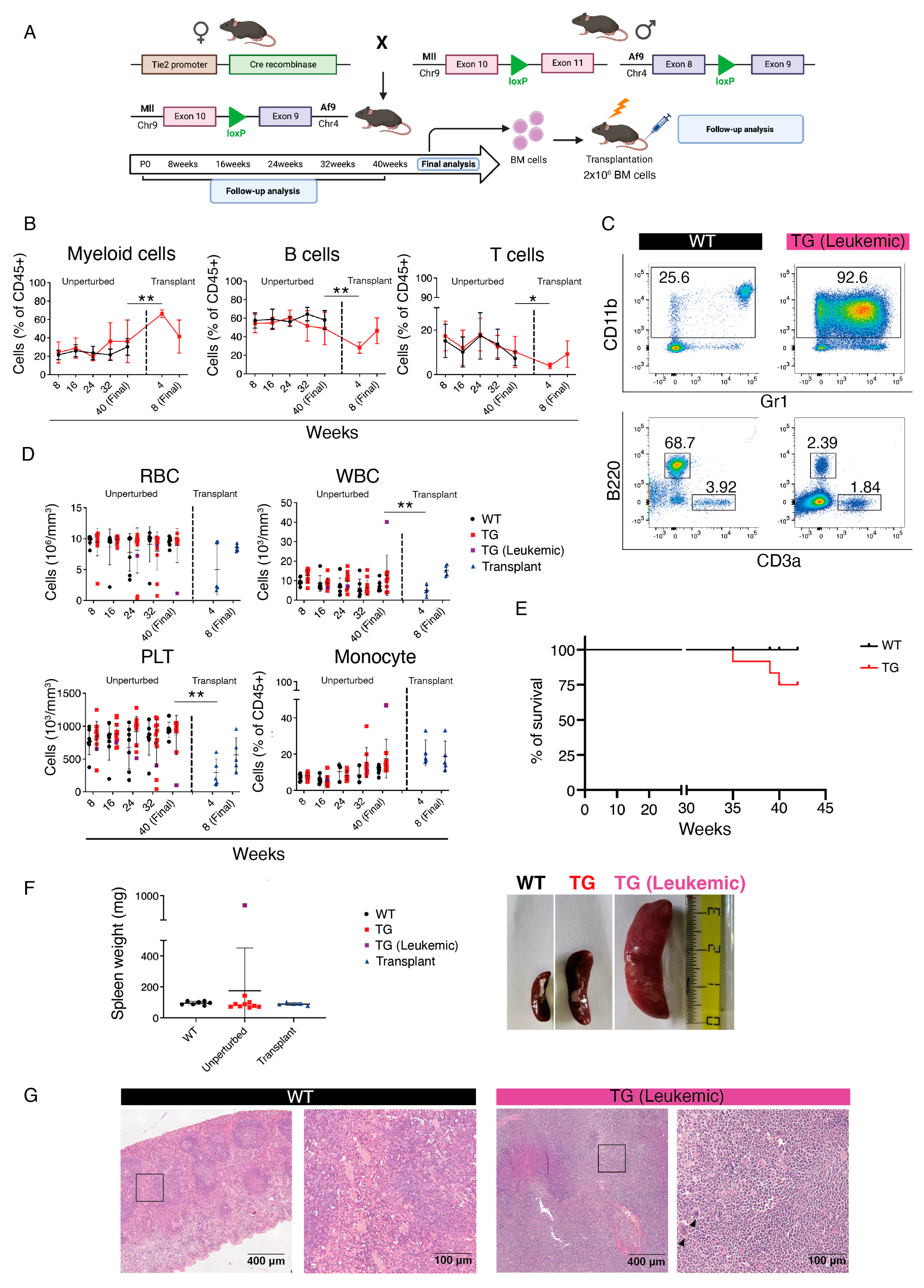
3.1. In Vivo Introduction of Mll-Af9 Inter-Chromosomal Translocation in Embryonic HSPCs Results in a Transplantable Myeloproliferative Disease
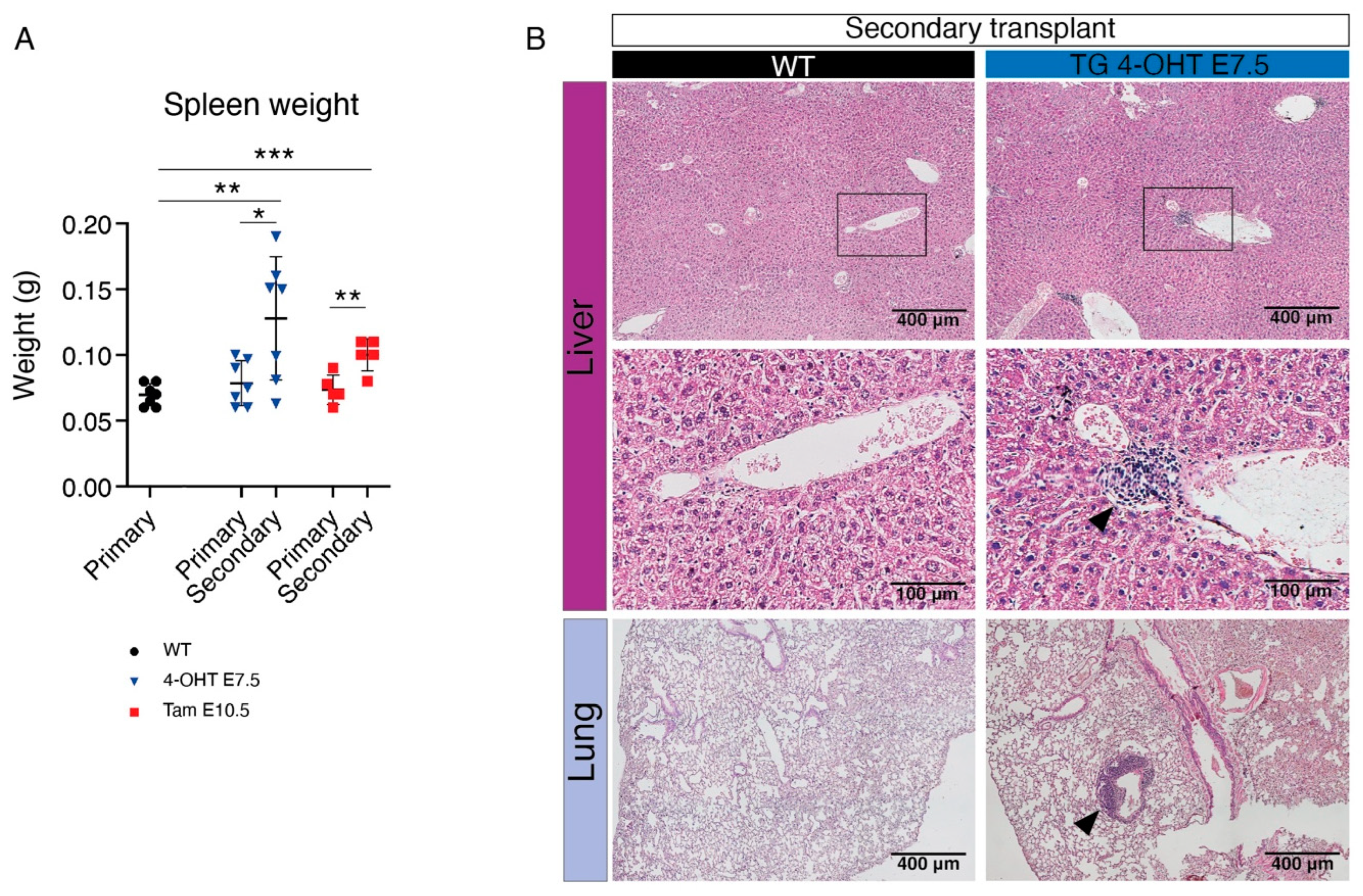
3.2. Transplantation of HSC-Independent Hematopoietic Progenitors Carrying Mll-Af9 Translocation Induces a Myeloid Neoplastic Disease
3.3. CRISPR/Cas9 Engineering of the Mll-Af9 Translocation in HSC-Independent Hematopoietic Progenitors Triggers an Aberrant Self-Renewal Program
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Biondi, A.; Cimino, G.; Pieters, R.; Pui, C.H. Biological and therapeutic aspects of infant leukemia. Blood 2000, 96, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Duguid, A.; Mattiucci, D.; Ottersbach, K. Infant leukaemia—Faithful models, cell of origin and the niche. Dis. Model. Mech. 2021, 14, dmm049189. [Google Scholar] [CrossRef] [PubMed]
- Ibrahimova, A.; Pommert, L.; Breese, E.H. Acute Leukemia in Infants. Curr. Oncol. Rep. 2021, 23, 27. [Google Scholar] [CrossRef] [PubMed]
- Marcotte, E.L.; Spector, L.G.; Mendes-de-Almeida, D.P.; Nelson, H.H. The Prenatal Origin of Childhood Leukemia: Potential Applications for Epidemiology and Newborn Screening. Front. Pediatr. 2021, 9, 639479. [Google Scholar] [CrossRef] [PubMed]
- Masetti, R.; Vendemini, F.; Zama, D.; Biagi, C.; Pession, A.; Locatelli, F. Acute myeloid leukemia in infants: Biology and treatment. Front. Pediatr. 2015, 3, 37. [Google Scholar] [CrossRef] [PubMed]
- Bolouri, H.; Farrar, J.E.; Triche, T., Jr.; Ries, R.E.; Lim, E.L.; Alonzo, T.A.; Ma, Y.; Moore, R.; Mungall, A.J.; Marra, M.A.; et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat. Med. 2018, 24, 103–112. [Google Scholar] [CrossRef]
- Cazzola, A.; Cazzaniga, G.; Biondi, A.; Meneveri, R.; Brunelli, S.; Azzoni, E. Prenatal Origin of Pediatric Leukemia: Lessons From Hematopoietic Development. Front. Cell Dev. Biol. 2020, 8, 618164. [Google Scholar] [CrossRef]
- Rice, S.; Roy, A. MLL-rearranged infant leukaemia: A ‘thorn in the side’ of a remarkable success story. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194564. [Google Scholar] [CrossRef]
- Mercher, T.; Schwaller, J. Pediatric Acute Myeloid Leukemia (AML): From Genes to Models Toward Targeted Therapeutic Intervention. Front. Pediatr. 2019, 7, 401. [Google Scholar] [CrossRef]
- Chaudhury, S.S.; Morison, J.K.; Gibson, B.E.; Keeshan, K. Insights into cell ontogeny, age, and acute myeloid leukemia. Exp. Hematol. 2015, 43, 745–755. [Google Scholar] [CrossRef]
- Horton, S.J.; Jaques, J.; Woolthuis, C.; van Dijk, J.; Mesuraca, M.; Huls, G.; Morrone, G.; Vellenga, E.; Schuringa, J.J. MLL-AF9-mediated immortalization of human hematopoietic cells along different lineages changes during ontogeny. Leukemia 2013, 27, 1116–1126. [Google Scholar] [CrossRef]
- Rowe, R.G.; Lummertz da Rocha, E.; Sousa, P.; Missios, P.; Morse, M.; Marion, W.; Yermalovich, A.; Barragan, J.; Mathieu, R.; Jha, D.K.; et al. The developmental stage of the hematopoietic niche regulates lineage in MLL-rearranged leukemia. J. Exp. Med. 2019, 216, 527–538. [Google Scholar] [CrossRef]
- Mendoza-Castrejon, J.; Magee, J.A. Layered immunity and layered leukemogenicity: Developmentally restricted mechanisms of pediatric leukemia initiation. Immunol. Rev. 2023, 315, 197–215. [Google Scholar] [CrossRef]
- Chen, W.; O’Sullivan, M.G.; Hudson, W.; Kersey, J. Modeling human infant MLL leukemia in mice: Leukemia from fetal liver differs from that originating in postnatal marrow. Blood 2011, 117, 3474–3475. [Google Scholar] [CrossRef]
- Sinha, R.; Porcheri, C.; d’Altri, T.; Gonzalez, J.; Ruiz-Herguido, C.; Rabbitts, T.; Espinosa, L.; Bigas, A. Development of embryonic and adult leukemia mouse models driven by MLL-ENL translocation. Exp. Hematol. 2020, 85, 13–19. [Google Scholar] [CrossRef]
- Okeyo-Owuor, T.; Li, Y.; Patel, R.M.; Yang, W.; Casey, E.B.; Cluster, A.S.; Porter, S.N.; Bryder, D.; Magee, J.A. The efficiency of murine MLL-ENL-driven leukemia initiation changes with age and peaks during neonatal development. Blood Adv. 2019, 3, 2388–2399. [Google Scholar] [CrossRef]
- Barone, C.; Orsenigo, R.; Meneveri, R.; Brunelli, S.; Azzoni, E. One Size Does Not Fit All: Heterogeneity in Developmental Hematopoiesis. Cells 2022, 11, 1061. [Google Scholar] [CrossRef]
- Soares-da-Silva, F.; Elsaid, R.; Peixoto, M.M.; Nogueira, G.; Pereira, P.; Bandeira, A.; Cumano, A. Assembling the layers of the hematopoietic system: A window of opportunity for thymopoiesis in the embryo. Immunol. Rev. 2023, 315, 54–70. [Google Scholar] [CrossRef]
- Soares-da-Silva, F.; Freyer, L.; Elsaid, R.; Burlen-Defranoux, O.; Iturri, L.; Sismeiro, O.; Pinto-do, O.P.; Gomez-Perdiguero, E.; Cumano, A. Yolk sac, but not hematopoietic stem cell-derived progenitors, sustain erythropoiesis throughout murine embryonic life. J. Exp. Med. 2021, 218, e20201729. [Google Scholar] [CrossRef]
- Ulloa, B.A.; Habbsa, S.S.; Potts, K.S.; Lewis, A.; McKinstry, M.; Payne, S.G.; Flores, J.C.; Nizhnik, A.; Feliz Norberto, M.; Mosimann, C.; et al. Definitive hematopoietic stem cells minimally contribute to embryonic hematopoiesis. Cell Rep. 2021, 36, 109703. [Google Scholar] [CrossRef]
- Yokomizo, T.; Ideue, T.; Morino-Koga, S.; Tham, C.Y.; Sato, T.; Takeda, N.; Kubota, Y.; Kurokawa, M.; Komatsu, N.; Ogawa, M.; et al. Independent origins of fetal liver haematopoietic stem and progenitor cells. Nature 2022, 609, 779–784. [Google Scholar] [CrossRef]
- Ema, H.; Nakauchi, H. Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood 2000, 95, 2284–2288. [Google Scholar] [CrossRef] [PubMed]
- Ganuza, M.; Hall, T.; Myers, J.; Nevitt, C.; Sanchez-Lanzas, R.; Chabot, A.; Ding, J.; Kooienga, E.; Caprio, C.; Finkelstein, D.; et al. Murine foetal liver supports limited detectable expansion of life-long haematopoietic progenitors. Nat. Cell Biol. 2022, 24, 1475–1486. [Google Scholar] [CrossRef] [PubMed]
- Azzoni, E.; Fantin, A. Fetal liver hematopoiesis revisited: A precast hierarchy. Nat. Cardiovasc. Res. 2022, 1, 872–873. [Google Scholar] [CrossRef]
- Kisanuki, Y.Y.; Hammer, R.E.; Miyazaki, J.; Williams, S.C.; Richardson, J.A.; Yanagisawa, M. Tie2-Cre transgenic mice: A new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 2001, 230, 230–242. [Google Scholar] [CrossRef]
- Collins, E.C.; Pannell, R.; Simpson, E.M.; Forster, A.; Rabbitts, T.H. Inter-chromosomal recombination of Mll and Af9 genes mediated by cre-loxP in mouse development. EMBO Rep. 2000, 1, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Drynan, L.F.; Pannell, R.; Forster, A.; Chan, N.M.; Cano, F.; Daser, A.; Rabbitts, T.H. Mll fusions generated by Cre-loxP-mediated de novo translocations can induce lineage reassignment in tumorigenesis. EMBO J. 2005, 24, 3136–3146. [Google Scholar] [CrossRef]
- Gentek, R.; Ghigo, C.; Hoeffel, G.; Bulle, M.J.; Msallam, R.; Gautier, G.; Launay, P.; Chen, J.; Ginhoux, F.; Bajenoff, M. Hemogenic Endothelial Fate Mapping Reveals Dual Developmental Origin of Mast Cells. Immunity 2018, 48, 1160–1171.e1165. [Google Scholar] [CrossRef]
- Gentek, R.; Ghigo, C.; Hoeffel, G.; Jorquera, A.; Msallam, R.; Wienert, S.; Klauschen, F.; Ginhoux, F.; Bajenoff, M. Epidermal gammadelta T cells originate from yolk sac hematopoiesis and clonally self-renew in the adult. J. Exp. Med. 2018, 215, 2994–3005. [Google Scholar] [CrossRef]
- Wang, Y.; Nakayama, M.; Pitulescu, M.E.; Schmidt, T.S.; Bochenek, M.L.; Sakakibara, A.; Adams, S.; Davy, A.; Deutsch, U.; Luthi, U.; et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 2010, 465, 483–486. [Google Scholar] [CrossRef]
- Platt, R.J.; Chen, S.; Zhou, Y.; Yim, M.J.; Swiech, L.; Kempton, H.R.; Dahlman, J.E.; Parnas, O.; Eisenhaure, T.M.; Jovanovic, M.; et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014, 159, 440–455. [Google Scholar] [CrossRef]
- Madisen, L.; Zwingman, T.A.; Sunkin, S.M.; Oh, S.W.; Zariwala, H.A.; Gu, H.; Ng, L.L.; Palmiter, R.D.; Hawrylycz, M.J.; Jones, A.R.; et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010, 13, 133–140. [Google Scholar] [CrossRef]
- Azzoni, E.; Frontera, V.; McGrath, K.E.; Harman, J.; Carrelha, J.; Nerlov, C.; Palis, J.; Jacobsen, S.E.W.; de Bruijn, M.F. Kit ligand has a critical role in mouse yolk sac and aorta-gonad-mesonephros hematopoiesis. EMBO Rep. 2018, 19, e45477. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, A.; Bailey, T.L. GT-Scan: Identifying unique genomic targets. Bioinformatics 2014, 30, 2673–2675. [Google Scholar] [CrossRef]
- Swiers, G.; Rode, C.; Azzoni, E.; de Bruijn, M.F. A short history of hemogenic endothelium. Blood Cells Mol. Dis. 2013, 51, 206–212. [Google Scholar] [CrossRef]
- Tang, Y.; Harrington, A.; Yang, X.; Friesel, R.E.; Liaw, L. The contribution of the Tie2+ lineage to primitive and definitive hematopoietic cells. Genesis 2010, 48, 563–567. [Google Scholar] [CrossRef]
- Barone, C. (School of Medicine and Surgery, University of Milano-Bicocca, 20900 Monza, Italy); Azzoni, E. (School of Medicine and Surgery, University of Milano-Bicocca, 20900 Monza, Italy). Personal communication, 2022.
- Chen, M.J.; Li, Y.; De Obaldia, M.E.; Yang, Q.; Yzaguirre, A.D.; Yamada-Inagawa, T.; Vink, C.S.; Bhandoola, A.; Dzierzak, E.; Speck, N.A. Erythroid/myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell 2011, 9, 541–552. [Google Scholar] [CrossRef]
- Martinez-Corral, I.; Makinen, T. Genetic Lineage Tracing of Lymphatic Endothelial Cells in Mice. Methods Mol. Biol. 2018, 1846, 37–53. [Google Scholar] [CrossRef]
- Jeong, J.; Jager, A.; Domizi, P.; Pavel-Dinu, M.; Gojenola, L.; Iwasaki, M.; Wei, M.C.; Pan, F.; Zehnder, J.L.; Porteus, M.H.; et al. High-efficiency CRISPR induction of t(9;11) chromosomal translocations and acute leukemias in human blood stem cells. Blood Adv. 2019, 3, 2825–2835. [Google Scholar] [CrossRef]
- Rice, S.; Jackson, T.; Crump, N.T.; Fordham, N.; Elliott, N.; O’Byrne, S.; Fanego, M.; Addy, D.; Crabb, T.; Dryden, C.; et al. A human fetal liver-derived infant MLL-AF4 acute lymphoblastic leukemia model reveals a distinct fetal gene expression program. Nat. Commun. 2021, 12, 6905. [Google Scholar] [CrossRef]
- Li, Y.; Kong, W.; Yang, W.; Patel, R.M.; Casey, E.B.; Okeyo-Owuor, T.; White, J.M.; Porter, S.N.; Morris, S.A.; Magee, J.A. Single-Cell Analysis of Neonatal HSC Ontogeny Reveals Gradual and Uncoordinated Transcriptional Reprogramming that Begins before Birth. Cell Stem Cell 2020, 27, 732–747.e7. [Google Scholar] [CrossRef]
- Bowie, M.B.; McKnight, K.D.; Kent, D.G.; McCaffrey, L.; Hoodless, P.A.; Eaves, C.J. Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J. Clin. Investig. 2006, 116, 2808–2816. [Google Scholar] [CrossRef]
- Lopez, C.K.; Noguera, E.; Stavropoulou, V.; Robert, E.; Aid, Z.; Ballerini, P.; Bilhou-Nabera, C.; Lapillonne, H.; Boudia, F.; Thirant, C.; et al. Ontogenic Changes in Hematopoietic Hierarchy Determine Pediatric Specificity and Disease Phenotype in Fusion Oncogene-Driven Myeloid Leukemia. Cancer Discov. 2019, 9, 1736–1753. [Google Scholar] [CrossRef]
- Mass, E.; Gentek, R. Fetal-Derived Immune Cells at the Roots of Lifelong Pathophysiology. Front. Cell Dev. Biol. 2021, 9, 648313. [Google Scholar] [CrossRef]
- McGrath, K.E.; Frame, J.M.; Fegan, K.H.; Bowen, J.R.; Conway, S.J.; Catherman, S.C.; Kingsley, P.D.; Koniski, A.D.; Palis, J. Distinct Sources of Hematopoietic Progenitors Emerge before HSCs and Provide Functional Blood Cells in the Mammalian Embryo. Cell Rep. 2015, 11, 1892–1904. [Google Scholar] [CrossRef]
- Barrett, N.A.; Malouf, C.; Kapeni, C.; Bacon, W.A.; Giotopoulos, G.; Jacobsen, S.E.W.; Huntly, B.J.; Ottersbach, K. Mll-AF4 Confers Enhanced Self-Renewal and Lymphoid Potential during a Restricted Window in Development. Cell Rep. 2016, 16, 1039–1054. [Google Scholar] [CrossRef]
- Symeonidou, V.; Jakobczyk, H.; Bashanfer, S.; Malouf, C.; Fotopoulou, F.; Kotecha, R.S.; Anderson, R.A.; Finch, A.J.; Ottersbach, K. Defining the fetal origin of MLL-AF4 infant leukemia highlights specific fatty acid requirements. Cell Rep. 2021, 37, 109900. [Google Scholar] [CrossRef]
- Boiers, C.; Carrelha, J.; Lutteropp, M.; Luc, S.; Green, J.C.; Azzoni, E.; Woll, P.S.; Mead, A.J.; Hultquist, A.; Swiers, G.; et al. Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell 2013, 13, 535–548. [Google Scholar] [CrossRef]
- Malouf, C.; Ottersbach, K. The fetal liver lymphoid-primed multipotent progenitor provides the prerequisites for the initiation of t(4;11) MLL-AF4 infant leukemia. Haematologica 2018, 103, e571–e574. [Google Scholar] [CrossRef]
- Ding, J.; Cardoso, A.A.; Yoshimoto, M.; Kobayashi, M. The Earliest T-Precursors in the Mouse Embryo Are Susceptible to Leukemic Transformation. Front. Cell Dev. Biol. 2021, 9, 634151. [Google Scholar] [CrossRef]
- Azzoni, E.; Frontera, V.; Anselmi, G.; Rode, C.; James, C.; Deltcheva, E.M.; Demian, A.S.; Brown, J.; Barone, C.; Patelli, A.; et al. The onset of circulation triggers a metabolic switch required for endothelial to hematopoietic transition. Cell Rep. 2021, 37, 110103. [Google Scholar] [CrossRef]
- Hadland, B.K.; Huppert, S.S.; Kanungo, J.; Xue, Y.; Jiang, R.; Gridley, T.; Conlon, R.A.; Cheng, A.M.; Kopan, R.; Longmore, G.D. A requirement for Notch1 distinguishes 2 phases of definitive hematopoiesis during development. Blood 2004, 104, 3097–3105. [Google Scholar] [CrossRef]
- Espin-Palazon, R.; Weijts, B.; Mulero, V.; Traver, D. Proinflammatory Signals as Fuel for the Fire of Hematopoietic Stem Cell Emergence. Trends Cell Biol. 2018, 28, 58–66. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barone, C.; Orsenigo, R.; Cazzola, A.; D’Errico, E.; Patelli, A.; Quattrini, G.; Vergani, B.; Bombelli, S.; De Marco, S.; D’Orlando, C.; et al. Hematopoietic Stem Cell (HSC)-Independent Progenitors Are Susceptible to Mll-Af9-Induced Leukemic Transformation. Cancers 2023, 15, 3624. https://doi.org/10.3390/cancers15143624
Barone C, Orsenigo R, Cazzola A, D’Errico E, Patelli A, Quattrini G, Vergani B, Bombelli S, De Marco S, D’Orlando C, et al. Hematopoietic Stem Cell (HSC)-Independent Progenitors Are Susceptible to Mll-Af9-Induced Leukemic Transformation. Cancers. 2023; 15(14):3624. https://doi.org/10.3390/cancers15143624
Chicago/Turabian StyleBarone, Cristiana, Roberto Orsenigo, Anna Cazzola, Elisabetta D’Errico, Arianna Patelli, Giulia Quattrini, Barbara Vergani, Silvia Bombelli, Sofia De Marco, Cristina D’Orlando, and et al. 2023. "Hematopoietic Stem Cell (HSC)-Independent Progenitors Are Susceptible to Mll-Af9-Induced Leukemic Transformation" Cancers 15, no. 14: 3624. https://doi.org/10.3390/cancers15143624
APA StyleBarone, C., Orsenigo, R., Cazzola, A., D’Errico, E., Patelli, A., Quattrini, G., Vergani, B., Bombelli, S., De Marco, S., D’Orlando, C., Bianchi, C., Leone, B. E., Meneveri, R., Biondi, A., Cazzaniga, G., Rabbitts, T. H., Brunelli, S., & Azzoni, E. (2023). Hematopoietic Stem Cell (HSC)-Independent Progenitors Are Susceptible to Mll-Af9-Induced Leukemic Transformation. Cancers, 15(14), 3624. https://doi.org/10.3390/cancers15143624







