Current Landscape of Genome-Wide Association Studies in Acute Myeloid Leukemia: A Review
Abstract
Simple Summary
Abstract
1. Introduction
2. Evolution of Genetic Studies in AML
3. Literature Search Strategy
4. Results
Summary of GWAS Studies
5. Future Directions
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- National Cancer Institute. SEER Cancer Stat Facts: Acute Myeloid Leukemia. 2022. Available online: https://seer.cancer.gov/statfacts/html/amyl.html (accessed on 6 January 2023).
- Levine, E.G.; Bloomfield, C.D. Leukemias and myelodysplastic syndromes secondary to drug, radiation, and environmental exposure. Semin. Oncol. 1992, 19, 47–84. [Google Scholar] [PubMed]
- McHale, C.M.; Zhang, L.; Smith, M.T. Current understanding of the mechanism of benzene-induced leukemia in humans: Implications for risk assessment. Carcinogenesis 2012, 33, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Colamesta, V.; D’Aguanno, S.; Breccia, M.; Bruffa, S.; Cartoni, C.; La Torre, G. Do the smoking intensity and duration, the years since quitting, the methodological quality and the year of publication of the studies affect the results of the meta-analysis on cigarette smoking and Acute Myeloid Leukemia (AML) in adults? Crit. Rev. Oncol. Hematol. 2016, 99, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Christen, F.; Hoyer, K.; Yoshida, K.; Hou, H.A.; Waldhueter, N.; Heuser, M.; Hills, R.K.; Chan, W.; Hablesreiter, R.; Blau, O.; et al. Genomic landscape and clonal evolution of acute myeloid leukemia with t(8;21): An international study on 331 patients. Blood 2019, 133, 1140–1151. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M.; Network, C.G.A.R. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Charrot, S.; Armes, H.; Rio-Machin, A.; Fitzgibbon, J. AML through the prism of molecular genetics. Br. J. Haematol. 2020, 188, 49–62. [Google Scholar] [CrossRef]
- Hahn, C.N.; Chong, C.E.; Carmichael, C.L.; Wilkins, E.J.; Brautigan, P.J.; Li, X.C.; Babic, M.; Lin, M.; Carmagnac, A.; Lee, Y.K.; et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat. Genet. 2011, 43, 1012–1017. [Google Scholar]
- Smith, M.L.; Cavenagh, J.D.; Lister, T.A.; Fitzgibbon, J. Mutation of CEBPA in familial acute myeloid leukemia. N. Engl. J. Med. 2004, 351, 2403–2407. [Google Scholar] [CrossRef]
- Pathak, A.; Seipel, K.; Pemov, A.; Dewan, R.; Brown, C.; Ravichandran, S.; Luke, B.T.; Malasky, M.; Suman, S.; Yeager, M.; et al. Whole exome sequencing reveals a C-terminal germline variant in CEBPA-associated acute myeloid leukemia: 45-year follow up of a large family. Haematologica 2016, 101, 846–852. [Google Scholar] [CrossRef]
- Rio-Machin, A.; Vulliamy, T.; Hug, N.; Walne, A.; Tawana, K.; Cardoso, S.; Ellison, A.; Pontikos, N.; Wang, J.; Tummala, H.; et al. The complex genetic landscape of familial MDS and AML reveals pathogenic germline variants. Nat. Commun. 2020, 11, 1044. [Google Scholar] [CrossRef]
- Simon, L.; Spinella, J.F.; Yao, C.Y.; Lavallée, V.P.; Boivin, I.; Boucher, G.; Audemard, E.; Bordeleau, M.E.; Lemieux, S.; Hébert, J.; et al. High frequency of germline RUNX1 mutations in patients with RUNX1-mutated AML. Blood 2020, 135, 1882–1886. [Google Scholar] [CrossRef] [PubMed]
- NCCN Clinical Practice Guidelines in Oncology. (NCCN Guidelines®) for Acute Myeloid Leukemia V 1.2023. © National Comprehensive Cancer Network, Inc. 2023. To View the Most Recent and Complete Version of the Guideline. Available online: NCCN.org (accessed on 8 March 2023).
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Babushok, D.V.; Bessler, M.; Olson, T.S. Genetic predisposition to myelodysplastic syndrome and acute myeloid leukemia in children and young adults. Leuk Lymphoma 2016, 57, 520–536. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Yun, W.; Lee, S.T.; Choi, J.R.; Yoo, K.H.; Koo, H.H.; Jung, C.W.; Kim, S.H. Prevalence and clinical implications of germline predisposition gene mutations in patients with acute myeloid leukemia. Sci. Rep. 2020, 10, 14297. [Google Scholar] [CrossRef] [PubMed]
- Tawana, K.; Wang, J.; Renneville, A.; Bödör, C.; Hills, R.; Loveday, C.; Savic, A.; Van Delft, F.W.; Treleaven, J.; Georgiades, P.; et al. Disease evolution and outcomes in familial AML with germline CEBPA mutations. Blood 2015, 126, 1214–1223. [Google Scholar] [CrossRef]
- Reich, D.E.; Lander, E.S. On the allelic spectrum of human disease. Trends Genet. 2001, 17, 502–510. [Google Scholar] [CrossRef]
- Sollis, E.; Mosaku, A.; Abid, A.; Buniello, A.; Cerezo, M.; Gil, L.; Groza, T.; Güneş, O.; Hall, P.; Hayhurst, J.; et al. The NHGRI-EBI GWAS Catalog: Knowledgebase and deposition resource. Nucleic. Acids. Res. 2022, 51, D977–D985. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef]
- Knight, J.A.; Skol, A.D.; Shinde, A.; Hastings, D.; Walgren, R.A.; Shao, J.; Tennant, T.R.; Banerjee, M.; Allan, J.M.; Le Beau, M.M.; et al. Genome-wide association study to identify novel loci associated with therapy-related myeloid leukemia susceptibility. Blood 2009, 113, 5575–5582. [Google Scholar] [CrossRef]
- Cao, S.; Yang, G.; Zhang, J.; Shen, Y.; Ma, H.; Qian, X.; Hu, Z. Replication analysis confirms the association of several variants with acute myeloid leukemia in Chinese population. J. Cancer Res. Clin. Oncol. 2016, 142, 149–155. [Google Scholar] [CrossRef]
- Lv, H.; Zhang, M.; Shang, Z.; Li, J.; Zhang, S.; Lian, D.; Zhang, R. Genome-wide haplotype association study identify the FGFR2 gene as a risk gene for acute myeloid leukemia. Oncotarget 2017, 8, 7891–7899. [Google Scholar] [CrossRef]
- Bargal, S.A.; Rafiee, R.; Crews, K.R.; Wu, H.; Cao, X.; Rubnitz, J.E.; Ribeiro, R.C.; Downing, J.R.; Pounds, S.B.; Lamba, J.K. Genome-wide association analysis identifies SNPs predictive of. Oncotarget 2018, 9, 34859–34875. [Google Scholar] [CrossRef]
- Walker, C.J.; Oakes, C.C.; Genutis, L.K.; Giacopelli, B.; Liyanarachchi, S.; Nicolet, D.; Eisfeld, A.K.; Scholz, M.; Brock, P.; Kohlschmidt, J.; et al. Genome-wide association study identifies an acute myeloid leukemia susceptibility locus near BICRA. Leukemia 2019, 33, 771–775. [Google Scholar] [CrossRef]
- Lin, W.Y.; Fordham, S.E.; Hungate, E.; Sunter, N.J.; Elstob, C.; Xu, Y.; Park, C.; Quante, A.; Strauch, K.; Gieger, C.; et al. Genome-wide association study identifies susceptibility loci for acute myeloid leukemia. Nat. Commun. 2021, 12, 6233. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Clay-Gilmour, A.I.; Karaesmen, E.; Rizvi, A.; Zhu, Q.; Yan, L.; Preus, L.; Liu, S.; Wang, Y.; Griffiths, E.; et al. Genome-Wide Association Analyses Identify Variants in. Front. Genet. 2021, 12, 554948. [Google Scholar] [CrossRef] [PubMed]
- Ramazzotti, G.; Billi, A.M.; Manzoli, L.; Mazzetti, C.; Ruggeri, A.; Erneux, C.; Kim, S.; Suh, P.G.; Cocco, L.; Faenza, I. IPMK and β-catenin mediate PLC-β1-dependent signaling in myogenic differentiation. Oncotarget 2016, 7, 84118–84127. [Google Scholar] [CrossRef] [PubMed]
- Malabanan, M.M.; Blind, R.D. Inositol polyphosphate multikinase (IPMK) in transcriptional regulation and nuclear inositide metabolism. Biochem. Soc. Trans. 2016, 44, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Beon, J.; Lee, S.; Park, J.; Kim, S. IPMK: A versatile regulator of nuclear signaling events. Adv. Biol. Regul. 2016, 61, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Li, S.J.; Guo, H.; Li, Z.H.; Fei, X.; Chang, S.M.; Yang, Q.C.; Cheng, D.D. Ebastine exerts antitumor activity and induces autophagy by activating AMPK/ULK1 signaling in an IPMK-dependent manner in osteosarcoma. Int. J. Biol. Sci. 2023, 19, 537–551. [Google Scholar] [CrossRef]
- Guha, P.; Tyagi, R.; Chowdhury, S.; Reilly, L.; Fu, C.; Xu, R.; Resnick, A.C.; Snyder, S.H. IPMK Mediates Activation of ULK Signaling and Transcriptional Regulation of Autophagy Linked to Liver Inflammation and Regeneration. Cell Rep. 2019, 26, 2692–2703.e7. [Google Scholar] [CrossRef]
- Tai, A.L.; Mak, W.; Ng, P.K.; Chua, D.T.; Ng, M.Y.; Fu, L.; Chu, K.K.; Fang, Y.; Qiang Song, Y.; Chen, M.; et al. High-throughput loss-of-heterozygosity study of chromosome 3p in lung cancer using single-nucleotide polymorphism markers. Cancer Res. 2006, 66, 4133–4138. [Google Scholar] [CrossRef] [PubMed]
- Borthakur, G.; Kantarjian, H. Core binding factor acute myelogenous leukemia-2021 treatment algorithm. Blood Cancer J. 2021, 11, 114. [Google Scholar] [CrossRef]
- Consortium, I.H. The International HapMap Project. Nature 2003, 426, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Aerts, S.; Lambrechts, D.; Maity, S.; Van Loo, P.; Coessens, B.; De Smet, F.; Tranchevent, L.C.; De Moor, B.; Marynen, P.; Hassan, B.; et al. Gene prioritization through genomic data fusion. Nat. Biotechnol. 2006, 24, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Sood, R.; Kamikubo, Y.; Liu, P. Role of RUNX1 in hematological malignancies. Blood 2017, 129, 2070–2082. [Google Scholar] [CrossRef] [PubMed]
- Fasouli, E.S.; Katsantoni, E. JAK-STAT in Early Hematopoiesis and Leukemia. Front. Cell Dev. Biol. 2021, 9, 669363. [Google Scholar] [CrossRef]
- Bain, B.J. Myeloid and lymphoid neoplasms with eosinophilia and abnormalities of PDGFRA, PDGFRB or FGFR1. Haematologica 2010, 95, 696–698. [Google Scholar] [CrossRef]
- Ardizzone, A.; Bova, V.; Casili, G.; Repici, A.; Lanza, M.; Giuffrida, R.; Colarossi, C.; Mare, M.; Cuzzocrea, S.; Esposito, E.; et al. Role of Basic Fibroblast Growth Factor in Cancer: Biological Activity, Targeted Therapies, and Prognostic Value. Cells 2023, 12, 1002. [Google Scholar] [CrossRef]
- Yu, S.; Ye, J.; Wang, Y.; Lu, T.; Liu, Y.; Liu, N.; Zhang, J.; Lu, F.; Ma, D.; Gale, R.P.; et al. DNA damage to bone marrow stromal cells by antileukemia drugs induces chemoresistance in acute myeloid leukemia via paracrine FGF10-FGFR2 signaling. J. Biol. Chem. 2023, 299, 102787. [Google Scholar] [CrossRef]
- Ji, B.; Feng, Y.; Sun, Y.; Ji, D.; Qian, W.; Zhang, Z.; Wang, Q.; Zhang, Y.; Zhang, C. GPR56 promotes proliferation of colorectal cancer cells and enhances metastasis via epithelial-mesenchymal transition through PI3K/AKT signaling activation. Oncol. Rep. 2018, 40, 1885–1896. [Google Scholar] [CrossRef]
- Daga, S.; Rosenberger, A.; Quehenberger, F.; Krisper, N.; Prietl, B.; Reinisch, A.; Zebisch, A.; Sill, H.; Wölfler, A. High GPR56 surface expression correlates with a leukemic stem cell gene signature in CD34-positive AML. Cancer Med. 2019, 8, 1771–1778. [Google Scholar] [CrossRef]
- Weisberg, E.; Nonami, A.; Chen, Z.; Nelson, E.; Chen, Y.; Liu, F.; Cho, H.; Zhang, J.; Sattler, M.; Mitsiades, C.; et al. Upregulation of IGF1R by mutant RAS in leukemia and potentiation of RAS signaling inhibitors by small-molecule inhibition of IGF1R. Clin. Cancer Res. 2014, 20, 5483–5495. [Google Scholar] [CrossRef] [PubMed]
- Alpsoy, A.; Dykhuizen, E.C. Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. J. Biol. Chem. 2018, 293, 3892–3903. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Yang, B.; Zhou, M.; Huang, Q.; Mai, M.; Huang, Z.; Lai, M.; Xu, E.; Zhang, H. GLTSCR1 coordinates alternative splicing and transcription elongation of ZO1 to regulate colorectal cancer progression. J. Mol. Cell. Biol. 2022, 14, mjac009. [Google Scholar] [CrossRef] [PubMed]
- Vinci, M.; Burford, A.; Molinari, V.; Kessler, K.; Popov, S.; Clarke, M.; Taylor, K.R.; Pemberton, H.N.; Lord, C.J.; Gutteridge, A.; et al. Functional diversity and cooperativity between subclonal populations of pediatric glioblastoma and diffuse intrinsic pontine glioma cells. Nat. Med. 2018, 24, 1204–1215. [Google Scholar] [CrossRef]
- Hamamoto, R.; Nakamura, Y. Dysregulation of protein methyltransferases in human cancer: An emerging target class for anticancer therapy. Cancer Sci 2016, 107, 377–384. [Google Scholar] [CrossRef]
- Võsa, U.; Claringbould, A.; Westra, H.J.; Bonder, M.J.; Deelen, P.; Zeng, B.; Kirsten, H.; Saha, A.; Kreuzhuber, R.; Yazar, S.; et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet. 2021, 53, 1300–1310. [Google Scholar] [CrossRef]
- Jiménez, P.; Cantón, J.; Collado, A.; Cabrera, T.; Serrano, A.; Real, L.M.; García, A.; Ruiz-Cabello, F.; Garrido, F. Chromosome loss is the most frequent mechanism contributing to HLA haplotype loss in human tumors. Int. J. Cancer 1999, 83, 91–97. [Google Scholar] [CrossRef]
- McGranahan, N.; Rosenthal, R.; Hiley, C.T.; Rowan, A.J.; Watkins, T.B.K.; Wilson, G.A.; Birkbak, N.J.; Veeriah, S.; Van Loo, P.; Herrero, J.; et al. Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution. Cell 2017, 171, 1259–1271.e11. [Google Scholar] [CrossRef]
- Pagliuca, S.; Gurnari, C.; Rubio, M.T.; Visconte, V.; Lenz, T.L. Individual HLA heterogeneity and its implications for cellular immune evasion in cancer and beyond. Front. Immunol. 2022, 13, 944872. [Google Scholar] [CrossRef]
- Hahn, T.; Sucheston-Campbell, L.E.; Preus, L.; Zhu, X.; Hansen, J.A.; Martin, P.J.; Yan, L.; Liu, S.; Spellman, S.; Tritchler, D.; et al. Establishment of Definitions and Review Process for Consistent Adjudication of Cause-specific Mortality after Allogeneic Unrelated-donor Hematopoietic Cell Transplantation. Biol. Blood Marrow. Transpl. 2015, 21, 1679–1686. [Google Scholar] [CrossRef] [PubMed]
- Karaesmen, E.; Rizvi, A.A.; Preus, L.M.; McCarthy, P.L.; Pasquini, M.C.; Onel, K.; Zhu, X.; Spellman, S.; Haiman, C.A.; Stram, D.O.; et al. Replication and validation of genetic polymorphisms associated with survival after allogeneic blood or marrow transplant. Blood 2017, 130, 1585–1596. [Google Scholar] [CrossRef] [PubMed]
- Visser, M.; Palstra, R.J.; Kayser, M. Allele-specific transcriptional regulation of IRF4 in melanocytes is mediated by chromatin looping of the intronic rs12203592 enhancer to the IRF4 promoter. Hum. Mol. Genet. 2015, 24, 2649–2661. [Google Scholar] [CrossRef] [PubMed]
- Treviño, L.R.; Yang, W.; French, D.; Hunger, S.P.; Carroll, W.L.; Devidas, M.; Willman, C.; Neale, G.; Downing, J.; Raimondi, S.C.; et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat. Genet. 2009, 41, 1001–1005. [Google Scholar] [CrossRef]
- Papaemmanuil, E.; Hosking, F.J.; Vijayakrishnan, J.; Price, A.; Olver, B.; Sheridan, E.; Kinsey, S.E.; Lightfoot, T.; Roman, E.; Irving, J.A.; et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat. Genet. 2009, 41, 1006–1010. [Google Scholar] [CrossRef]
- Bolouri, H.; Farrar, J.E.; Triche, T.; Ries, R.E.; Lim, E.L.; Alonzo, T.A.; Ma, Y.; Moore, R.; Mungall, A.J.; Marra, M.A.; et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat. Med. 2018, 24, 103–112. [Google Scholar] [CrossRef]
- Rehm, H.L.; Page, A.J.H.; Smith, L.; Adams, J.B.; Alterovitz, G.; Babb, L.J.; Barkley, M.P.; Baudis, M.; Beauvais, M.J.S.; Beck, T.; et al. GA4GH: International policies and standards for data sharing across genomic research and healthcare. Cell Genom. 2021, 1, 100029. [Google Scholar] [CrossRef]
- Burd, A.; Levine, R.L.; Ruppert, A.S.; Mims, A.S.; Borate, U.; Stein, E.M.; Patel, P.; Baer, M.R.; Stock, W.; Deininger, M.; et al. Precision medicine treatment in acute myeloid leukemia using prospective genomic profiling: Feasibility and preliminary efficacy of the Beat AML Master Trial. Nat. Med. 2020, 26, 1852–1858. [Google Scholar] [CrossRef]
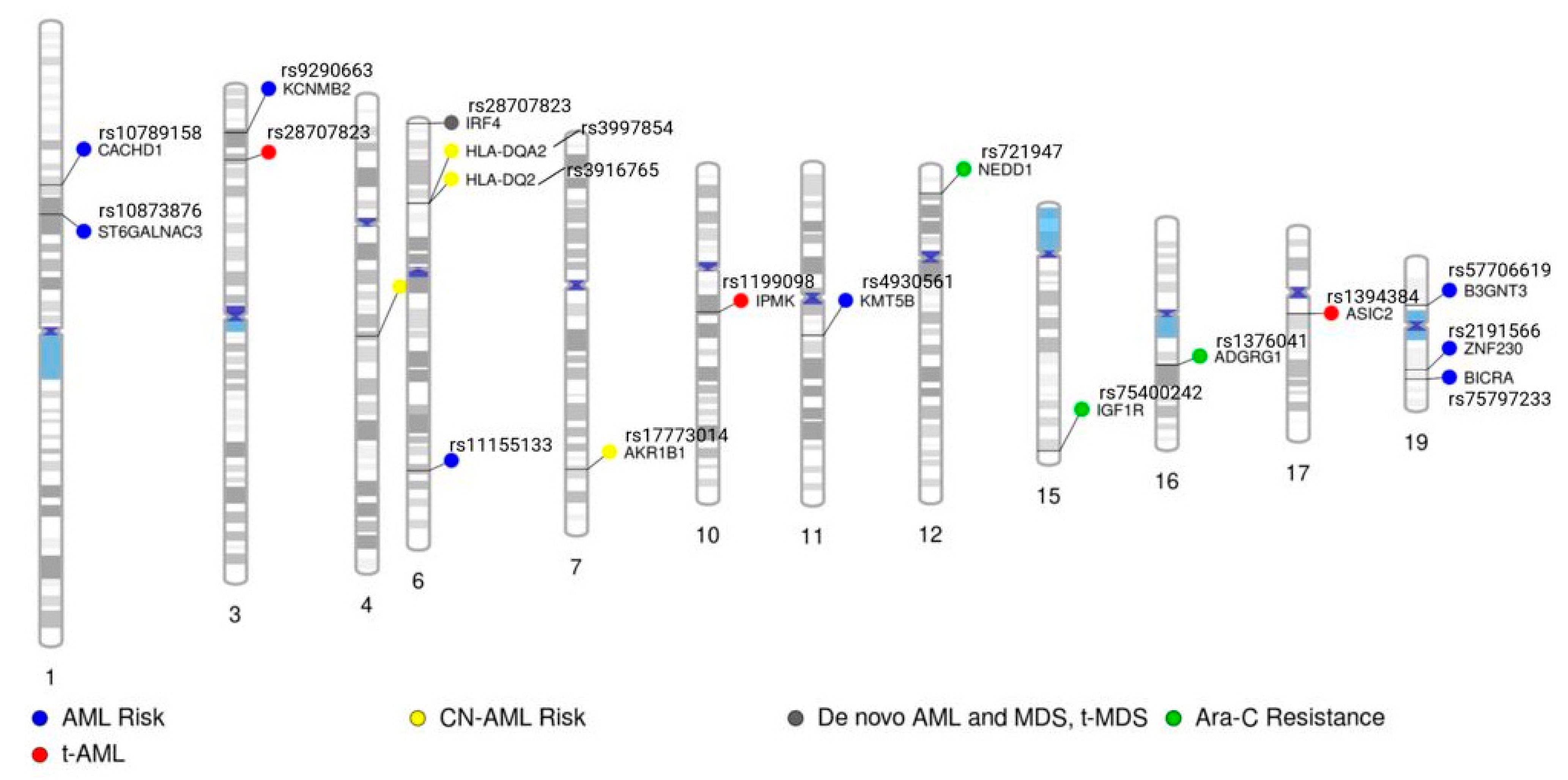
| Reference | Phenotype | SNP(s) | Gene or Nearest Gene Symbol | Gene Information and Reference to Any Known Association with Cancer/Leukemia |
|---|---|---|---|---|
| [21] | Therapy-related AML | rs1199098 | IPMK | Involved in inositol–trisphosphate metabolic process and necroptotic process. Associated with various cancers, such as breast, prostate, osteosarcoma, and papillary thyroid cancer. (PMIDs: 35864548, 37193176, 36632464, 36186479) |
| rs1381392 | LINC00693 | RNA gene of lncRNA class | ||
| rs1394384 | ASIC2 | Predicted to enable ligand-gated sodium channel activity. Increased expression in colorectal cancer is associated with poor prognosis (PMID: 28927426) | ||
| [22] | AML disease risk | rs2191566 | ZNF230 | Predicted to enable DNA-binding transcription factor activity, as well as RNA polymerase II-specific and RNA polymerase II cis-regulatory region sequence-specific DNA binding activity. Predicted to be involved in regulation of transcription via RNA polymerase II. Predicted to be active in nucleus. |
| rs10873876 | ST6GALNAC3 | ST6GALNAC3 belongs to a family of sialyltransferases that transfer sialic acids from CMP-sialic acid to terminal positions of carbohydrate groups in glycoproteins and glycolipids. Associated with hepatocellular carcinoma and prostate cancer (PMIDs: 35321246, 29465788) | ||
| rs9290663 | KCNMB2 | Encodes a protein important in regulating the flow of potassium ions across cell membranes and plays a role in various physiological processes, such as neuronal excitability, smooth muscle contraction, and hormone secretion. Associated with following cancers: non-small cell lung cancer, esophageal cancer, bladder cancer, cervical cancer, and endometrial cancer. (PMIDs: 35979065, 34516362, 34367236, 34026626, 33028109, 31539276) | ||
| rs11155133 | LOC102723724 | RNA gene of ncRNA class | ||
| [23] | AML disease risk | rs7090018 | FGFR2 | Gene encodes a protein that engages with fibroblast growth factors, initiating a series of subsequent signals that ultimately impact cell division and differentiation. Associated with AML, T-cell ALL, mixed phenotype acute leukemia, and chronic myeloid leukemia. (PMIDs: 30668205, 36333298, 33049052, 31502137) |
| rs2912759 | ||||
| [24] | In vitro Ara-C chemosensitivity | rs1376041 | ADGRG1 | Receptor involved in cell adhesion and probably involved in cell–cell interactions. Mediates cell matrix adhesion in developing neurons and hematopoietic stem cells. Reported to play a role in leukemogenesis (PMID: 23478665) and is part of a 17-gene stemness score that aids in determining leukemia risk (PMID: 27926740) |
| rs721947 | 93kb 3′ of NEDD1 | Predicted to be involved in protein localization to centrosome. Associated with B-cell lymphoma, renal cell carcinoma, and cell cycle arrest. PMIDs: 36840486, 36347549, 23106787) | ||
| rs75400242 | IGF1R | Receptor binds insulin-like growth factor with a high affinity. Highly overexpressed in most malignant tissues, where it functions as an anti-apoptotic agent by enhancing cell survival. Associated with B-cell ALL, regulation of proliferation of AML stem cells, and T-cell ALL. (PMIDs: 37298628, 30013477, 27532210) | ||
| [25] | AML disease risk | rs75797233 | BICRA | Enables transcription regulator activator activity. Involved in positive regulation of transcription, DNA-templated. Associated with colorectal cancer and lung cancer risk. (PMID: 30291333, 30128886) |
| [26] | AML and cytogenetically normal AML | rs4930561 | KMT5B | This gene encodes a protein that contains a SET domain. SET domains appear to be protein–protein interaction domains that mediate interactions among a family of proteins that display similarity to dual-specificity phosphatases (dsPTPases). The function of this gene has not been determined. Associated with glioblastoma, sarcomas, and tumor recurrence. (PMIDs: 29967352, 36376321) |
| rs3916765 | LOC102725019 | RNA gene of lncRNA class | ||
| [27] | De novo AML with and without abnormal cytogenetics, de novo MDS, and therapy-related AML and MDS | rs12203592 | IRF4 | Plays a role in regulation of interferons in response to infection by a virus and interferon-inducible genes. Lymphocyte specifically and negatively regulates Toll-like-receptor (TLR) signaling, which is important in the activation of innate and adaptive immune systems. Associated with B-cell malignancies, adult T-cell leukemia, childhood acute leukemias, and chronic myeloid leukemia (PMIDs: 36495369, 36446869, 34775495, 11013272) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marrero, R.J.; Lamba, J.K. Current Landscape of Genome-Wide Association Studies in Acute Myeloid Leukemia: A Review. Cancers 2023, 15, 3583. https://doi.org/10.3390/cancers15143583
Marrero RJ, Lamba JK. Current Landscape of Genome-Wide Association Studies in Acute Myeloid Leukemia: A Review. Cancers. 2023; 15(14):3583. https://doi.org/10.3390/cancers15143583
Chicago/Turabian StyleMarrero, Richard J., and Jatinder K. Lamba. 2023. "Current Landscape of Genome-Wide Association Studies in Acute Myeloid Leukemia: A Review" Cancers 15, no. 14: 3583. https://doi.org/10.3390/cancers15143583
APA StyleMarrero, R. J., & Lamba, J. K. (2023). Current Landscape of Genome-Wide Association Studies in Acute Myeloid Leukemia: A Review. Cancers, 15(14), 3583. https://doi.org/10.3390/cancers15143583






