The Landscape of Small Leucine-Rich Proteoglycan Impact on Cancer Pathogenesis with a Focus on Biglycan and Lumican
Abstract
Simple Summary
Abstract
1. Introduction
2. The Tumor ECM
3. SLRPs Structure and Roles
3.1. Biglycan Structure
3.2. Lumican Structure
4. Biglycan and Lumican Expression and Roles in Cancerogenesis
5. SLRPs Modulate Cancer Cell Adhesion, Migration, and Invasion
5.1. Biglycan
5.2. Lumican
6. SLRPs Affect Tumor Cell Growth and Cell Cycle Regulation
6.1. Biglycan
6.2. Lumican
7. Biglycan Effects on Cancer-Associated Inflammation
8. Biglycan Affects Tumor Angiogenesis
9. Biglycan and Lumican Affect Cell Death Mechanisms and Chemoresistance
10. SLRPs as Therapy Targets
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Tzanakakis, G.; Neagu, M.; Tsatsakis, A.; Nikitovic, D. Proteoglycans and Immunobiology of Cancer-Therapeutic Implications. Front. Immunol. 2019, 10, 875. [Google Scholar] [CrossRef] [PubMed]
- Tzanakakis, G.; Giatagana, E.M.; Kuskov, A.; Berdiaki, A.; Tsatsakis, A.M.; Neagu, M.; Nikitovic, D. Proteoglycans in the Pathogenesis of Hormone-Dependent Cancers: Mediators and Effectors. Cancers 2020, 12, 2401. [Google Scholar] [CrossRef]
- Joshi, R.S.; Kanugula, S.S.; Sudhir, S.; Pereira, M.P.; Jain, S.; Aghi, M.K. The Role of Cancer-Associated Fibroblasts in Tumor Progression. Cancers 2021, 13, 1399. [Google Scholar] [CrossRef]
- Suveges, S.; Eftimie, R.; Trucu, D. Directionality of Macrophages Movement in Tumour Invasion: A Multiscale Moving-Boundary Approach. Bull. Math. Biol. 2020, 82, 148. [Google Scholar] [CrossRef]
- Nikitovic, D.; Tzardi, M.; Berdiaki, A.; Tsatsakis, A.; Tzanakakis, G.N. Cancer microenvironment and inflammation: Role of hyaluronan. Front. Immunol. 2015, 6, 169. [Google Scholar] [CrossRef]
- Nikitovic, D.; Berdiaki, A.; Spyridaki, I.; Krasanakis, T.; Tsatsakis, A.; Tzanakakis, G.N. Proteoglycans-Biomarkers and Targets in Cancer Therapy. Front. Endocrinol. 2018, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Pickup, M.W.; Mouw, J.K.; Weaver, V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014, 15, 1243–1253. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Han, F.; Fu, M.; Wang, Z. High expression of VCAN is an independent predictor of poor prognosis in gastric cancer. J Int. Med. Res. 2020, 48, 300060519891271. [Google Scholar] [CrossRef] [PubMed]
- Verginadis, I.I.; Avgousti, H.; Monslow, J.; Skoufos, G.; Chinga, F.; Kim, K.; Leli, N.M.; Karagounis, I.V.; Bell, B.I.; Velalopoulou, A.; et al. A stromal Integrated Stress Response activates perivascular cancer-associated fibroblasts to drive angiogenesis and tumour progression. Nat. Cell Biol. 2022, 24, 940–953. [Google Scholar] [CrossRef]
- Hua, S.H.; Viera, M.; Yip, G.W.; Bay, B.H. Theranostic Applications of Glycosaminoglycans in Metastatic Renal Cell Carcinoma. Cancers 2022, 15, 266. [Google Scholar] [CrossRef]
- Hynes, R.O. The extracellular matrix: Not just pretty fibrils. Science 2009, 326, 1216–1219. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Dunn, G.P.; Old, L.J.; Schreiber, R.D. The immunobiology of cancer immunosurveillance and immunoediting. Immunity 2004, 21, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Iozzo, R.V. The family of the small leucine-rich proteoglycans: Key regulators of matrix assembly and cellular growth. Crit. Rev. Biochem. Mol. Biol. 1997, 32, 141–174. [Google Scholar] [CrossRef] [PubMed]
- Kram, V.; Shainer, R.; Jani, P.; Meester, J.A.N.; Loeys, B.; Young, M.F. Biglycan in the Skeleton. J. Histochem. Cytochem. 2020, 68, 747–762. [Google Scholar] [CrossRef]
- Iozzo, R.V.; Schaefer, L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015, 42, 11–55. [Google Scholar] [CrossRef]
- Schaefer, L.; Schaefer, R.M. Proteoglycans: From structural compounds to signaling molecules. Cell Tissue Res. 2010, 339, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Kram, V.; Kilts, T.M.; Bhattacharyya, N.; Li, L.; Young, M.F. Small leucine rich proteoglycans, a novel link to osteoclastogenesis. Sci. Rep. 2017, 7, 12627. [Google Scholar] [CrossRef]
- Chan, W.L.; Steiner, M.; Witkos, T.; Egerer, J.; Busse, B.; Mizumoto, S.; Pestka, J.M.; Zhang, H.; Hausser, I.; Khayal, L.A.; et al. Impaired proteoglycan glycosylation, elevated TGF-beta signaling, and abnormal osteoblast differentiation as the basis for bone fragility in a mouse model for gerodermia osteodysplastica. PLoS Genet. 2018, 14, e1007242. [Google Scholar] [CrossRef]
- Sarbu, M.; Ica, R.; Sharon, E.; Clemmer, D.E.; Zamfir, A.D. Glycomics by ion mobility tandem mass spectrometry of chondroitin sulfate disaccharide domain in biglycan. J. Mass Spectrom. 2023, 58, e4908. [Google Scholar] [CrossRef]
- Diehl, V.; Huber, L.S.; Trebicka, J.; Wygrecka, M.; Iozzo, R.V.; Schaefer, L. The Role of Decorin and Biglycan Signaling in Tumorigenesis. Front. Oncol. 2021, 11, 801801. [Google Scholar] [CrossRef]
- Zhao, S.F.; Yin, X.J.; Zhao, W.J.; Liu, L.C.; Wang, Z.P. Biglycan as a potential diagnostic and prognostic biomarker in multiple human cancers. Oncol. Lett. 2020, 19, 1673–1682. [Google Scholar] [CrossRef] [PubMed]
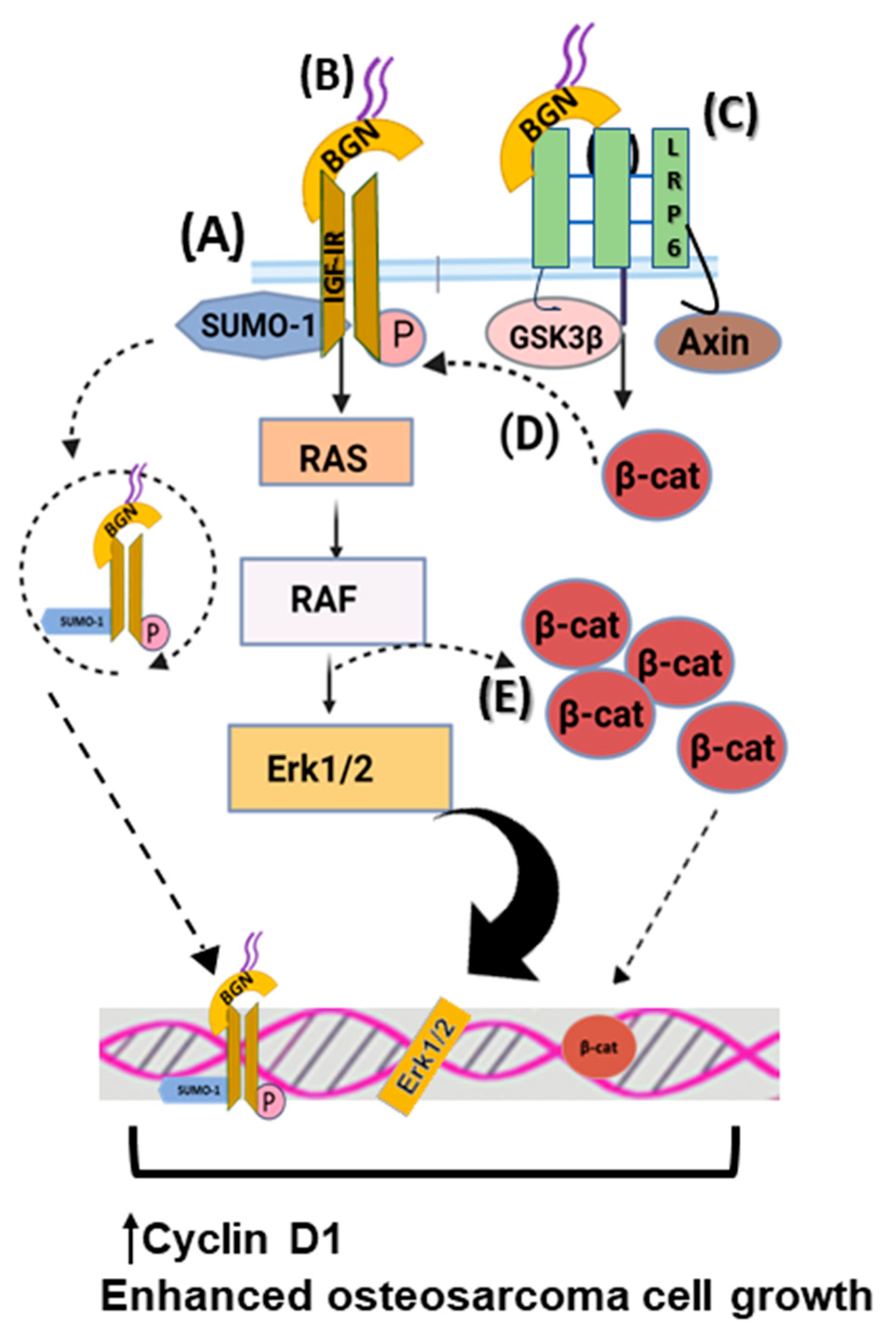
- Aggelidakis, J.; Berdiaki, A.; Nikitovic, D.; Papoutsidakis, A.; Papachristou, D.J.; Tsatsakis, A.M.; Tzanakakis, G.N. Biglycan Regulates MG63 Osteosarcoma Cell Growth Through a LPR6/beta-Catenin/IGFR-IR Signaling Axis. Front. Oncol. 2018, 8, 470. [Google Scholar] [CrossRef]
- Giatagana, E.M.; Berdiaki, A.; Tsatsakis, A.; Tzanakakis, G.N.; Nikitovic, D. Lumican in Carcinogenesis-Revisited. Biomolecules 2021, 11, 1319. [Google Scholar] [CrossRef] [PubMed]
- Giatagana, E.M.; Berdiaki, A.; Gaardlos, M.; Samsonov, S.A.; Tzanakakis, G.N.; Nikitovic, D. Biglycan Interacts with Type I Insulin-like Receptor (IGF-IR) Signaling Pathway to Regulate Osteosarcoma Cell Growth and Response to Chemotherapy. Cancers 2022, 14, 1196. [Google Scholar] [CrossRef]
- Papoutsidakis, A.; Giatagana, E.M.; Berdiaki, A.; Spyridaki, I.; Spandidos, D.A.; Tsatsakis, A.; Tzanakakis, G.N.; Nikitovic, D. Lumican mediates HTB94 chondrosarcoma cell growth via an IGF-IR/Erk1/2 axis. Int. J. Oncol. 2020, 57, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, L. Complexity of danger: The diverse nature of damage-associated molecular patterns. J. Biol. Chem. 2014, 289, 35237–35245. [Google Scholar] [CrossRef]
- Schaefer, L.; Tredup, C.; Gubbiotti, M.A.; Iozzo, R.V. Proteoglycan neofunctions: Regulation of inflammation and autophagy in cancer biology. FEBS J. 2017, 284, 10–26. [Google Scholar] [CrossRef]
- Giatagana, E.M.; Berdiaki, A.; Gaardlos, M.; Tsatsakis, A.M.; Samsonov, S.A.; Nikitovic, D. Rapamycin-induced autophagy in osteosarcoma cells is mediated via the biglycan/Wnt/beta-catenin signaling axis. Am. J. Physiol. Cell Physiol. 2022, 323, C1740–C1756. [Google Scholar] [CrossRef]
- Jarvelainen, H.; Sainio, A.; Koulu, M.; Wight, T.N.; Penttinen, R. Extracellular matrix molecules: Potential targets in pharmacotherapy. Pharmacol. Rev. 2009, 61, 198–223. [Google Scholar] [CrossRef]
- Kavasi, R.M.; Neagu, M.; Constantin, C.; Munteanu, A.; Surcel, M.; Tsatsakis, A.; Tzanakakis, G.N.; Nikitovic, D. Matrix Effectors in the Pathogenesis of Keratinocyte-Derived Carcinomas. Front. Med. 2022, 9, 879500. [Google Scholar] [CrossRef]
- Chen, G.Y.; Nunez, G. Sterile inflammation: Sensing and reacting to damage. Nat. Rev. Immunol. 2010, 10, 826–837. [Google Scholar] [CrossRef]
- Sorensen, H.T.; Friis, S.; Olsen, J.H.; Thulstrup, A.M.; Mellemkjaer, L.; Linet, M.; Trichopoulos, D.; Vilstrup, H.; Olsen, J. Risk of liver and other types of cancer in patients with cirrhosis: A nationwide cohort study in Denmark. Hepatology 1998, 28, 921–925. [Google Scholar] [CrossRef]
- Morris, B.A.; Burkel, B.; Ponik, S.M.; Fan, J.; Condeelis, J.S.; Aguirre-Ghiso, J.A.; Castracane, J.; Denu, J.M.; Keely, P.J. Collagen Matrix Density Drives the Metabolic Shift in Breast Cancer Cells. EBioMedicine 2016, 13, 146–156. [Google Scholar] [CrossRef]
- Reszegi, A.; Tatrai, P.; Regos, E.; Kovalszky, I.; Baghy, K. Syndecan-1 in liver pathophysiology. Am. J. Physiol. Cell Physiol. 2022, 323, C289–C294. [Google Scholar] [CrossRef]
- Mytilinaiou, M.; Nikitovic, D.; Berdiaki, A.; Kostouras, A.; Papoutsidakis, A.; Tsatsakis, A.M.; Tzanakakis, G.N. Emerging roles of syndecan 2 in epithelial and mesenchymal cancer progression. IUBMB Life 2017, 69, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, N.; Miyashita, H.; Kretsinger, R.H. Sequence features, structure, ligand interaction, and diseases in small leucine rich repeat proteoglycans. J. Cell Commun. Signal. 2021, 15, 519–531. [Google Scholar] [CrossRef]
- McEwan, P.A.; Scott, P.G.; Bishop, P.N.; Bella, J. Structural correlations in the family of small leucine-rich repeat proteins and proteoglycans. J. Struct. Biol. 2006, 155, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Kobe, B.; Kajava, A.V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001, 11, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Scott, P.G.; McEwan, P.A.; Dodd, C.M.; Bergmann, E.M.; Bishop, P.N.; Bella, J. Crystal structure of the dimeric protein core of decorin, the archetypal small leucine-rich repeat proteoglycan. Proc. Natl. Acad. Sci. USA 2004, 101, 15633–15638. [Google Scholar] [CrossRef]
- Schaefer, L.; Iozzo, R.V. Biological functions of the small leucine-rich proteoglycans: From genetics to signal transduction. J. Biol. Chem. 2008, 283, 21305–21309. [Google Scholar] [CrossRef]
- Dellett, M.; Hu, W.; Papadaki, V.; Ohnuma, S. Small leucine rich proteoglycan family regulates multiple signalling pathways in neural development and maintenance. Dev. Growth Differ. 2012, 54, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Henry, S.P.; Takanosu, M.; Boyd, T.C.; Mayne, P.M.; Eberspaecher, H.; Zhou, W.; de Crombrugghe, B.; Hook, M.; Mayne, R. Expression pattern and gene characterization of asporin. a newly discovered member of the leucine-rich repeat protein family. J. Biol. Chem. 2001, 276, 12212–12221. [Google Scholar] [CrossRef] [PubMed]
- Nikitovic, D.; Katonis, P.; Tsatsakis, A.; Karamanos, N.K.; Tzanakakis, G.N. Lumican, a small leucine-rich proteoglycan. IUBMB Life 2008, 60, 818–823. [Google Scholar] [CrossRef]
- Sanders, E.J.; Walter, M.A.; Parker, E.; Aramburo, C.; Harvey, S. Opticin binds retinal growth hormone in the embryonic vitreous. Investig. Ophthalmol. Vis. Sci. 2003, 44, 5404–5409. [Google Scholar] [CrossRef]
- Neame, P.J.; Sommarin, Y.; Boynton, R.E.; Heinegård, D. The structure of a 38-kDa leucine-rich protein (chondroadherin) isolated from bovine cartilage. J. Biol. Chem. 1994, 269, 21547–21554. [Google Scholar] [CrossRef]
- Raspanti, M.; Viola, M.; Forlino, A.; Tenni, R.; Gruppi, C.; Tira, M.E. Glycosaminoglycans show a specific periodic interaction with type I collagen fibrils. J. Struct. Biol. 2008, 164, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Douglas, T.; Heinemann, S.; Bierbaum, S.; Scharnweber, D.; Worch, H. Fibrillogenesis of collagen types I, II, and III with small leucine-rich proteoglycans decorin and biglycan. Biomacromolecules 2006, 7, 2388–2393. [Google Scholar] [CrossRef]
- Johnstone, B.; Markopoulos, M.; Neame, P.; Caterson, B. Identification and characterization of glycanated and non-glycanated forms of biglycan and decorin in the human intervertebral disc. Biochem. J. 1993, 292 Pt 3, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Roughley, P.J.; White, R.J.; Magny, M.C.; Liu, J.; Pearce, R.H.; Mort, J.S. Non-proteoglycan forms of biglycan increase with age in human articular cartilage. Biochem. J. 1993, 295 (Pt 2), 421–426. [Google Scholar] [CrossRef]
- Rivet, R.; Rao, R.M.; Nizet, P.; Belloy, N.; Huber, L.; Dauchez, M.; Ramont, L.; Baud, S.; Brezillon, S. Differential MMP-14 targeting by biglycan, decorin, fibromodulin, and lumican unraveled by in silico approach. Am. J. Physiol. Cell Physiol. 2023, 324, C353–C365. [Google Scholar] [CrossRef] [PubMed]
- Nikitovic, D.; Berdiaki, A.; Zafiropoulos, A.; Katonis, P.; Tsatsakis, A.; Karamanos, N.K.; Tzanakakis, G.N. Lumican expression is positively correlated with the differentiation and negatively with the growth of human osteosarcoma cells. FEBS J. 2008, 275, 350–361. [Google Scholar] [CrossRef]
- Nikitovic, D.; Aggelidakis, J.; Young, M.F.; Iozzo, R.V.; Karamanos, N.K.; Tzanakakis, G.N. The biology of small leucine-rich proteoglycans in bone pathophysiology. J. Biol. Chem. 2012, 287, 33926–33933. [Google Scholar] [CrossRef]
- Tatara, Y.; Kakizaki, I.; Suto, S.; Ishioka, H.; Negishi, M.; Endo, M. Chondroitin sulfate cluster of epiphycan from salmon nasal cartilage defines binding specificity to collagens. Glycobiology 2015, 25, 557–569. [Google Scholar] [CrossRef]
- Monfort, J.; Tardif, G.; Reboul, P.; Mineau, F.; Roughley, P.; Pelletier, J.P.; Martel-Pelletier, J. Degradation of small leucine-rich repeat proteoglycans by matrix metalloprotease-13: Identification of a new biglycan cleavage site. Arthritis Res. Ther. 2006, 8, R26. [Google Scholar] [CrossRef]
- McBride, O.W.; Fisher, L.W.; Young, M.F. Localization of PGI (biglycan, BGN) and PGII (decorin, DCN, PG-40) genes on human chromosomes Xq13-qter and 12q, respectively. Genomics 1990, 6, 219–225. [Google Scholar] [CrossRef]
- Jongwattanapisan, P.; Terajima, M.; Miguez, P.A.; Querido, W.; Nagaoka, H.; Sumida, N.; Gurysh, E.G.; Ainslie, K.M.; Pleshko, N.; Perera, L.; et al. Identification of the effector domain of biglycan that facilitates BMP-2 osteogenic function. Sci. Rep. 2018, 8, 7022. [Google Scholar] [CrossRef] [PubMed]
- Roughley, P.J.; White, R.J. Dermatan sulphate proteoglycans of human articular cartilage. The properties of dermatan sulphate proteoglycans I and II. Biochem. J. 1989, 262, 823–827. [Google Scholar] [CrossRef]
- Neame, P.J.; Choi, H.U.; Rosenberg, L.C. The primary structure of the core protein of the small, leucine-rich proteoglycan (PG I) from bovine articular cartilage. J. Biol. Chem. 1989, 264, 8653–8661. [Google Scholar] [CrossRef]
- Blochberger, T.C.; Cornuet, P.K.; Hassell, J.R. Isolation and partial characterization of lumican and decorin from adult chicken corneas. A keratan sulfate-containing isoform of decorin is developmentally regulated. J. Biol. Chem. 1992, 267, 20613–20619. [Google Scholar] [CrossRef] [PubMed]
- Schrecengost, P.K.; Blochberger, T.C.; Hassell, J.R. Identification of chick corneal keratan sulfate proteoglycan precursor protein in whole corneas and in cultured corneal fibroblasts. Arch. Biochem. Biophys. 1992, 292, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Chakravarti, S.; Stallings, R.L.; SundarRaj, N.; Cornuet, P.K.; Hassell, J.R. Primary structure of human lumican (keratan sulfate proteoglycan) and localization of the gene (LUM) to chromosome 12q21.3-q22. Genomics 1995, 27, 481–488. [Google Scholar] [CrossRef]
- Dunlevy, J.R.; Neame, P.J.; Vergnes, J.P.; Hassell, J.R. Identification of the N-linked oligosaccharide sites in chick corneal lumican and keratocan that receive keratan sulfate. J. Biol. Chem. 1998, 273, 9615–9621. [Google Scholar] [CrossRef]
- Melching, L.I.; Roughley, P.J. Modulation of keratan sulfate synthesis on lumican by the action of cytokines on human articular chondrocytes. Matrix Biol. 1999, 18, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Roughley, P.J.; White, R.J.; Cs-Szabo, G.; Mort, J.S. Changes with age in the structure of fibromodulin in human articular cartilage. Osteoarthr. Cartil. 1996, 4, 153–161. [Google Scholar] [CrossRef]
- Cornuet, P.K.; Blochberger, T.C.; Hassell, J.R. Molecular polymorphism of lumican during corneal development. Investig. Ophthalmol. Vis. Sci. 1994, 35, 870–877. [Google Scholar]
- Grover, J.; Chen, X.N.; Korenberg, J.R.; Roughley, P.J. The human lumican gene. Organization, chromosomal location, and expression in articular cartilage. J. Biol. Chem. 1995, 270, 21942–21949. [Google Scholar] [CrossRef] [PubMed]
- Chakravarti, S.; Magnuson, T.; Lass, J.H.; Jepsen, K.J.; LaMantia, C.; Carroll, H. Lumican regulates collagen fibril assembly: Skin fragility and corneal opacity in the absence of lumican. J. Cell Biol. 1998, 141, 1277–1286. [Google Scholar] [CrossRef]
- Schaefer, L.; Babelova, A.; Kiss, E.; Hausser, H.J.; Baliova, M.; Krzyzankova, M.; Marsche, G.; Young, M.F.; Mihalik, D.; Gotte, M.; et al. The matrix component biglycan is proinflammatory and signals through Toll-like receptors 4 and 2 in macrophages. J. Clin. Investig. 2005, 115, 2223–2233. [Google Scholar] [CrossRef]
- Zeng-Brouwers, J.; Beckmann, J.; Nastase, M.V.; Iozzo, R.V.; Schaefer, L. De novo expression of circulating biglycan evokes an innate inflammatory tissue response via MyD88/TRIF pathways. Matrix Biol. 2014, 35, 132–142. [Google Scholar] [CrossRef]
- Liu, B.; Xu, T.; Xu, X.; Cui, Y.; Xing, X. Biglycan promotes the chemotherapy resistance of colon cancer by activating NF-kappaB signal transduction. Mol. Cell. Biochem. 2018, 449, 285–294. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, S.; Guo, J.; Sun, L.; Wang, S.; Wang, Y.; Qiu, W.; Lv, J. Identification and Analysis of Key Genes Driving Gastric Cancer Through Bioinformatics. Genet. Test. Mol. Biomark. 2021, 25, 1–11. [Google Scholar] [CrossRef]
- Chen, W.; Yang, Z. Identification of Differentially Expressed Genes Reveals BGN Predicting Overall Survival and Tumor Immune Infiltration of Gastric Cancer. Comput. Math. Methods Med. 2021, 2021, 5494840. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, W.; Piao, S.; Shen, Y.; Li, Z.; Ding, W.; Li, J.; Saiyin, W. Relationship of biglycan and decorin expression with clinicopathological features and prognosis in patients with oral squamous cell carcinoma. J. Oral Pathol. Med. 2023, 52, 20–28. [Google Scholar] [CrossRef]
- Wei, Z.; Zhongqiu, T.; Lu, S.; Zhang, F.; Xie, W.; Wang, Y. Gene coexpression analysis offers important modules and pathway of human lung adenocarcinomas. J. Cell Physiol. 2020, 235, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Schulz, G.B.; Grimm, T.; Sers, C.; Riemer, P.; Elmasry, M.; Kirchner, T.; Stief, C.G.; Karl, A.; Horst, D. Prognostic value and association with epithelial-mesenchymal transition and molecular subtypes of the proteoglycan biglycan in advanced bladder cancer. Urol. Oncol. 2019, 37, 530.e9–530.e18. [Google Scholar] [CrossRef]
- Appunni, S.; Anand, V.; Khandelwal, M.; Seth, A.; Mathur, S.; Sharma, A. Altered expression of small leucine-rich proteoglycans (Decorin, Biglycan and Lumican): Plausible diagnostic marker in urothelial carcinoma of bladder. Tumour Biol. 2017, 39, 1010428317699112. [Google Scholar] [CrossRef]
- Zhu, Y.H.; Yang, F.; Zhang, S.S.; Zeng, T.T.; Xie, X.; Guan, X.Y. High expression of biglycan is associated with poor prognosis in patients with esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2013, 6, 2497–2505. [Google Scholar]
- Gu, X.; Ma, Y.; Xiao, J.; Zheng, H.; Song, C.; Gong, Y.; Xing, X. Up-regulated biglycan expression correlates with the malignancy in human colorectal cancers. Clin. Exp. Med. 2012, 12, 195–199. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Lin, J.; Chen, C.; Chen, Y.; Yang, S.; Cai, X.; He, Y.; Liu, S. Identification of BGN and THBS2 as metastasis-specific biomarkers and poor survival key regulators in human colon cancer by integrated analysis. Clin. Transl. Med. 2022, 12, e973. [Google Scholar] [CrossRef]
- Jacobsen, F.; Kraft, J.; Schroeder, C.; Hube-Magg, C.; Kluth, M.; Lang, D.S.; Simon, R.; Sauter, G.; Izbicki, J.R.; Clauditz, T.S.; et al. Up-regulation of Biglycan is Associated with Poor Prognosis and PTEN Deletion in Patients with Prostate Cancer. Neoplasia 2017, 19, 707–715. [Google Scholar] [CrossRef]
- Furumido, J.; Maishi, N.; Yanagawa-Matsuda, A.; Kikuchi, H.; Matsumoto, R.; Osawa, T.; Abe, T.; Matsuno, Y.; Shinohara, N.; Hida, Y.; et al. Stroma biglycan expression can be a prognostic factor in prostate cancers. Int. J. Urol. 2023, 30, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Culp, M.B.; Soerjomataram, I.; Efstathiou, J.A.; Bray, F.; Jemal, A. Recent Global Patterns in Prostate Cancer Incidence and Mortality Rates. Eur. Urol. 2020, 77, 38–52. [Google Scholar] [CrossRef]
- Zheng, S.; Liang, J.Y.; Tang, Y.; Xie, J.; Zou, Y.; Yang, A.; Shao, N.; Kuang, X.; Ji, F.; Liu, X.; et al. Dissecting the role of cancer-associated fibroblast-derived biglycan as a potential therapeutic target in immunotherapy resistance: A tumor bulk and single-cell transcriptomic study. Clin. Transl. Med. 2023, 13, e1189. [Google Scholar] [CrossRef]
- Huang, H.C.; Cai, B.H.; Suen, C.S.; Lee, H.Y.; Hwang, M.J.; Liu, F.T.; Kannagi, R. BGN/TLR4/NF-B Mediates Epigenetic Silencing of Immunosuppressive Siglec Ligands in Colon Cancer Cells. Cells 2020, 9, 397. [Google Scholar] [CrossRef] [PubMed]
- Niedworok, C.; Rock, K.; Kretschmer, I.; Freudenberger, T.; Nagy, N.; Szarvas, T.; Vom Dorp, F.; Reis, H.; Rubben, H.; Fischer, J.W. Inhibitory role of the small leucine-rich proteoglycan biglycan in bladder cancer. PLoS ONE 2013, 8, e80084. [Google Scholar] [CrossRef]
- Thiesen, A.P.; Mielczarski, B.; Savaris, R.F. Deep learning neural network image analysis of immunohistochemical protein expression reveals a significantly reduced expression of biglycan in breast cancer. PLoS ONE 2023, 18, e0282176. [Google Scholar] [CrossRef] [PubMed]
- Silva Neto, P.C.D.; Kunst, R.; Barbosa, J.L.V.; Leindecker, A.P.T.; Savaris, R.F. Breast cancer dataset with biomarker Biglycan. Data Brief 2023, 47, 108978. [Google Scholar] [CrossRef]
- Onyeisi, J.O.S.; Lopes, C.C.; Gotte, M. Role of syndecan-4 in breast cancer pathophysiology. Am. J. Physiol. Cell Physiol. 2022, 323, C1345–C1354. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.; Zuo, Y.; Ma, F.; Song, H. Lumican expression in gastric cancer and its association with biological behavior and prognosis. Oncol. Lett. 2017, 14, 5235–5240. [Google Scholar] [CrossRef]
- Chen, S.; Guo, D.; Lei, B.; Bi, J.; Yang, H. Biglycan protects human neuroblastoma cells from nitric oxide-induced death by inhibiting AMPK-mTOR mediated autophagy and intracellular ROS level. Biotechnol. Lett. 2020, 42, 657–668. [Google Scholar] [CrossRef]
- Chen, X.; Li, X.; Hu, X.; Jiang, F.; Shen, Y.; Xu, R.; Wu, L.; Wei, P.; Shen, X. LUM Expression and Its Prognostic Significance in Gastric Cancer. Front. Oncol. 2020, 10, 605. [Google Scholar] [CrossRef]
- Pocas, J.; Marques, C.; Gomes, C.; Otake, A.H.; Pinto, F.; Ferreira, M.; Silva, T.; Faria-Ramos, I.; Matos, R.; Ribeiro, A.R.; et al. Syndecan-4 is a maestro of gastric cancer cell invasion and communication that underscores poor survival. Proc. Natl. Acad. Sci. USA 2023, 120, e2214853120. [Google Scholar] [CrossRef] [PubMed]
- Seya, T.; Tanaka, N.; Shinji, S.; Yokoi, K.; Koizumi, M.; Teranishi, N.; Yamashita, K.; Tajiri, T.; Ishiwata, T.; Naito, Z. Lumican expression in advanced colorectal cancer with nodal metastasis correlates with poor prognosis. Oncol. Rep. 2006, 16, 1225–1230. [Google Scholar] [CrossRef]
- Leygue, E.; Snell, L.; Dotzlaw, H.; Hole, K.; Hiller-Hitchcock, T.; Roughley, P.J.; Watson, P.H.; Murphy, L.C. Expression of lumican in human breast carcinoma. Cancer Res. 1998, 58, 1348–1352. [Google Scholar] [PubMed]
- Karamanou, K.; Franchi, M.; Onisto, M.; Passi, A.; Vynios, D.H.; Brezillon, S. Evaluation of lumican effects on morphology of invading breast cancer cells, expression of integrins and downstream signaling. FEBS J. 2020, 287, 4862–4880. [Google Scholar] [CrossRef] [PubMed]
- Karamanou, K.; Franchi, M.; Vynios, D.; Brezillon, S. Epithelial-to-mesenchymal transition and invadopodia markers in breast cancer: Lumican a key regulator. Semin. Cancer Biol. 2020, 62, 125–133. [Google Scholar] [CrossRef]
- Brezillon, S.; Venteo, L.; Ramont, L.; D’Onofrio, M.F.; Perreau, C.; Pluot, M.; Maquart, F.X.; Wegrowski, Y. Expression of lumican, a small leucine-rich proteoglycan with antitumour activity, in human malignant melanoma. Clin. Exp. Dermatol. 2007, 32, 405–416. [Google Scholar] [CrossRef]
- Karamanou, K.; Franchi, M.; Proult, I.; Rivet, R.; Vynios, D.; Brezillon, S. Lumican Inhibits In Vivo Melanoma Metastasis by Altering Matrix-Effectors and Invadopodia Markers. Cells 2021, 10, 841. [Google Scholar] [CrossRef]
- Sifaki, M.; Assouti, M.; Nikitovic, D.; Krasagakis, K.; Karamanos, N.K.; Tzanakakis, G.N. Lumican, a small leucine-rich proteoglycan substituted with keratan sulfate chains is expressed and secreted by human melanoma cells and not normal melanocytes. IUBMB Life 2006, 58, 606–610. [Google Scholar] [CrossRef] [PubMed]
- Suhail, Y.; Cain, M.P.; Vanaja, K.; Kurywchak, P.A.; Levchenko, A.; Kalluri, R. Kshitiz Systems Biology of Cancer Metastasis. Cell Syst. 2019, 9, 109–127. [Google Scholar] [CrossRef]
- Condeelis, J.; Segall, J.E. Intravital imaging of cell movement in tumours. Nat. Rev. Cancer 2003, 3, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Wang, X.; Zhang, Y.; Che, X.; Liu, Z.; Zhang, L.; Qiu, C.; Lv, Q.; Jiang, J. Biglycan enhances the ability of migration and invasion in endometrial cancer. Arch. Gynecol. Obstet. 2016, 293, 429–438. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, R.; Feng, L.; Ma, H.; Fang, J. LINC00460 Promotes Cell Proliferation, Migration, Invasion, and Epithelial-Mesenchymal Transition of Head and Neck Squamous Cell Carcinoma via miR-320a/BGN Axis. Onco Targets Ther. 2021, 14, 2279–2291. [Google Scholar] [CrossRef]
- Hu, L.; Duan, Y.T.; Li, J.F.; Su, L.P.; Yan, M.; Zhu, Z.G.; Liu, B.Y.; Yang, Q.M. Biglycan enhances gastric cancer invasion by activating FAK signaling pathway. Oncotarget 2014, 5, 1885–1896. [Google Scholar] [CrossRef] [PubMed]
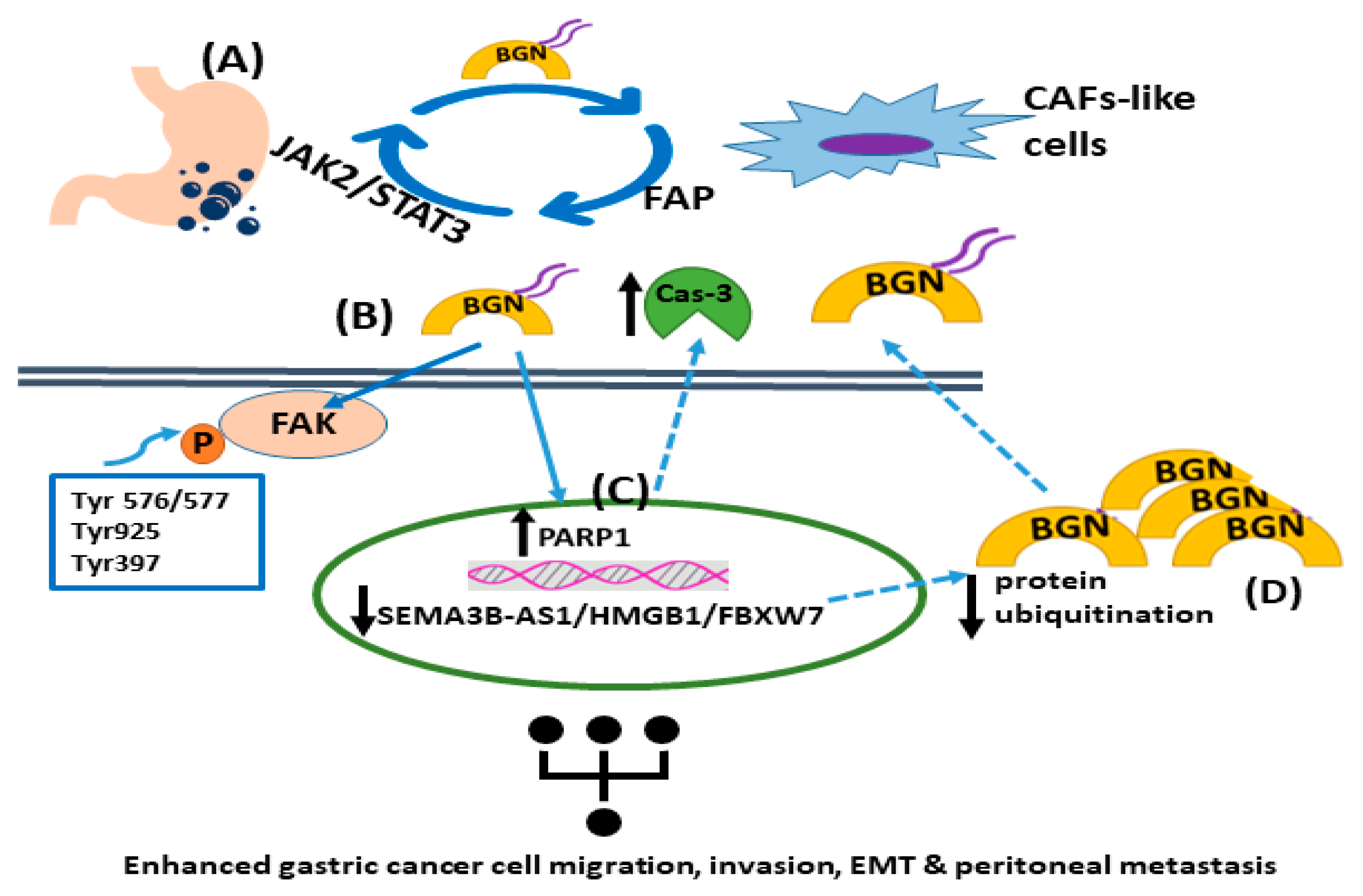
- Wu, H.; Xiang, Z.; Huang, G.; He, Q.; Song, J.; Dou, R.; Yang, C.; Wang, S.; Xiong, B. BGN/FAP/STAT3 positive feedback loop mediated mutual interaction between tumor cells and mesothelial cells contributes to peritoneal metastasis of gastric cancer. Int. J. Biol. Sci. 2023, 19, 465–483. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Xiang, Z.; Wu, H.; He, Q.; Dou, R.; Yang, C.; Song, J.; Huang, S.; Wang, S.; Xiong, B. The lncRNA SEMA3B-AS1/HMGB1/FBXW7 Axis Mediates the Peritoneal Metastasis of Gastric Cancer by Regulating BGN Protein Ubiquitination. Oxid. Med. Cell Longev. 2022, 2022, 5055684. [Google Scholar] [CrossRef]
- Pinto, F.; Santos-Ferreira, L.; Pinto, M.T.; Gomes, C.; Reis, C.A. The Extracellular Small Leucine-Rich Proteoglycan Biglycan Is a Key Player in Gastric Cancer Aggressiveness. Cancers 2021, 13, 1330. [Google Scholar] [CrossRef]
- Guo, D.; Zhang, W.; Yang, H.; Bi, J.; Xie, Y.; Cheng, B.; Wang, Y.; Chen, S. Celastrol Induces Necroptosis and Ameliorates Inflammation via Targeting Biglycan in Human Gastric Carcinoma. Int. J. Mol. Sci. 2019, 20, 5716. [Google Scholar] [CrossRef] [PubMed]
- Manupati, K.; Paul, R.; Hao, M.; Haas, M.; Bian, Z.C.; Holm, T.M.; Guan, J.L.; Yeo, S.K. Biglycan Promotes Cancer Stem Cell Properties, NFkappaB Signaling and Metastatic Potential in Breast Cancer Cells. Cancers 2022, 14, 455. [Google Scholar] [CrossRef]
- Recktenwald, C.V.; Leisz, S.; Steven, A.; Mimura, K.; Muller, A.; Wulfanger, J.; Kiessling, R.; Seliger, B. HER-2/neu-mediated down-regulation of biglycan associated with altered growth properties. J. Biol. Chem. 2012, 287, 24320–24329. [Google Scholar] [CrossRef] [PubMed]
- Subbarayan, K.; Massa, C.; Leisz, S.; Steven, A.; Bethmann, D.; Biehl, K.; Wickenhauser, C.; Seliger, B. Biglycan as a potential regulator of tumorgenicity and immunogenicity in K-RAS-transformed cells. Oncoimmunology 2022, 11, 2069214. [Google Scholar] [CrossRef]
- Datsis, G.A.; Berdiaki, A.; Nikitovic, D.; Mytilineou, M.; Katonis, P.; Karamanos, N.K.; Tzanakakis, G.N. Parathyroid hormone affects the fibroblast growth factor-proteoglycan signaling axis to regulate osteosarcoma cell migration. FEBS J. 2011, 278, 3782–3792. [Google Scholar] [CrossRef]
- Coulson-Thomas, V.J.; Coulson-Thomas, Y.M.; Gesteira, T.F.; de Paula, C.A.A.; Mader, A.M.; Waisberg, J.; Pinhal, M.A.; Friedl, A.; Toma, L.; Nader, H.B. Colorectal cancer desmoplastic reaction up-regulates collagen synthesis and restricts cancer cell invasion. Cell Tissue Res. 2011, 346, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Kunz-Schughart, L.A.; Knuechel, R. Tumor-associated fibroblasts (part I): Active stromal participants in tumor development and progression? Histol. Histopathol. 2002, 17, 599–621. [Google Scholar] [CrossRef]
- Apte, M.V.; Park, S.; Phillips, P.A.; Santucci, N.; Goldstein, D.; Kumar, R.K.; Ramm, G.A.; Buchler, M.; Friess, H.; McCarroll, J.A.; et al. Desmoplastic reaction in pancreatic cancer: Role of pancreatic stellate cells. Pancreas 2004, 29, 179–187. [Google Scholar] [CrossRef]
- Otterbein, H.; Lehnert, H.; Ungefroren, H. Negative Control of Cell Migration by Rac1b in Highly Metastatic Pancreatic Cancer Cells Is Mediated by Sequential Induction of Nonactivated Smad3 and Biglycan. Cancers 2019, 11, 1959. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.L.; Condeelis, J.S. Cytoplasmic structure and contractility in amoeboid cells. Int. Rev. Cytol. 1979, 56, 57–144. [Google Scholar] [CrossRef]
- Genot, E.; Gligorijevic, B. Invadosomes in their natural habitat. Eur. J. Cell Biol. 2014, 93, 367–379. [Google Scholar] [CrossRef]
- Di Martino, J.; Henriet, E.; Ezzoukhry, Z.; Goetz, J.G.; Moreau, V.; Saltel, F. The microenvironment controls invadosome plasticity. J. Cell Sci. 2016, 129, 1759–1768. [Google Scholar] [CrossRef]
- Coulson-Thomas, V.J.; Coulson-Thomas, Y.M.; Gesteira, T.F.; Andrade de Paula, C.A.; Carneiro, C.R.; Ortiz, V.; Toma, L.; Kao, W.W.; Nader, H.B. Lumican expression, localization and antitumor activity in prostate cancer. Exp. Cell Res. 2013, 319, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Radwanska, A.; Baczynska, D.; Nowak, D.; Brezillon, S.; Popow, A.; Maquart, F.X.; Wegrowski, Y.; Malicka-Blaszkiewicz, M. Lumican affects actin cytoskeletal organization in human melanoma A375 cells. Life Sci. 2008, 83, 651–660. [Google Scholar] [CrossRef]
- Stasiak, M.; Boncela, J.; Perreau, C.; Karamanou, K.; Chatron-Colliet, A.; Proult, I.; Przygodzka, P.; Chakravarti, S.; Maquart, F.X.; Kowalska, M.A.; et al. Lumican Inhibits SNAIL-Induced Melanoma Cell Migration Specifically by Blocking MMP-14 Activity. PLoS ONE 2016, 11, e0150226. [Google Scholar] [CrossRef]
- Zeltz, C.; Brezillon, S.; Kapyla, J.; Eble, J.A.; Bobichon, H.; Terryn, C.; Perreau, C.; Franz, C.M.; Heino, J.; Maquart, F.X.; et al. Lumican inhibits cell migration through alpha2beta1 integrin. Exp. Cell Res. 2010, 316, 2922–2931. [Google Scholar] [CrossRef]
- Karamanou, K.; Franchi, M.; Piperigkou, Z.; Perreau, C.; Maquart, F.X.; Vynios, D.H.; Brezillon, S. Lumican effectively regulates the estrogen receptors-associated functional properties of breast cancer cells, expression of matrix effectors and epithelial-to-mesenchymal transition. Sci. Rep. 2017, 7, 45138. [Google Scholar] [CrossRef]
- Mao, W.; Luo, M.; Huang, X.; Wang, Q.; Fan, J.; Gao, L.; Zhang, Y.; Geng, J. Knockdown of Lumican Inhibits Proliferation and Migration of Bladder Cancer. Transl. Oncol. 2019, 12, 1072–1078. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, K.C.; Chu, P.Y.; Chang, G.C.; Liu, K.J. Elevated Expression of Lumican in Lung Cancer Cells Promotes Bone Metastasis through an Autocrine Regulatory Mechanism. Cancers 2020, 12, 233. [Google Scholar] [CrossRef]
- Radwanska, A.; Litwin, M.; Nowak, D.; Baczynska, D.; Wegrowski, Y.; Maquart, F.X.; Malicka-Blaszkiewicz, M. Overexpression of lumican affects the migration of human colon cancer cells through up-regulation of gelsolin and filamentous actin reorganization. Exp. Cell Res. 2012, 318, 2312–2323. [Google Scholar] [CrossRef]
- Nikitovic, D.; Chalkiadaki, G.; Berdiaki, A.; Aggelidakis, J.; Katonis, P.; Karamanos, N.K.; Tzanakakis, G.N. Lumican regulates osteosarcoma cell adhesion by modulating TGFbeta2 activity. Int. J. Biochem. Cell Biol. 2011, 43, 928–935. [Google Scholar] [CrossRef]
- Salcher, S.; Spoden, G.; Huber, J.M.; Golderer, G.; Lindner, H.; Ausserlechner, M.J.; Kiechl-Kohlendorfer, U.; Geiger, K.; Obexer, P. Repaglinide Silences the FOXO3/Lumican Axis and Represses the Associated Metastatic Potential of Neuronal Cancer Cells. Cells 2019, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Ishiwata, T.; Cho, K.; Kawahara, K.; Yamamoto, T.; Fujiwara, Y.; Uchida, E.; Tajiri, T.; Naito, Z. Role of lumican in cancer cells and adjacent stromal tissues in human pancreatic cancer. Oncol. Rep. 2007, 18, 537–543. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Berendsen, A.D.; Fisher, L.W.; Kilts, T.M.; Owens, R.T.; Robey, P.G.; Gutkind, J.S.; Young, M.F. Modulation of canonical Wnt signaling by the extracellular matrix component biglycan. Proc. Natl. Acad. Sci. USA 2011, 108, 17022–17027. [Google Scholar] [CrossRef]
- Mirzaei, R.; D’Mello, C.; Liu, M.; Nikolic, A.; Kumar, M.; Visser, F.; Bose, P.; Gallo, M.; Yong, V.W. Single-Cell Spatial Analysis Identifies Regulators of Brain Tumor-Initiating Cells. Cancer Res. 2023, 83, 1725–1741. [Google Scholar] [CrossRef]
- Xing, X.; Gu, X.; Ma, T. Knockdown of biglycan expression by RNA interference inhibits the proliferation and invasion of, and induces apoptosis in, the HCT116 colon cancer cell line. Mol. Med. Rep. 2015, 12, 7538–7544. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Gu, X.; Ma, T.; Ye, H. Biglycan up-regulated vascular endothelial growth factor (VEGF) expression and promoted angiogenesis in colon cancer. Tumour Biol. 2015, 36, 1773–1780. [Google Scholar] [CrossRef]
- Zeng, S.; Zhou, F.; Wang, Y.; Zhai, Z.; Xu, L.; Wang, H.; Chen, X.; Luo, S.; Cheng, M. Aberrant expression of the extracellular matrix component Biglycan regulated by Hedgehog signalling promotes colorectal cancer cell proliferation. Acta Biochim. Biophys. Sin. 2022, 54, 243–251. [Google Scholar] [CrossRef]
- Weber, C.K.; Sommer, G.; Michl, P.; Fensterer, H.; Weimer, M.; Gansauge, F.; Leder, G.; Adler, G.; Gress, T.M. Biglycan is overexpressed in pancreatic cancer and induces G1-arrest in pancreatic cancer cell lines. Gastroenterology 2001, 121, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, Q.; Yu, Z.; Wu, X.; Chen, X.; Li, J.; Li, C.; Yan, M.; Zhu, Z.; Liu, B.; et al. Cancer-associated fibroblast-derived Lumican promotes gastric cancer progression via the integrin β1-FAK signaling pathway. Int. J. Cancer 2017, 141, 998–1010. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.T.; Hsu, P.C.; Chow, S.E. Downregulation of lumican enhanced mitotic defects and aneuploidy in lung cancer cells. Cell Cycle 2020, 19, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Appunni, S.; Anand, V.; Khandelwal, M.; Gupta, N.; Rubens, M.; Sharma, A. Small Leucine Rich Proteoglycans (decorin, biglycan and lumican) in cancer. Clin. Chim. Acta 2019, 491, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, Z.; Chan, D.; Chung, P.Y.; Wang, Y.; Chan, A.S.C.; Law, S.; Lam, K.H.; Tang, J.C.O. The Anticancer Effect of a Novel Quinoline Derivative 91b1 through Downregulation of Lumican. Int. J. Mol. Sci. 2022, 23, 13181. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Truty, M.A.; Kang, Y.; Chopin-Laly, X.; Zhang, R.; Roife, D.; Chatterjee, D.; Lin, E.; Thomas, R.M.; Wang, H.; et al. Extracellular lumican inhibits pancreatic cancer cell growth and is associated with prolonged survival after surgery. Clin. Cancer Res. 2014, 20, 6529–6540. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kang, Y.; Roife, D.; Lee, Y.; Pratt, M.; Perez, M.R.; Dai, B.; Koay, E.J.; Fleming, J.B. Prolonged exposure to extracellular lumican restrains pancreatic adenocarcinoma growth. Oncogene 2017, 36, 5432–5438. [Google Scholar] [CrossRef] [PubMed]
- Jeanne, A.; Untereiner, V.; Perreau, C.; Proult, I.; Gobinet, C.; Boulagnon-Rombi, C.; Terryn, C.; Martiny, L.; Brezillon, S.; Dedieu, S. Lumican delays melanoma growth in mice and drives tumor molecular assembly as well as response to matrix-targeted TAX2 therapeutic peptide. Sci. Rep. 2017, 7, 7700. [Google Scholar] [CrossRef]
- Dvorak, H.F. Tumors: Wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 1986, 315, 1650–1659. [Google Scholar] [CrossRef]
- Zeng-Brouwers, J.; Huber, L.S.; Merline, R.; Trebicka, J.; Wygrecka, M.; Schaefer, L. Evaluation of the In Vitro and In Vivo Effects of Biglycan in Innate Immunity. Methods Mol. Biol. 2023, 2619, 109–124. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Zou, Y.; Tang, Y.; Yang, A.; Liang, J.Y.; Wu, L.; Tian, W.; Xiao, W.; Xie, X.; Yang, L.; et al. Landscape of cancer-associated fibroblasts identifies the secreted biglycan as a protumor and immunosuppressive factor in triple-negative breast cancer. Oncoimmunology 2022, 11, 2020984. [Google Scholar] [CrossRef] [PubMed]
- Roedig, H.; Nastase, M.V.; Wygrecka, M.; Schaefer, L. Breaking down chronic inflammatory diseases: The role of biglycan in promoting a switch between inflammation and autophagy. FEBS J. 2019, 286, 2965–2979. [Google Scholar] [CrossRef]
- Roedig, H.; Nastase, M.V.; Frey, H.; Moreth, K.; Zeng-Brouwers, J.; Poluzzi, C.; Hsieh, L.T.; Brandts, C.; Fulda, S.; Wygrecka, M.; et al. Biglycan is a new high-affinity ligand for CD14 in macrophages. Matrix Biol. 2019, 77, 4–22. [Google Scholar] [CrossRef]
- Moreth, K.; Frey, H.; Hubo, M.; Zeng-Brouwers, J.; Nastase, M.V.; Hsieh, L.T.; Haceni, R.; Pfeilschifter, J.; Iozzo, R.V.; Schaefer, L. Biglycan-triggered TLR-2- and TLR-4-signaling exacerbates the pathophysiology of ischemic acute kidney injury. Matrix Biol. 2014, 35, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yang, H.; Xiang, X.; Liu, L.; Huang, H.; Tang, G. BGN May be a Potential Prognostic Biomarker and Associated With Immune Cell Enrichment of Gastric Cancer. Front. Genet. 2022, 13, 765569. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liang, J.; Zhong, W.; Han, C.; Liu, D.; Chen, X. Expression and prognostic analysis of BGN in head and neck squamous cell carcinoma. Gene 2022, 827, 146461. [Google Scholar] [CrossRef] [PubMed]
- Vaxevanis, C.K.; Bauer, M.; Subbarayan, K.; Friedrich, M.; Massa, C.; Biehl, K.; Al-Ali, H.K.; Wickenhauser, C.; Seliger, B. Biglycan as a mediator of proinflammatory response and target for MDS and sAML therapy. Oncoimmunology 2023, 12, 2152998. [Google Scholar] [CrossRef] [PubMed]
- Hatano, S.; Watanabe, H. Regulation of Macrophage and Dendritic Cell Function by Chondroitin Sulfate in Innate to Antigen-Specific Adaptive Immunity. Front. Immunol. 2020, 11, 232. [Google Scholar] [CrossRef] [PubMed]
- Mittal, K.; Ebos, J.; Rini, B. Angiogenesis and the tumor microenvironment: Vascular endothelial growth factor and beyond. Semin. Oncol. 2014, 41, 235–251. [Google Scholar] [CrossRef]
- Aguilar-Cazares, D.; Chavez-Dominguez, R.; Carlos-Reyes, A.; Lopez-Camarillo, C.; Hernadez de la Cruz, O.N.; Lopez-Gonzalez, J.S. Contribution of Angiogenesis to Inflammation and Cancer. Front. Oncol. 2019, 9, 1399. [Google Scholar] [CrossRef]
- Lapeyre-Prost, A.; Terme, M.; Pernot, S.; Pointet, A.L.; Voron, T.; Tartour, E.; Taieb, J. Immunomodulatory Activity of VEGF in Cancer. Int. Rev. Cell Mol. Biol. 2017, 330, 295–342. [Google Scholar] [CrossRef]
- Arroyo, A.G.; Iruela-Arispe, M.L. Extracellular matrix, inflammation, and the angiogenic response. Cardiovasc. Res. 2010, 86, 226–235. [Google Scholar] [CrossRef]
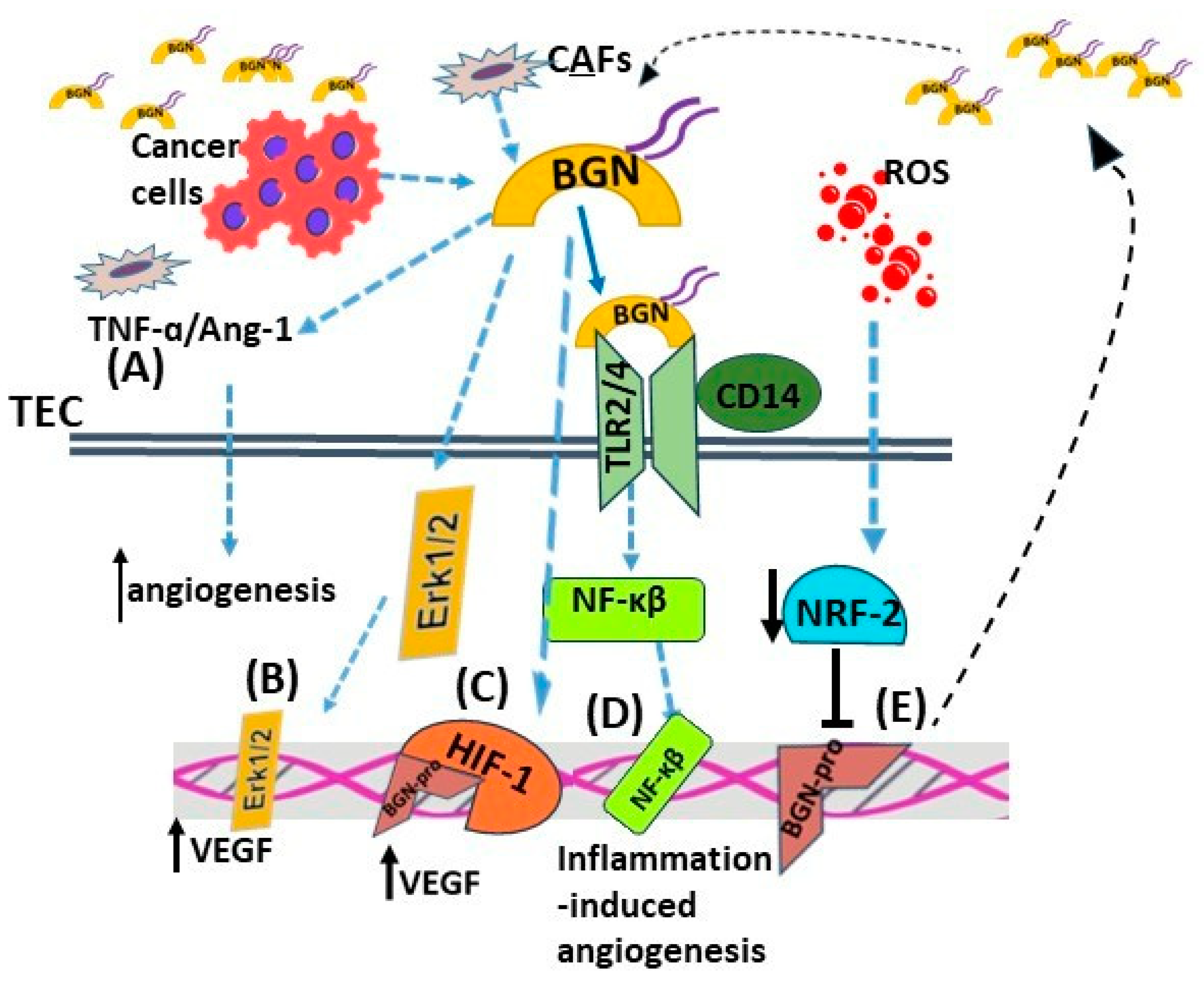
- Yamamoto, K.; Ohga, N.; Hida, Y.; Maishi, N.; Kawamoto, T.; Kitayama, K.; Akiyama, K.; Osawa, T.; Kondoh, M.; Matsuda, K.; et al. Biglycan is a specific marker and an autocrine angiogenic factor of tumour endothelial cells. Br. J. Cancer 2012, 106, 1214–1223. [Google Scholar] [CrossRef]
- Maishi, N.; Ohba, Y.; Akiyama, K.; Ohga, N.; Hamada, J.; Nagao-Kitamoto, H.; Alam, M.T.; Yamamoto, K.; Kawamoto, T.; Inoue, N.; et al. Tumour endothelial cells in high metastatic tumours promote metastasis via epigenetic dysregulation of biglycan. Sci. Rep. 2016, 6, 28039. [Google Scholar] [CrossRef]
- Cong, L.; Maishi, N.; Annan, D.A.; Young, M.F.; Morimoto, H.; Morimoto, M.; Nam, J.M.; Hida, Y.; Hida, K. Inhibition of stromal biglycan promotes normalization of the tumor microenvironment and enhances chemotherapeutic efficacy. Breast Cancer Res. 2021, 23, 51. [Google Scholar] [CrossRef]
- Maishi, N.; Sakurai, Y.; Hatakeyama, H.; Umeyama, Y.; Nakamura, T.; Endo, R.; Alam, M.T.; Li, C.; Annan, D.A.; Kikuchi, H.; et al. Novel antiangiogenic therapy targeting biglycan using tumor endothelial cell-specific liposomal siRNA delivery system. Cancer Sci. 2022, 113, 1855–1867. [Google Scholar] [CrossRef]
- Hu, L.; Zang, M.D.; Wang, H.X.; Li, J.F.; Su, L.P.; Yan, M.; Li, C.; Yang, Q.M.; Liu, B.Y.; Zhu, Z.G. Biglycan stimulates VEGF expression in endothelial cells by activating the TLR signaling pathway. Mol. Oncol. 2016, 10, 1473–1484. [Google Scholar] [CrossRef] [PubMed]
- Roedig, H.; Damiescu, R.; Zeng-Brouwers, J.; Kutija, I.; Trebicka, J.; Wygrecka, M.; Schaefer, L. Danger matrix molecules orchestrate CD14/CD44 signaling in cancer development. Semin. Cancer Biol. 2020, 62, 31–47. [Google Scholar] [CrossRef] [PubMed]
- Hojo, T.; Maishi, N.; Towfik, A.M.; Akiyama, K.; Ohga, N.; Shindoh, M.; Hida, Y.; Minowa, K.; Fujisawa, T.; Hida, K. ROS enhance angiogenic properties via regulation of NRF2 in tumor endothelial cells. Oncotarget 2017, 8, 45484–45495. [Google Scholar] [CrossRef]
- Chavez-Dominguez, R.; Perez-Medina, M.; Lopez-Gonzalez, J.S.; Galicia-Velasco, M.; Aguilar-Cazares, D. The Double-Edge Sword of Autophagy in Cancer: From Tumor Suppression to Pro-tumor Activity. Front. Oncol. 2020, 10, 578418. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, G.E.; Akim, A.M.; Sung, Y.Y.; Muhammad, T.S.T. Cancer and Apoptosis. Methods Mol. Biol. 2022, 2543, 191–210. [Google Scholar] [CrossRef]
- Poluzzi, C.; Nastase, M.V.; Zeng-Brouwers, J.; Roedig, H.; Hsieh, L.T.; Michaelis, J.B.; Buhl, E.M.; Rezende, F.; Manavski, Y.; Bleich, A.; et al. Biglycan evokes autophagy in macrophages via a novel CD44/Toll-like receptor 4 signaling axis in ischemia/reperfusion injury. Kidney Int. 2019, 95, 540–562. [Google Scholar] [CrossRef]
- Schulz, M.; Diehl, V.; Trebicka, J.; Wygrecka, M.; Schaefer, L. Biglycan: A regulator of hepatorenal inflammation and autophagy. Matrix Biol. 2021, 100–101, 150–161. [Google Scholar] [CrossRef]
- Mintz, M.B.; Sowers, R.; Brown, K.M.; Hilmer, S.C.; Mazza, B.; Huvos, A.G.; Meyers, P.A.; Lafleur, B.; McDonough, W.S.; Henry, M.M.; et al. An expression signature classifies chemotherapy-resistant pediatric osteosarcoma. Cancer Res. 2005, 65, 1748–1754. [Google Scholar] [CrossRef]
- Fang, D.; Lai, Z.; Wang, Y. Overexpression of Biglycan is Associated with Resistance to Rapamycin in Human WERI-Rb-1 Retinoblastoma Cells by Inducing the Activation of the Phosphatidylinositol 3-Kinases (PI3K)/Akt/Nuclear Factor kappa B (NF-kappaB) Signaling Pathway. Med. Sci. Monit. 2019, 25, 6639–6648. [Google Scholar] [CrossRef]
- Li, X.; Lee, Y.; Kang, Y.; Dai, B.; Perez, M.R.; Pratt, M.; Koay, E.J.; Kim, M.; Brekken, R.A.; Fleming, J.B. Hypoxia-induced autophagy of stellate cells inhibits expression and secretion of lumican into microenvironment of pancreatic ductal adenocarcinoma. Cell Death Differ. 2019, 26, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Vij, N.; Roberts, L.; Joyce, S.; Chakravarti, S. Lumican suppresses cell proliferation and aids Fas-Fas ligand mediated apoptosis: Implications in the cornea. Exp. Eye Res. 2004, 78, 957–971. [Google Scholar] [CrossRef] [PubMed]
- Vuillermoz, B.; Khoruzhenko, A.; D’Onofrio, M.F.; Ramont, L.; Venteo, L.; Perreau, C.; Antonicelli, F.; Maquart, F.X.; Wegrowski, Y. The small leucine-rich proteoglycan lumican inhibits melanoma progression. Exp. Cell Res. 2004, 296, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Brezillon, S.; Untereiner, V.; Mohamed, H.T.; Ahallal, E.; Proult, I.; Nizet, P.; Boulagnon-Rombi, C.; Sockalingum, G.D. Label-Free Infrared Spectral Histology of Skin Tissue Part II: Impact of a Lumican-Derived Peptide on Melanoma Growth. Front. Cell Dev. Biol. 2020, 8, 377. [Google Scholar] [CrossRef]
- Yu, Z.; Liu, L.; Shu, Q.; Li, D.; Wang, R. Leukemia stem cells promote chemoresistance by inducing downregulation of lumican in mesenchymal stem cells. Oncol. Lett. 2019, 18, 4317–4327. [Google Scholar] [CrossRef]
- Li, X.; Roife, D.; Kang, Y.; Dai, B.; Pratt, M.; Fleming, J.B. Extracellular lumican augments cytotoxicity of chemotherapy in pancreatic ductal adenocarcinoma cells via autophagy inhibition. Oncogene 2016, 35, 4881–4890. [Google Scholar] [CrossRef]
- Morimoto, H.; Hida, Y.; Maishi, N.; Nishihara, H.; Hatanaka, Y.; Li, C.; Matsuno, Y.; Nakamura, T.; Hirano, S.; Hida, K. Biglycan, tumor endothelial cell secreting proteoglycan, as possible biomarker for lung cancer. Thorac. Cancer 2021, 12, 1347–1357. [Google Scholar] [CrossRef]
- Babelova, A.; Moreth, K.; Tsalastra-Greul, W.; Zeng-Brouwers, J.; Eickelberg, O.; Young, M.F.; Bruckner, P.; Pfeilschifter, J.; Schaefer, R.M.; Grone, H.J.; et al. Biglycan, a danger signal that activates the NLRP3 inflammasome via toll-like and P2X receptors. J. Biol. Chem. 2009, 284, 24035–24048. [Google Scholar] [CrossRef]
- Schaefer, L.; Iozzo, R.V. Small leucine-rich proteoglycans, at the crossroad of cancer growth and inflammation. Curr. Opin. Genet. Dev. 2012, 22, 56–57. [Google Scholar] [CrossRef] [PubMed]
- Farnebo, L.; Shahangian, A.; Lee, Y.; Shin, J.H.; Scheeren, F.A.; Sunwoo, J.B. Targeting Toll-like receptor 2 inhibits growth of head and neck squamous cell carcinoma. Oncotarget 2015, 6, 9897–9907. [Google Scholar] [CrossRef] [PubMed]
- Dajon, M.; Iribarren, K.; Cremer, I. Toll-like receptor stimulation in cancer: A pro- and anti-tumor double-edged sword. Immunobiology 2017, 222, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Moody, S.E.; Perez, D.; Pan, T.C.; Sarkisian, C.J.; Portocarrero, C.P.; Sterner, C.J.; Notorfrancesco, K.L.; Cardiff, R.D.; Chodosh, L.A. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 2005, 8, 197–209. [Google Scholar] [CrossRef]
- Perez, S.; Makshakova, O.; Angulo, J.; Bedini, E.; Bisio, A.; de Paz, J.L.; Fadda, E.; Guerrini, M.; Hricovini, M.; Lisacek, F.; et al. Glycosaminoglycans: What Remains To Be Deciphered? JACS Au 2023, 3, 628–656. [Google Scholar] [CrossRef] [PubMed]
| Role | References |
|---|---|
| Biglycan, which is attached to the matrix, is cleaved by proteases and released under pathological conditions. | [21,69] |
| Activated macrophages can produce de novo biglycan. | [70] |
| Overexpression of biglycan mRNA has been detected in the vast majority of cancer tissues and 28 cancer types using the Oncomine database. | [22] |
| Biglycan is overexpressed in human endometrial cancer, both in the parenchyma and mesenchyme compartments. | [71] |
| Biglycan’s gene expression was correlated with gastric cancer metastases. | [72] |
| Biglycan facilitates the incidence of lung adenocarcinoma through ECM–receptor interaction. | [75] |
| Biglycan was identified as a metastasis-specific biomarker in human colon cancer using integrated analysis. | [80] |
| Overexpressed biglycan was strongly correlated with prostate cancer development. | [81] |
| Biglycan expressed in the cancerous prostate stroma was suggested as a prognostic factor. | [82] |
| Bladder cancer patients displaying high biglycan expression exhibited reduced tumor cell growth and enhanced ten-year survival in patients. | [86] |
| High biglycan expression in oral squamous cell carcinoma was correlated with poor overall survival, as well as tumor-specific survival. | [74] |
| Biglycan protein expression was reduced in breast cancer tissue compared to normal tissue. | [87] |
| Single-cell RNA sequencing pan-cancer cohorts showed that enhanced biglycan production by CAF was negatively correlated with patient survival and response to therapy. | [84] |
| Role | References |
|---|---|
| Lumican protein was overexpressed in gastric cancer tissues compared to healthy tissues. Lumican is suggested to be an independent prognostic factor, which needs to be verified in prospective studies. | [90,91,92] |
| Lumican mRNA present in the tumor stroma of breast cancer was correlated with higher tumor grade, lower expression of estrogen receptors (ERs), and the patient’s younger age. | [95] |
| In breast cancer cell lines, lumican affected cancer cell functions and modulated the expression of ECM molecules, abrogating EMT. | [97] |
| In colon cancer, lumican expression was associated with metastasis in lymph nodes, tumor invasion, and a lower survival rate. | [94] |
| In melanoma, lumican was expressed in the dermis and peritumoral stroma, but not in melanoma cells. | [98] |
| Lumican significantly reduced melanoma lung metastasis in vivo and cell invasion in vitro. | [99] |
| Lumican is expressed and secreted by some melanoma cell lines, suggesting its possible involvement in the progression of the disease. | [100] |
| Lumican Action | Type of Cancer | References |
|---|---|---|
| Inhibition of invadopodia and lamellipodia formation. | Prostate | [121] |
| Attenuation of invadopodia formation. | Melanoma | [122] |
| Regulation of Snail-dependent MMP-14 action. | Melanoma | [123] |
| Reduction in migration capacity via core protein binding to α2β1 integrin. | Melanoma | [124] |
| Modulation of ECM molecule expression and reduction in MMP-releasing invadopodia. | Lung | [99] |
| Modulation of cell morphology and EMT, reduction in ECM regulators, like MMPs, and inhibition of migration and invasion. | Breast | [125] |
| Inhibition of lumican reduced migration via the MAPK signaling pathway. | Bladder | [126] |
| Induction of adhesion, cell migration, invasion, and osteogenic metastasis. | Lung | [127] |
| Increased formation and migration of podosome-like protrusions. | Colon | [128] |
| Enhancement of cell migration, chemotactic response to fibronectin, and TGFβ2-dependent adhesion to fibronectin. | Osteosarcoma | [44,52,129] |
| Enhanced migration through a FOX-3-dependent increase in lumican expression. | Neuroblastoma | [130] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berdiaki, A.; Giatagana, E.-M.; Tzanakakis, G.; Nikitovic, D. The Landscape of Small Leucine-Rich Proteoglycan Impact on Cancer Pathogenesis with a Focus on Biglycan and Lumican. Cancers 2023, 15, 3549. https://doi.org/10.3390/cancers15143549
Berdiaki A, Giatagana E-M, Tzanakakis G, Nikitovic D. The Landscape of Small Leucine-Rich Proteoglycan Impact on Cancer Pathogenesis with a Focus on Biglycan and Lumican. Cancers. 2023; 15(14):3549. https://doi.org/10.3390/cancers15143549
Chicago/Turabian StyleBerdiaki, Aikaterini, Eirini-Maria Giatagana, George Tzanakakis, and Dragana Nikitovic. 2023. "The Landscape of Small Leucine-Rich Proteoglycan Impact on Cancer Pathogenesis with a Focus on Biglycan and Lumican" Cancers 15, no. 14: 3549. https://doi.org/10.3390/cancers15143549
APA StyleBerdiaki, A., Giatagana, E.-M., Tzanakakis, G., & Nikitovic, D. (2023). The Landscape of Small Leucine-Rich Proteoglycan Impact on Cancer Pathogenesis with a Focus on Biglycan and Lumican. Cancers, 15(14), 3549. https://doi.org/10.3390/cancers15143549








